Introduction
Although the incidence of cervical cancer has
decreased due to more regular cytological screening programs and
the success of human papillomavirus vaccinations, it remains the
third most common type of gynecological cancer worldwide (1). The International Agency for Research on
Cancer estimated there were 528,000 new cases and 266,000 deaths in
2012, and a total of ~9/10 (87%) of cervical cancer mortalities
occur in less developed regions (2).
At present, radiotherapy, as an adjuvant or primary treatment,
remains the most common and effective therapeutic intervention for
cervical cancer. Several international clinical trials have
reported that adjuvant radiotherapy or concurrent chemoradiation
therapy can improve disease-free survival and overall survival
outcomes in patients with pathological risk factors for recurrence
(3–5).
However, radiotherapy ultimately fails in >20% of patients with
locally advanced cervical cancer (FIGO stage IB2-IVA), the main
cause of which is radio-resistance (6,7).
Clinically, the identification and treatment of radio-resistant
cervical cancer remains a challenge.
Eukaryotic initiation factor (eIF) 4F is composed of
ATP-dependent RNA helicase eIF4A1, 5′ cap mRNA-binding protein
eIF4E and the scaffolding protein eIF4G, which scans mRNAs through
the 5′ untranslated region (5′UTR), and unwinds the mRNA secondary
structure to expose the translation initiation codon and initiates
translation (8,9). Assembly of the eIF4F complex is
rate-limiting step for translation initiation. Increased eIF4F
complex formation elevates the translation of all cap-dependent
mRNAs, thereby increasing global protein synthesis rates. However,
mRNAs vary widely in their inherent translatability, largely as a
function of differences in the length and structure of their
5′UTRs. Cellular mRNAs that are the most sensitive to alterations
in eIF4F complex formation include weak mRNAs, which are GC rich
and highly-structured 5′UTRs (8,9). c-Myc,
vascular endothelial growth factor, ornithine decarboxylase and
survivin typically encode growth and survival factors (8,9).
Therefore, eIF4A1 may perform an essential role in the translation
of weak mRNAs, indicating that eIF4A1 is an important oncogenic
protein. This has been verified by numerous prior studies (10–13). Our
previous study (10) also revealed
that eIF4A1 expression is associated with certain
clinicopathological variables of cervical cancer, including the
International Federation of Gynecology and Obstetrics (FIGO) stage
(14), histological type, pelvic
lymph node metastasis status, parametrial invasion and deep stromal
invasion. In addition, the decreased expression of eIF4A1 following
brachytherapy may predict improved radiosensitivity and
tumor-specific survival. These findings indicate that eIF4A1 may be
of importance in cervical cancer, particularly in cancer cell
radiosensitivity. Therefore, the present study analyzed the
function of eIF4A1 in cervical cancer and attempted to explore the
underlying mechanisms.
Materials and methods
Cell lines and cell culture
The HeLa and SiHa cervical cancer cell lines and the
293T cell line were obtained from the American Type Culture
Collection (Manassas, VA, USA). Cells were maintained in Dulbecco's
modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) supplemented with 10% heat-inactivated
fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) and
penicillin (100 µg/ml) and streptomycin (100 U/ml), and incubated
in a humidified incubator at 37°C in an atmosphere containing 5%
CO2.
RNA interference and eIF4A1
downregulated cells
The two selected short hairpin RNA (shRNA/shR)
sequences for eIF4A1 were as follows: P1 sense,
5′-CCGGGCCGTGTGTTTGATATGCTTACTCGAGTAAGCATATCAAACACACGGCTTTTTG-3′
and antisense,
5′-AATTCAAAAAGCCGTGTGTTTGATATGCTTACTCGAGTAAGCATATCAAACACACGGC-3′;
P2 sense,
5′-CCGGGCCGTAAAGGTGTGGCTATTACTCGAGTAATAGCCACACCTTTACGGCTTTTTG-3′
and antisense,
5′-AATTCAAAAAGCCGTAAAGGTGTGGCTATTACTCGAGTAATAGCCACACCTTTACGGC-3′.
These sequences were synthesized by Sangon Biotech Co., Ltd.
