Introduction
Autophagy serves an important role in maintaining
cell metabolism and homeostasis (1).
Autophagy is a process of endogenous substrate digestion in cells;
autophagosomes are formed, and mature proteins or damaged
organelles in the cytoplasm are encased by lysosomes, in which
lysosomal proteases degrade them.
During tumorigenesis and tumor progression,
autophagy exerts its role as a tumor suppressor by removing
abnormally folded proteins and dysfunctional organelles such as
mitochondria, inhibiting cell stress responses (2). However, in instances of nutritional
deficiencies and hypoxia, autophagy supports tumor cell survival,
which promotes cell proliferation and suppresses cell death
(3). Compromised autophagy promotes
chromosomal instability, including increased DNA damage, gene
amplification and aneuploidy (4).
As a key autophagy regulator associated with
apoptosis and differentiation, the autophagy-associated protein
Beclin-1 has been demonstrated to be involved in many types of
cancer, including ovarian carcinoma (5), hepatocellular carcinoma (6), melanoma (7), rectal cancer (8) and tongue squamous cell carcinoma
(9). It has been suggested that the
endogenous Beclin-1 protein expression is frequently low in human
breast epithelial carcinoma cell lines and tissues, whereas it is
expressed ubiquitously at high levels in normal breast epithelia
(10). Concomitantly, the
autophagy-promoting activity in the MCF7 breast cancer cell line
following transfection with Beclin-1 was observed to inhibit MCF7
proliferation, clonogenicity and tumorigenesis (10). A previous in vivo study
demonstrated that mice with inactivated or deleted Beclin-1 were
susceptible to tumors including lymphoma, lung cancer and liver
cancer (11,12).
Beclin-1 modulates cancer initiation and progression
by affecting a wide range of pathological events, including
extracellular matrix degradation, epithelial-to-mesenchymal
transition, tumor angiogenesis and alterations to the tumor
microenvironment (13). However, the
effect of Beclin-1 in cancer development is complex, as a number of
reports have indicated the pro-neoplastic and anti-neoplastic
functions for Beclin-1, as reviewed by Ozpolat and Benbrook
(14). The present study aimed to
address Beclin-1 expression and its clinical significance in
gastric cancer, a type of cancer with one of the highest incidence
rates worldwide, and to explore its primary potential
mechanism.
Materials and methods
Clinical specimens and patient
data
A total of 60 specimens of gastric carcinoma tumors
and para-carcinoma tissues were sampled from patients during
resection in Heilongjiang Province Land Reclamation Headquarter
General Hospital (Harbin, China) between January 2014 and February
2016. The clinicopathological data of all cases were reviewed,
including patient age, sex, tumor size, differentiation status,
tumor node metastasis (TNM) stage, lymph node metastasis and
invasion status (Table I) (15). No patients had received preoperative
chemotherapy and radiotherapy, and those who succumbed to other
diseases or accidents were excluded from the study. All specimens
were fixed in 10% neutral formaldehyde at room temperature for
24–48 h, and embedded in paraffin at 65°C and cut into 5-µm
sections. All specimens were subjected to hematoxylin staining for
5 min and eosin staining for 1 min at room temperature and
diagnosed as gastric carcinoma by two pathologists. Informed
consent was obtained from all patients or their relatives. The
experimental protocol was approved by the Ethics Committee of the
King Medical Diagnostics Center (Shanghai, China).
 | Table I.Expression of Beclin-1 in gastric
carcinoma and para-carcinoma tissue samples. |
Table I.
Expression of Beclin-1 in gastric
carcinoma and para-carcinoma tissue samples.
| Group | Cases | Low expression | High expression |
|---|
| Para-carcinoma
tissue | 60 | 14 | 46 |
| Gastric carcinoma
tissue | 60 | 38 | 22a |
Cell culture and transfection
MKN-45, MKN-28, and SGC-7901 gastric cancer cell
lines (Cancer Cell Bank, Chinese Academy of Medical Sciences,
Beijing, China) were cultured in Dulbecco's modified Eagle's medium
(DMEM; PAN Biotech GmbH, Aidenbach, Germany) supplemented with 10%
fetal bovine serum (FBS; Hyclone; GE Healthcare Life Sciences,
Logan, UT, USA) at 37°C with 5% CO2. Western blot
analysis determined that Beclin-1 expression was relatively low in
the MKN-45 cell line (data not shown), thus the MKN-45 cell line
was selected for all follow-up overexpression experiments.