(Shanghai, China), annealed and ligated into the pLKO.1 lentiviral
shRNA vector linearized by AgeI and EcoRI (New
England Biolabs, Inc., Ipswich, MA, USA). eIF4A1 downregulated
cells were established using the following process: The recombinant
lentivirus with shR-eIF4A1 was produced by co-transfecting 293T
cells with the plasmids, psPAX2 and pMD2.G using
Lipofectamine® 2000 transfection reagent (Invitrogen;
Thermo Fisher Scientific, Inc.), according to the manufacturer's
protocol. After transfection for 48 h, Lentivirus-containing
supernatants were centrifuged at 400 × g for 5 min at room
temperature and filtered through a 0.45-µm cellulose acetate filter
(Merck KGaA, Darmstadt, Germany), and stored at 4°C. Subsequently,
SiHa and HeLa cells which obtained 30% confluency were cultured in
a humidified incubator at 37°C with 5% CO2 with the
shR-eIF4A1 lentivirus-containing supernatant, diluted with
serum-free DMEM at a ratio of 1:1. After 24 h, the culture medium
was removed and fresh medium was added to the cells. Finally, 2
mg/ml puromycin (Sigma-Aldrich; Merck KGaA) was added to the
medium. Following antibiotic selection for 48 h,
eIF4A1-downregulated cells (HeLa-shR1, 2; SiHa-shR1, 2) were
obtained.
Cell proliferation assay
A Cell Counting kit-8 (CCK)-8 (Dojindo Molecular
Technologies, Inc., Kumamoto, Japan) was used to evaluate cell
proliferation. The shR-eIF4A1-infected cells and parental cells
were seeded at a density of 1,000 cells/well on a 96-well plate.
After 6 h and 1, 2, 3 and 4 days, 10 µl CCK-8 reagent was added to
each well and incubated for 2 h at 37°C with 5% CO2. The
absorbance at 450 nm was then measured using a Tecan Sunrise
Microplate Reader (Tecan Group, Ltd., Männedorf, Switzerland) to
determine the cell number.
Cell invasion and migration
assays
The eIF4A1-downregulated cells and parental cells
were resuspended in serum-free DMEM. Subsequently, the upper
Transwell chambers (8-mm pore size; BD Biosciences, Franklin Lakes,
USA) containing Matrigel coatings were seeded with 100,000 cells,
which were cultured with serum-free medium. A total of 50,000 cells
were added to the upper Transwell chambers without Matrigel
coatings and cultured with serum-free medium. The lower chambers
contained medium supplemented with 10% FBS. After 24 h of
incubation, the non-migrated cells were gently removed with cotton
swabs, and the migrated cells on the bottom surface of the
membranes were fixed and then stained with 1% crystal violet in
100% ethanol for 15 min at room temperature. Cells were then
visualized using an Olympus light microscope (Olympus Corporation,
Tokyo, Japan) at magnification, ×20. The invasive/migratory cells
were counted from five random fields of view.
Cell apoptosis
Apoptosis was determined using an Annexin V and
propidium iodide (PI) staining-based Fluorescein
Isothiocyanate-Annexin V Apoptosis Detection kit (BD Biosciences)
according to the manufacturer's protocol. The eIF4A1-downregulated
cells and parental cells were seeded into a 6-well plate
(2×105 cells/well) and incubated for 24 h at 37°C with
5% CO2. Cells were then resuspended and labeled using
the Apoptosis Detection kit, and the apoptotic cells were
determined using a FACScan cytofluorometer from BD Biosciences
(Franklin Lakes, NJ, USA) with Cell Quest software version 5.1 (BD
Biosciences).
Cervical cancer radiosensitivity assay
in vitro and in vivo
A cell colony formation assay was applied for
cervical cancer cell radiosensitivity analyses. Exponential growth
phase eIF4A1-downregulated cells and parental cells (200–8,000)
were seeded onto 6-well plates. Following incubation for 6–8 h, the
plates were irradiated with 137Cs (Nordion, Ottawa, ON, Canada).
The dose rate was 1.25 Gy/min, with doses of 0, 2, 4, 6, 8 Gy given
in a single fraction. Following incubation at 37°C with 5%
CO2 for 10–13 days, the cells were fixed and then
stained with 1% crystal violet in 100% ethanol for 15 min at room
temperature. Colonies containing ≥50 cells were counted. The
plating efficiency (PE)=(the number of clones/the number of planted
cells) ×100%, and the surviving fraction (SF) was the number of
colonies formed/(the number of cells plated × the plating
efficiency) (15,16).
The in vivo radiosensitivity assay was
approved by the Department of Laboratory Animal Science, Fudan
University (Shanghai, China). The mice were housed under controlled
12 h light-dark cycles, constant temperature (22–24°C) and humidity
(55–60%), and were given sterilized food and tap water ad
libitum. The mice were used when they were 6-weeks and weighed
~25 g. SiHa-shR1 cells or SiHa cells (1×106/mouse) were
subcutaneous injected into the right thigh of the 32 BALB/c female
nude mice (Shanghai SLAC Laboratory Animal Co., Shanghai, China).