The Beclin-1 sequence was cloned into a PCDNA3.0
plasmid (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA,
USA). For transfection, the cells were seeded at a density of
1×105/well in a 24-well plate and grown to >50-70%
confluency. A total of 1 µl Lipofectamine 2000®
(Invitrogen; Thermo Fisher Scientific, Inc.) was added to 50 µl
serum-free Opti-MEMI medium (Invitrogen; Thermo Fisher Scientific,
Inc.) and mixed gently at room temperature for 5 min; 2 µg Beclin-1
plasmid was added to 50 µl serum-free Opti-MEMI medium and mixed
gently. After 5 min, the diluted Lipofectamine 2000® was
gently mixed with the diluted Beclin-1 plasmid at room temperature
for 20 min. Subsequent to transfection with Beclin-1 or control
plasmids (empty PCDNA3.0 vector), the 24-well plate was incubated
at 37°C with 5% CO2 for 6 h, following which the culture
medium was replaced with complete medium.
Immunohistochemical staining
The paraffin sections were incubated at 65°C for 2 h
and dewaxed in xylene for 20 min at room temperature. The specimens
were washed with distilled water three times, incubated in 3%
H2O2 at room temperature for 10 min and
washed with distilled water again. Antigen retrieval was performed
using Tris-EDTA buffer (Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany) in a pressure cooker at medium to high pressure for 3 min.
Subsequent to washing with PBS, the specimens were incubated with
monoclonal primary antibody against Beclin-1 (1:500; cat. no.
ab210498; Abcam, Cambridge, UK) at 4°C overnight. The specimens
were then rinsed with PBS buffer, and incubated in a 1:1,000
solution of biotin-labeled anti-Human Immunoglobulin G (IgG)
secondary antibody (cat. no. SAB3701279; Sigma-Aldrich; Merck KGaA)
at room temperature for 60 min. The specimens were washed with PBS
buffer and treated with 3,3-Diaminobenzidine (Sigma-Aldrich; Merck,
KGaA) for 2 min at room temperature. Slides were counterstained
with hematoxylin for 1 min at room temperature, washed with
distilled water, dehydrated sequentially with a graded ethanol
series (80, 95 and 100% ethanol), transparentized with xylene and
sealed with neutral gum at room temperature.
Immunohistochemical staining
analysis
For each section, 10 high-power fields at
magnification, ×400 were randomly selected, and ~500 cells were
counted in each field. Scores between 0–3 were assigned according
to staining intensity and the number of positive cells. The cells
were scored as follows: No staining, 0; light yellow, 1; pale
brown, 2; dark brown, 3. The number of positive cells was scored as
follows: <10%, 0; 10–45%, 1; 45–70%, 2; and >70%, 3. The two
scores were totaled and the sum was divided into 4 levels: A total
score of 1 was considered negative (−), a score of 2 as weakly
positive (+), a score of 3–4 as positive (++), and a score of 5–6
as strongly positive (+++). Negative and weakly positive were
regarded as low expression, while positive and strongly positive as
high expression.
Western blotting analysis
Tissues were lysed in buffer containing 50 mM Tris
(pH 7.4), 150 mM NaCl, 1% nonidet P-40, 0.25% sodium deoxycholate,
1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride and 1% protease
cocktail inhibitor, followed by centrifugation at 12,000 × g for 10
min at 4°C. The tissue lysis supernatants were collected and the
protein concentrations were determined using BCA Protein
Quantification kit (cat. no. 23227; Thermo Fisher Scientific,
Inc.). A total of 20 µg protein samples were loaded in each well
and separated by electrophoresis in a 10% SDS-polyacrylamide gel
and transferred to a polyvinylidene fluoride membrane (EMD
Millipore, Billerica, MA, USA). Subsequent to blocking with 5% milk
in TBS containing 0.1% Tween-20 (TBST) at room temperature for 2 h,
the membrane was incubated with primary antibodies against Beclin-1
(1:1,000; cat. no. ab210498; Abcam), light chain (LC) 3 II/I
(1:500; cat. no. ab128025; Abcam), B-cell lymphoma-extra large
(Bcl-xL; 1:800; cat. no. ab2568; Abcam) and β-actin (1:4,000; cat.
no. A5441; Sigma-Aldrich; Thermo Fisher Scientific, Inc.).