When tumors reached 8 mm in diameter (~14 days post-injection), all
mice were randomly divided into two groups. One group received
radiation treatment (16 Gy with 6 MV of X-rays), whilst the other
group served as the controls. Tumor diameters were measured every 3
days for ≤37 days to calculate the tumor volume (V) using the
following equation V=(LxW2) ×0.5, where L was the length
of the long side of the tumor and W was the length of the short
side.
Western blot analysis
The cells were collected and then lysed with RIPA
lysis buffer (1% NP-40, 0.1% SDS, 0.5% sodium deoxycholate, 150
mmol/l NaCl and 10 mmol/l Tris-HCl) containing 1/100
phenylmethanesulfonyl fluoride solution. The total protein
concentration was determined using a BCA protein assay kit
(Beyotime Institute of Biotechnology, Haimen, China). Aliquots (30
µg) of each sample were separated by 12% polyacrylamide gel
electrophoresis and transferred to PVDF membranes (EMD Millipore).
The membranes were blocked with 5% non-fat milk at room temperature
for 2 h. Subsequently, the membranes were probed with primary
antibodies against eIF4A1 (1:1,000 dilution; cat no. T2192;
Epitomics; Abcam, Cambridge, UK) or β-actin (1:2,000 dilution; cat
no. 4967S; Cell Signaling Technology, Inc., Danvers, MA, USA) or
γ-H2AX (1:1,000 dilution; cat no. 2577; Cell Signaling Technology,
Inc.) in TBS-Tween-20 (TBST) containing 5% non-fat milk
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) at 4°C overnight,
followed by three washes in TBST for 10 min per wash. Subsequently,
the membranes were incubated with the goat anti-rabbit horseradish
peroxidase conjugated-secondary antibody (1:3,000 dilution; cat no.
ab97051; Abcam) for 1 h at room temperature. Immunoreactive bands
were visualized using an enhanced chemiluminescence reaction
(Pierce; Thermo Fisher Scientific, Inc.). β-actin was used as an
internal control.
Statistical analysis
Data are presented as the mean ± standard error.
SPSS software (version 19.0 for Windows; IBM Corp., Armonk, NY,
USA) was used for statistical analysis. The χ2 test,
Fisher's exact test and Student's t-test were applied to assess
statistical significance. All tests were two-sided, and P<0.05
was considered to indicate a statistically significant
difference.
Results
Downregulated eIF4A1 inhibits cervical
cancer cell proliferation and migration, and promotes cell
apoptosis
To assess the effects of eIF4A1 in cervical cancer
cells, eIF4A1-downregulated cells (HeLa-shR1, 2; SiHa-shR1, 2) were
established and western blotting was used to verify the success of
the transfection and knockdown (Fig.
1A). A CCK-8 assay determined that the downregulation of eIF4A1
in HeLa and SiHa cells notably attenuated cell proliferation
(Fig. 1B; P<0.001). The effect of
eIF4A1 on cervical cancer cell invasion was examined using a
Matrigel-based invasion assay with a Transwell chamber. The number
of HeLa-shR1, 2 and SiHa-shR1, 2 cells (Fig. 1C) that passed through the Matrigel
basement membrane matrix was markedly decreased compared with the
parental cells (P<0.001). Likewise, the migration ability of
HeLa and SiHa cells decreased significantly after the silence of
eIF4A1 (Fig. 1D; P<0.001).
However, flow cytometry revealed that HeLa-shR1, 2 and SiHa-shR1, 2
cells had a larger proportion of apoptotic cells than the parental
cells (Fig. 1E; P<0.05). These
results revealed that downregulated eIF4A1 markedly repressed
cervical cancer cell viability and promote cell apoptosis.
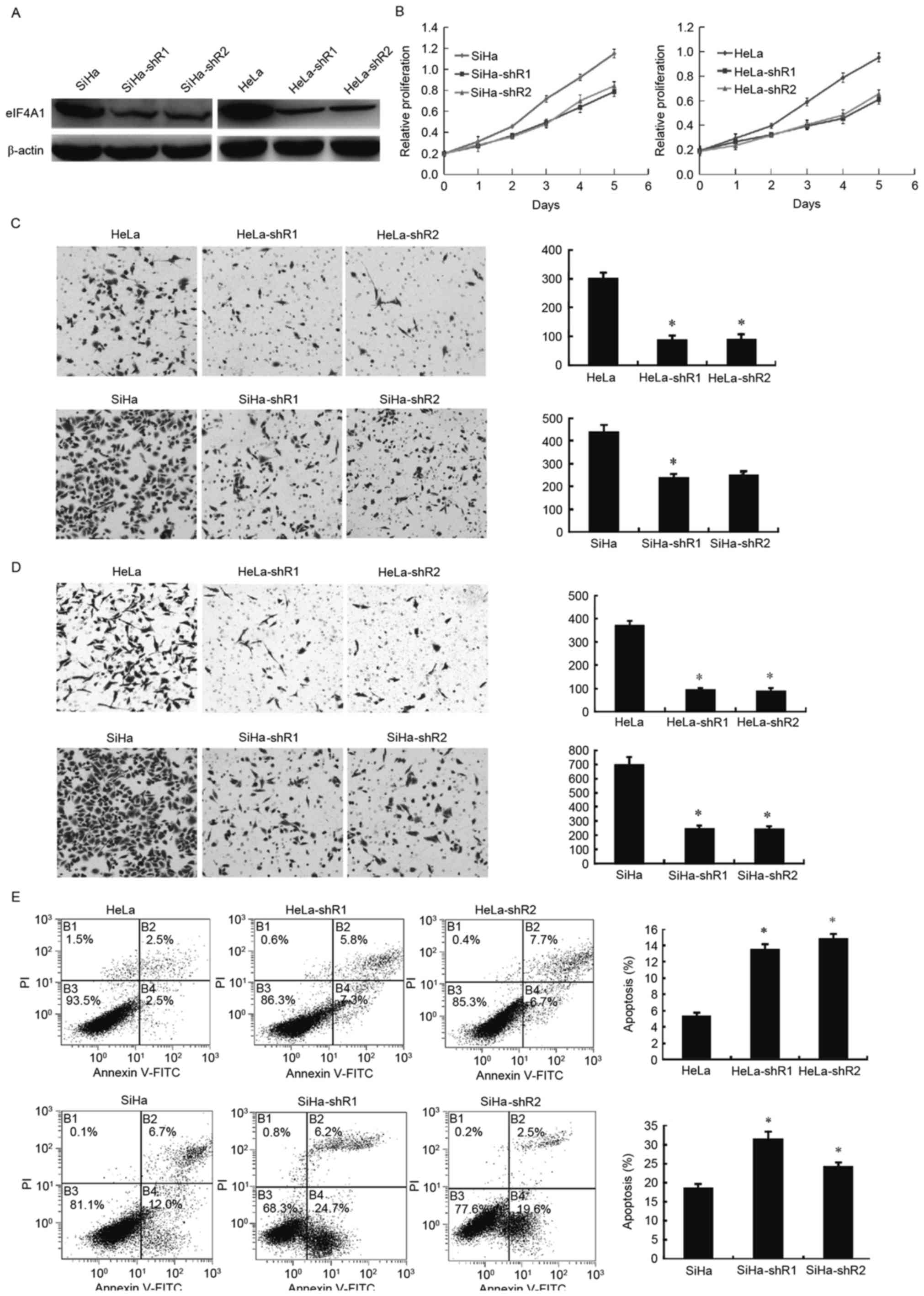 | Figure 1.Downregulation of eIF4A1 in cervical
cancer cells significantly affected cell proliferation, viability
and apoptosis. (A) Construction and validation of downregulated
eIF4A1 in HeLa and SiHa cell lines. pLKO.1-TRC cloning vector and
two pairs of shRNA for eIF4A1 were used to construct
stable-knockdown eIF4A1 cells. Western blot analysis revealed that
eIF4A1 was significantly downregulated in cervical cancer cells
(HeLa-shR1, 2; SiHa-shR1, 2). (B) Effects of eIF4A1-knockdown on
cervical cancer cell proliferation. Downregulated eIF4A1 inhibited
HeLa and SiHa cell growth, as measured by a Cell Counting kit-8
assay. This inhibition effect was significant by day 5
(P<0.001). (C) The effect of eIF4A1 on cervical cancer cell
invasion was examined using a Matrigel-based invasion assay with
Transwell chambers. The number of HeLa-shR1, 2 and SiHa-shR1, 2
cells that passed through the Matrigel basement membrane matrix was
markedly lower compared with the parental cells (P<0.001). (D)
The effect of eIF4A1 on cervical cancer cell migration was examined