Following washing, membranes were incubated with a 1:3,000 solution
of goat anti-rabbit IgG HRP-conjugated secondary antibody (cat. no.
7074; Cell Signaling Technology, Inc., Danvers, MA, USA) for 2 h at
room temperature. The antigen-antibody complexes were detected
using Western Lightening Plus enhanced chemiluminescence reagent
(cat. no. NEL103E001EA; PerkinElmer, Inc., Waltham, MA, USA) with
β-actin as the internal control protein. For immunoblotting
quantification, the band intensity of the target protein was
normalized to the internal control protein band from the same lane
(ImageJ software, version 1.6; National Institutes of Health,
Bethesda, MD, USA). Data were presented as the mean ± standard
deviation of ≥3 replications for each group.
Apoptosis assay
At 48 h post-transfection, the cells were washed
twice with cold PBS and dual staining was performed using the
Annexin V-fluorescein isothiocyanate (FITC) Apoptosis
Staining/Detection kit (cat. no. 55654; BD Biosciences, Franklin
Lanes, NJ, USA). Briefly, cells were resuspended in 1X binding
buffer at a concentration of 1×106 cells/ml. The 100 µl
solution was transferred to a 5-ml culture tube. A total of 5 µl
Annexin V-FITC and 5 µl propidium iodide were added to the tube and
incubated for 15 min at 37°C. A total of 400 µl 1X binding buffer
was added to each tube. The cells were evaluated by flow cytometer
(BD Biosciences) and data were analyzed by FCS Express software
(version 3; De Novo Software, Los Angeles, CA, USA).
Cell migration assay
Following transfection, the cells were collected and
resuspended in serum-free DMEM at a concentration of
1×105 cells/ml. The lower chambers of Transwells (8 µm
pore size; Corning Incorporated, Corning, NY, USA) were filled with
800 µl DMEM with 10% FBS, and 400 µl cell suspension was added to
the upper chamber. Subsequent to incubation at 37°C with 5%
CO2 in air for 48 h, cells on the lower surface were
fixed with 75% ethanol for 30 min at room temperature and stained
with 0.1% crystal violet for 2 min at room temperature. The
migrated cells were counted at magnification, ×60 using a light
microscope (BX60; Olympus Corporation, Tokyo, Japan). Cells were
counted in 3–5 fields of interest, which were randomly selected on
each membrane, and the average number of cells was calculated.
Statistical analysis
Statistical analyses were performed using SPSS 19.0
software (IBM Corp., Armonk, NY, USA) and GraphPad Prism 5
(GraphPad Software, Inc., La Jolla, CA, USA). P<0.05 was
considered to indicate a statistically significant difference. The
results were analyzed with χ2 tests.
Results
Detection of Beclin-1 protein
expression in gastric tumor tissues
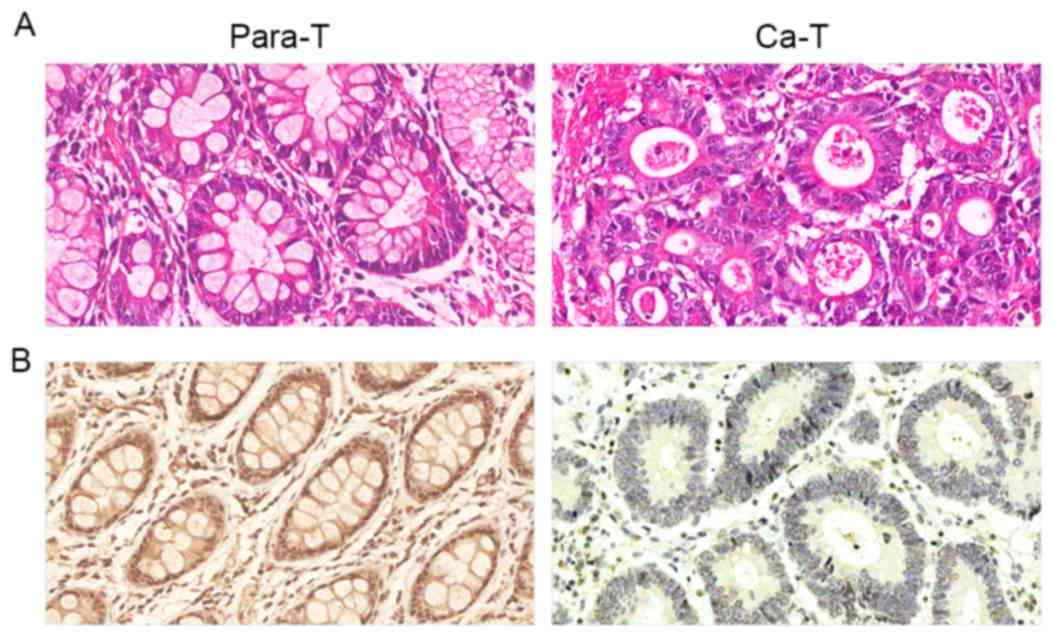
The immunohistochemical results demonstrated that
positive Beclin-1 staining was primarily localized in the cytoplasm
and occasionally in the nuclei (Fig.