using a Transwell chamber assay. The migration ability of HeLa and
SiHa cells decreased significantly after the silence of eIF4A1
(P<0.001). (E) Flow cytometric analysis of cells stained with
Annexin V/PI. The percentages of cells in each quadrant are
indicated. Knockdown of eIF4A1 significantly induced apoptosis in
HeLa and SiHa cells (P<0.05). (Q1, necrotic cells; Q2, cells in
late apoptosis; Q3, normal cells; and Q4, cells in early
apoptosis.). PI, propidium iodide; FITC, fluorescein
isothiocyanate; eIF4A1, eukaryotic initiation factor 4A1; shR,
short hairpin RNA. *P<0.05 vs. the control. |
Downregulation of eIF4A1 improves
cervical cancer radiosensitivity in vitro and in vivo
Our previous study (10) demonstrated that the alteration of
eIF4A1 expression in response to brachytherapy was associated with
cervical cancer radiosensitivity. In the present study, a cervical
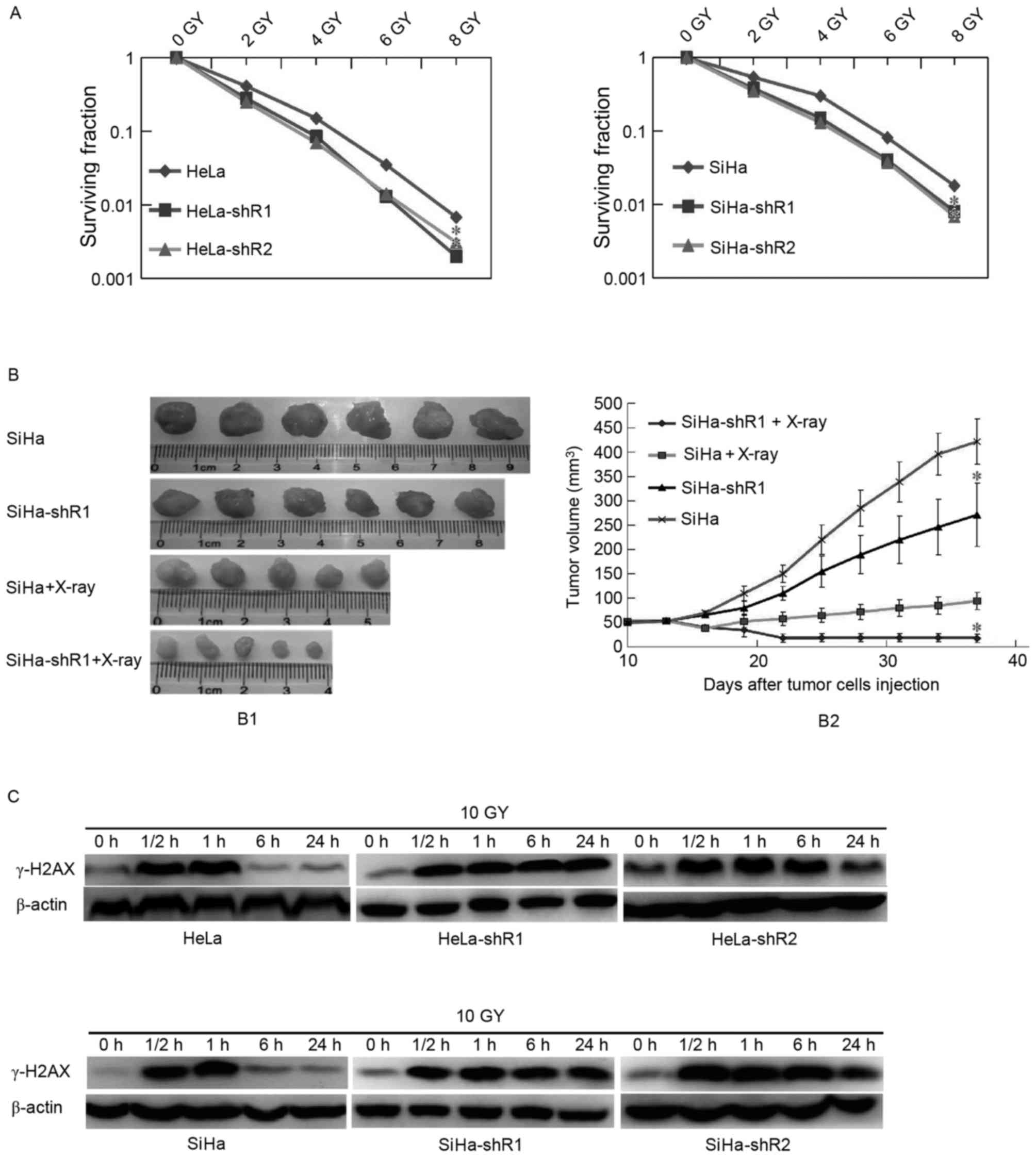
cancer cell colony formation assay with radiation was conducted. As
presented in Fig. 2A, decreased
expression of eIF4A1 in HeLa and SiHa cells increased their
radiosensitivity in vitro (P<0.05).
A tumorigenesis assay in nude mice was then
performed, which revealed that the SiHa-shR1 cell tumor volumes
following X-ray irradiation were significantly smaller than those
formed by the parental cells (Fig.
2B; P<0.05), indicating that the downregulation of eIF4A1
improved cervical cancer radiosensitivity.
Downregulation of eIF4A1 improves
cervical cancer radiosensitivity by delaying cancer cell DSB
repair
To disclose how decreased eIF4A1 expression improved
cervical cancer cell radiosensitivity, DNA DSB repair, which
measured by the expression of γ-H2AX, was examined. In the present
study, the expression of γ-H2AX was detected by western blot
analysis at 0, 0.5, 1, 6 and 24 h following irradiation of the
cancer cells. As presented in Fig.
2C, 0.5 and 1 h following irradiation, which means a period of
DNA DSB, the band concentration of γ-H2AX between eIF4A1
downregulation and parental cells were all at a high level, and no
significant differences were observed. These results indicated that
eIF4A1 has no notable effect on the initial level of
radiation-induced DSB. However, at 6 and 24 h post-irradiation,
γ-H2AX protein levels in eIF4A1 downregulated cells were markedly
increased than in parental cells, which illustrated that
eIF4A1-knockdown, causes the inhibition of radiation-induced DNA
DSB repair. Overall, it was considered that downregulated eIF4A1
improves cervical cancer cell radiosensitivity by delaying cell DSB
repair.
Discussion
eIF4A1 is a canonical DEAD-box helicase that
exhibits ATP-dependent RNA helicase activity in mRNA translation
(17–19), particularly for oncoproteins or
proteins implicated in cell growth, death or proliferation
(20,21). Elevated expression of eIF4A1 has been
reported in primary hepatocellular carcinomas, melanoma cell lines
and congenital melanocytic nevi (11,12).
Furthermore, targeting eIF4A1 has been determined to reverse
lymphoma cancer cell chemo-resistance (13). In our previous study (10), it was demonstrated that eIF4A1
overexpression promotes cervical cancer. In the present study,
downregulated eIF4A1 was revealed to inhibit cervical cancer cell
proliferation and induce apoptosis in vitro. In addition,
SiHa-shR1 cells generated xenografts were statistically smaller
than their parental SiHa cells (Fig.
2B; P<0.05). These results confirmed that eIF4A1 promotes
cervical cancer and may be used as a therapeutic target.
The clinical findings from our previous study
(10) also indicated that eIF4A1
expression is associated with cervical cancer lymph node
metastasis. Ji et al (22)
reported that eIF4A1 is an early marker of distant metastasis of
non-small cell lung cancer. Therefore, Transwell experiments were
performed in vitro to verify this association. As predicted,
the results of the current study revealed that the invasion and
migration abilities of eIF4A1 downregulated cells were
significantly suppressed compared with the parental cells. These
findings indicated that eIF4A1 performs an important role in
cervical cancer viability.
To determine the association between eIF4A1
expression and cervical cancer radiosensitivity, a colony formation
assay was conducted with X-ray radiation in vitro and in
vivo. Compared with parental cells, the colony-forming ability
of eIF4A1 downregulated cells was significantly lower (Fig. 2A; P<0.05). Furthermore, studies
in vivo have also observed that the radiosensitivity of
SiHa-shR cells was markedly enhanced. Thus, to the best of our
knowledge, the current study confirmed for the first time that the
reduced expression of eIF4A1 improved cervical cancer
radiosensitivity, thus presenting a potential novel target for
cervical cancer therapy.
Radiation is known to cause damage to DNA, including
base damage, DNA strand breaks (single strand breaks and DSB) and
DNA strand cross-linking (23–25). Base
damage, DNA strand cross-linking and single strand breaks are often
repaired successfully, however, DSB repair tends to result in gene
mutations or genomic instability (26–28). It
has been reported that γ-H2AX forms at the DSB site in response to
DNA damage caused by ionizing radiation, and that each foci of
γ-H2AX corresponds to one DSB (29–34).
Subsequently, DSB repair would occur within 30 min, 75% of which
would fade away within 5 h, in which an almost complete γ-H2AX loss
is estimated to occur within 7–8 h (35–38).
Therefore, in the present study, western blot analysis was used to
detect the expression of γ-H2AX within 24 h. For the parental HeLa
and SiHa cells, γ-H2AX expression was barely detectable within 6 h,
suggesting that DSB repair was successful. However, for eIF4A1
downregulated cells, γ-H2AX remained at a high level at 6 h and at
24 h, which indicated postponed DSB repair. Therefore, it was
concluded that downregulated eIF4A1 increased cervical cancer
radiosensitivity by delaying cancer cell DSB repair. However, the
mechanism by which downregulated eIF4A1 delays DSB repair remains
under investigation.