1). Beclin-1 protein expression was positive in the majority of
para-carcinoma tissue samples, with a positive expression rate of
76.67% (46/60). Gastric cancer tissues were primarily negative or
weakly positive for Beclin-1, with a positive expression rate of
36.67% (22/60). The frequency of positive Beclin-1 expression in
gastric carcinoma tissue samples was significantly lower compared
with that in para-carcinoma samples (P<0.01; Table I).
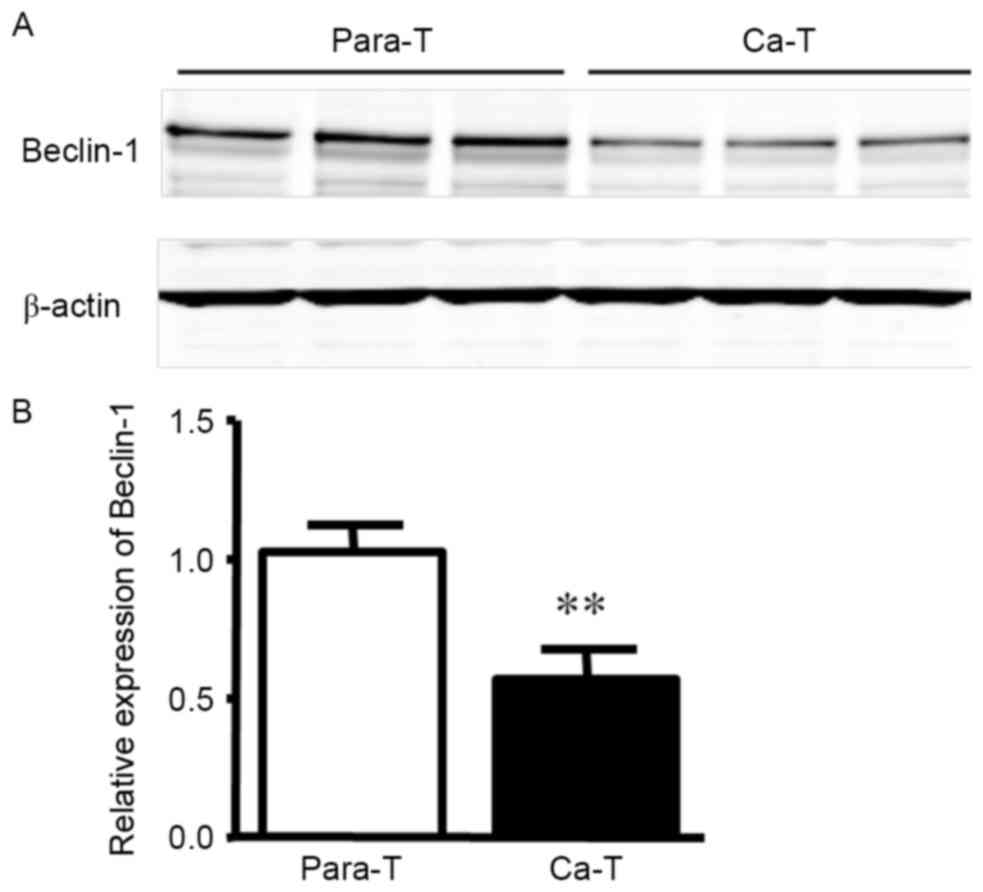
Detection of Beclin-1 protein
expression in gastric carcinoma tissues by western blotting
Western blot analysis indicated that Beclin-1
protein expression in para-carcinoma tissue samples was markedly
higher than in tumor tissue (Fig. 2;
1.024±0.097 vs. 0.572±0.102; P<0.01).