In conclusion, the present study confirmed that
downregulated eIF4A1 significantly inhibited cervical cancer cell
proliferation and viability, and induced cell apoptosis. In
addition, the present study provided evidence that cervical cancer
radiosensitivity may be enhanced by downregulated eIF4A1 due to
delayed cancer cell DSB repair, indicating that eIF4A1 may be a
promising target for increasing cervical cancer
radiosensitivity.
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sedlis A, Bundy BN, Rotman MZ, Lentz SS,
Muderspach LI and Zaino RJ: A randomized trial of pelvic radiation
therapy versus no further therapy in selected patients with stage
IB carcinoma of the cervix after radical hysterectomy and pelvic
lymphadenectomy: A gynecologic oncology group study. Gynecol Oncol.
73:177–183. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Monk BJ, Wang J, Im S, Stock RJ, Peters WA
III, Liu PY, Barrett RJ II, Berek JS, Souhami L, Grigsby PW, et al:
Rethinking the use of radiation and chemotherapy after radical
hysterectomy: A clinical-pathologic analysis of a gynecologic
oncology group/southwest oncology group/radiation therapy oncology
group trial. Gynecol Oncol. 96:721–728. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rotman M, Sedlis A, Piedmonte MR, Bundy B,
Lentz SS, Muderspach LI and Zaino RJ: A phase III randomized trial
of postoperative pelvic irradiation in Stage IB cervical carcinoma
with poor prognostic features: Follow-up of a gynecologic oncology
group study. Int J Radiat Oncol Biol Phys. 65:169–176. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Eifel PJ, Winter K, Morris M, Levenback C,
Grigsby PW, Cooper J, Rotman M, Gershenson D and Mutch DG: Pelvic
irradiation with concurrent chemotherapy versus pelvic and
para-aortic irradiation for high-risk cervical cancer: An update of
radiation therapy oncology group trial (RTOG) 90–01. J Clin Oncol.
22:872–880. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Legge F, Chiantera V, Macchia G, Fagotti
A, Fanfani F, Ercoli A, Gallotta V, Morganti AG, Valentini V,
Scambia G and Ferrandina G: Clinical outcome of recurrent locally
advanced cervical cancer (LACC) submitted to primary multimodality
therapies. Gynecol Oncol. 138:83–88. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
De Benedetti A and Graff JR: eIF4E
expression and its role in malignancies and metastases. Oncogene.
23:3189–3199. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Graff JR, Konicek BW, Carter JH and
Marcusson EG: Targeting the eukaryotic translation initiation
factor 4E for cancer therapy. Cancer Res. 68:631–634. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liang S, Zhou Y, Chen Y, Ke G, Wen H and
Wu X: Decreased expression of EIF4A1 after preoperative
brachytherapy predicts better tumor-specific survival in cervical
cancer. Int J Gynecol Cancer. 24:908–915. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shuda M, Kondoh N, Tanaka K, Ryo A,
Wakatsuki T, Hada A, Goseki N, Igari T, Hatsuse K, Aihara T, et al:
Enhanced expression of translation factor mRNAs in hepatocellular
carcinoma. Anticancer Res. 20:2489–2494. 2000.PubMed/NCBI
|
|
12
|
Eberle J, Krasagakis K and Orfanos CE:
Translation initiation factor eIF-4A1 mRNA is consistently
overexpressed in human melanoma cells in vitro. Int J Cancer.
71:396–401. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bordeleau ME, Robert F, Gerard B,
Lindqvist L, Chen SM, Wendel HG, Brem B, Greger H, Lowe SW, Porco
JA Jr and Pelletier J: Therapeutic suppression of translation
initiation modulates chemosensitivity in a mouse lymphoma model. J
Clin Invest. 118:2651–2660. 2008.PubMed/NCBI
|
|
14
|
Pecorelli S: Revised FIGO staging for
carcinoma of the vulva, cervix, and endometrium. Int J Gynaecol
Obstet. 105:103–104. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Puck TT and Marcus PI: Rapid method for
viable cell titration and clone production with HELA cells in
tissue culture: The use of X-irradiated cells to supply
conditioning factors. Proc Natl Acad Sci USA. 41:pp. 432–437. 1955;
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Puck TT and Marcus PI: Action of X rays on
mammalian cells. J Exp Med. 103:653–666. 1956. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Linder P, Tanner NK and Banroques J: From
RNA helicases to RNPases. Trends Biochem Sci. 26:339–341. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cordin O, Banroques J, Tanner NK and
Linder P: The DEAD-box protein family of RNA helicases. Gene.
367:17–37. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Pyle AM: Translocation and unwinding
mechanisms of RNA and DNA helicases. Annu Rev Biophys. 37:317–336.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Abdelhaleem M: Do human RNA helicases have
a role in cancer? Biochim Biophys Acta. 1704:37–46. 2004.PubMed/NCBI
|
|
21
|
Kozak M: An analysis of 5′-noncoding
sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res.
15:8125–8148. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ji P, Diederichs S, Wang W, Böing S,
Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et
al: MALAT-1, a novel noncoding RNA, and thymosin beta4 predict
metastasis and survival in early-stage non-small cell lung cancer.
Oncogene. 22:8031–8041. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Painter RB: DNA damage and repair in
eukaryotic cells. Genetics. 78:139–148. 1974.PubMed/NCBI
|
|
24
|
O'Neill P and Wardman P: Radiation
chemistry comes before radiation biology. Int J Radiat Biol.
85:9–25. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nocentini S: Rejoining kinetics of DNA
single- and double-strand breaks in normal and DNA ligase-deficient
cells after exposure to ultraviolet C and gamma radiation: an
evaluation of ligating activities involved in different DNA repair
processes. Radiat Res. 151:423–432. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jeggo PA and Löbrich M: How cancer cells
hijack DNA double-strand break repair pathways to gain genomic
instability. Biochem J. 471:1–11. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Iliakis G, Wang H, Perrault AR, Boecker W,
Rosidi B, Windhofer F, Wu W, Guan J, Terzoudi G and Pantelias G:
Mechanisms of DNA double strand break repair and chromosome
aberration formation. Cytogenet Genome Res. 104:14–20. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mladenov E and Iliakis G: Induction and
repair of DNA double strand breaks: The increasing spectrum of
non-homologous end joining pathways. Mutat Res. 711:61–72. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sánchez-Flores M, Pásaro E, Bonassi S,
Laffon B and Valdiglesias V: γH2AX assay as DNA damage biomarker
for human population studies: Defining experimental conditions.
Toxicol Sci. 144:406–413. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hamer G, Roepers-Gajadien HL, van
Duyn-Goedhart A, Gademan IS, Kal HB, van Buul PP and de Rooij DG:
DNA double-strand breaks and gamma-H2AX signaling in the testis.
Biol Reprod. 68:628–634. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Paull TT, Rogakou EP, Yamazaki V,
Kirchgessner CU, Gellert M and Bonner WM: A critical role for
histone H2AX in recruitment of repair factors to nuclear foci after
DNA damage. Curr Biol. 10:886–895. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sedelnikova OA, Rogakou EP, Panyutin IG
and Bonner WM: Quantitative detection of (125)IdU-induced DNA
double-strand breaks with gamma-H2AX antibody. Radiat Res.
158:486–492. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Pilch DR, Sedelnikova OA, Redon C, Celeste
A, Nussenzweig A and Bonner WM: Characteristics of gamma-H2AX foci
at DNA double-strand breaks sites. Biochem Cell Biol. 81:123–129.
2003. View
Article : Google Scholar : PubMed/NCBI
|
|
34
|
Rogakou EP, Boon C, Redon C and Bonner WM:
Megabase chromatin domains involved in DNA double-strand breaks in
vivo. J Cell Biol. 146:905–916. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Nazarov IB, Smirnova AN, Krutilina RI,
Svetlova MP, Solovjeva LV, Nikiforov AA, Oei SL, Zalenskaya IA, Yau
PM, Bradbury EM and Tomilin NV: Dephosphorylation of histone
gamma-H2AX during repair of DNA double-strand breaks in mammalian
cells and its inhibition by calyculin A. Radiat Res. 160:309–317.
2003. View
Article : Google Scholar : PubMed/NCBI
|
|
36
|
Svetlova M, Solovjeva L, Nishi K, Nazarov
I, Siino J and Tomilin N: Elimination of radiation-induced
gamma-H2AX foci in mammalian nucleus can occur by histone exchange.
Biochem Biophys Res Commun. 358:650–654. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Solovjeva LV, Pleskach NM, Firsanov DV,
Svetlova MP, Serikov VB and Tomilin NV: Forskolin decreases
phosphorylation of histone H2AX in human cells induced by ionizing
radiation. Radiat Res. 171:419–424. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Löbrich M, Shibata A, Beucher A, Fisher A,
Ensminger M, Goodarzi AA, Barton O and Jeggo PA: gammaH2AX foci
analysis for monitoring DNA double-strand break repair: Strengths,
limitations and optimization. Cell Cycle. 9:662–669. 2010.
View Article : Google Scholar : PubMed/NCBI
|