Correlation between Beclin-1 protein
expression and clinical characteristics
Based on the Beclin-1 protein expression levels
detected by immunohistochemical staining, the gastric carcinoma
specimens were divided into a low expression group (−, +) and a
high expression group (++, +++). The clinical data of 60 cases were
reviewed, and it was demonstrated that Beclin-1 protein expression
was not associated with age, sex, tumor size, differentiation or
lymph node metastasis, whereas it was associated with TNM stage
(P=0.008) and invasion status (Table
II; P=0.035).
 | Table II.Association between Beclin-1
expression and clinical parameters in gastric carcinoma tissue
samples. |
Table II.
Association between Beclin-1
expression and clinical parameters in gastric carcinoma tissue
samples.
| Clinical
parameters | Cases | Low expression (−,
+) | High expression (++,
+++) | χ2 | P-value |
|---|
| Age, years |
|
|
| 1.279 | 0.258 |
|
<60 | 27 | 15 | 12 |
|
|
| ≥60 | 33 | 23 | 10 |
|
|
| Sex |
|
|
| 0.745 | 0.388 |
| Male | 37 | 25 | 12 |
|
|
|
Female | 23 | 13 | 10 |
|
|
| Tumor size, cm |
|
|
| 0.191 | 0.662 |
|
<5 | 36 | 22 | 14 |
|
|
| ≥5 | 24 | 16 | 8 |
|
|
|
Differentiation |
|
|
| 0.269 | 0.604 |
|
Low | 38 | 25 | 13 |
|
|
|
Moderate-high | 22 | 13 | 9 |
|
|
| Tumor node
metastasis stage |
|
|
| 6.969 | 0.008a |
|
I/II | 39 | 20 | 19 |
|
|
|
III/IV | 21 | 18 | 3 |
|
|
| Lymph node
metastasis |
|
|
|
|
|
|
Negative | 36 | 26 | 10 | 3.060 | 0.080 |
|
Positive | 24 | 12 | 12 |
|
|
| Invasion
status |
|
|
|
|
|
| Mucosa
and muscle | 42 | 23 | 19 | 4.429 | 0.035b |
|
Serosa | 18 | 15 | 3 |
|
|
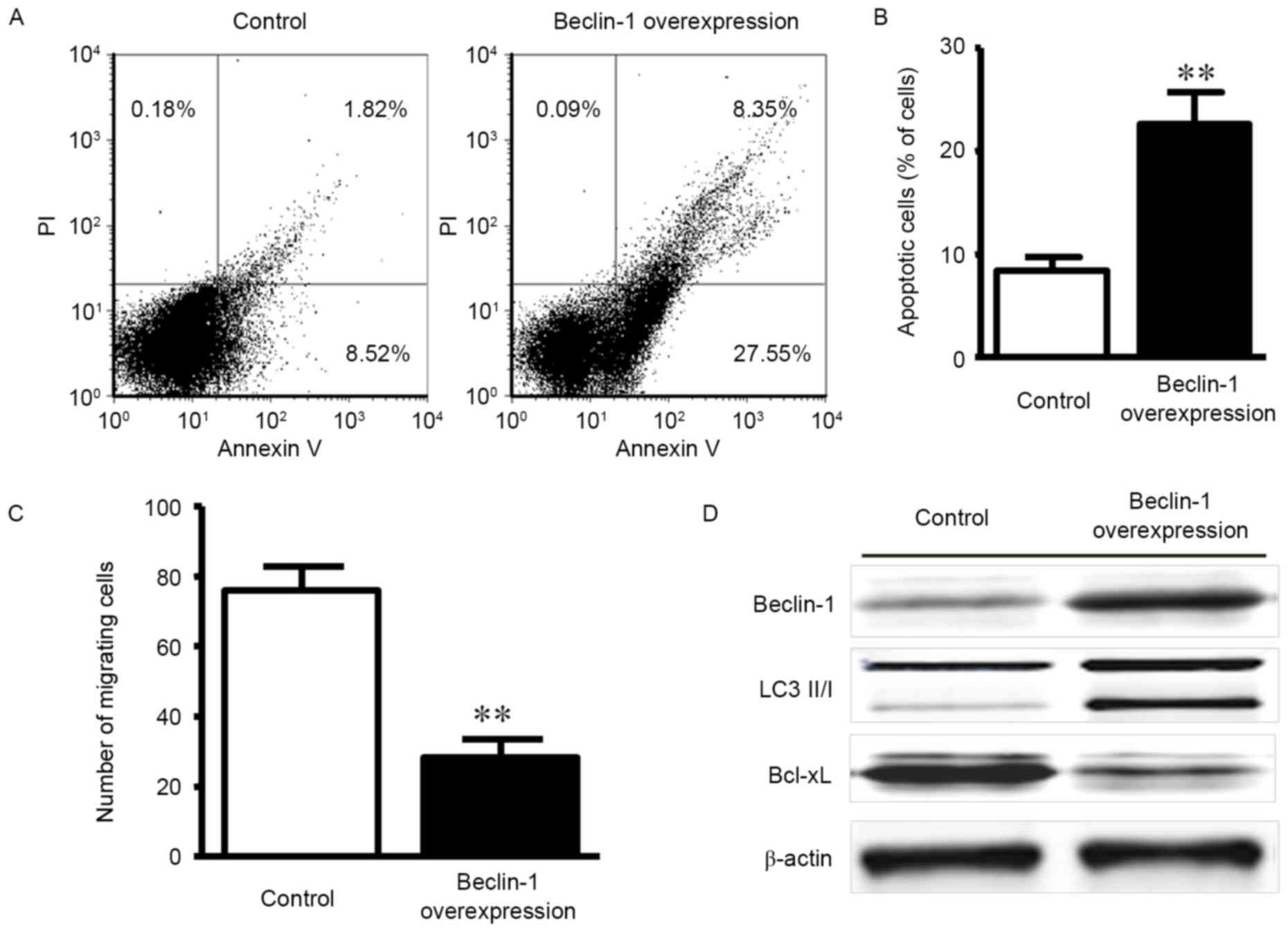
Gastric cancer cell line apoptosis and
migration in Beclin-1 overexpression
As Beclin-1 expression was low in the gastric
carcinoma tissues and correlated with the TNM stage and invasion
status of patients, the present study aimed to characterize the
role of Beclin-1 in gastric cancer. A previous study indicated that
Beclin-1 expression was associated with tumor cell apoptosis and
migration (6); therefore, we
hypothesized that Beclin-1 acted through a similar mechanism in
gastric cancer. Beclin-1-overexpressing gastric cancer cells were
produced by the plasmid transfection of MKN-45 gastric cancer
cells. As indicated in Fig. 3A, the
apoptosis rate was increased in the overexpression group
(22.6±3.1%) compared with the control transfection group (8.4±1.4%)
at 48 h post-transfection. A significant difference in the
apoptosis rate between the groups was observed (Fig. 3B; P<0.01).
A Transwell assay was employed to evaluate the
effect of Beclin-1 on MKN-45 cell invasion. The numbers of
migrating cells in the Beclin-1 overexpression and control groups
were 28.4±5.1 and 75.9±6.9, respectively, which was demonstrated to
be a statistically significant difference (Fig. 3C; P<0.01).
To further explore the mechanisms for
Beclin-1-regulated apoptosis resistance, alterations to the protein
expression levels of the downstream autophagy effector LC 3 II/I
and the pro-apoptotic factor Bcl-xL were examined by western
blotting. Beclin-1 overexpression was associated with the increased
expression of the autophagy effector LC 3 II/I and the reduced
expression of the anti-apoptotic factor Bcl-xL (Fig. 3D). The data indicated that Beclin-1
promoted autophagy and apoptosis.
Discussion
The present study explored whether tumor Beclin-1
expression was associated with the clinical features of patients
with gastric cancer. Reduced Beclin-1 expression levels were
identified in gastric carcinoma tumor tissue, suggesting that
Beclin-1 expression may be associated with the occurrence and
development of gastric cancer. However, the association between
Beclin-1 expression and the clinicopathological characteristics of
different tumors is complex, and under debate. A number of studies
have demonstrated that the high expression of Beclin-1, as
confirmed by immunohistochemistry, is correlated with unfavorable
clinicopathological parameters (3,16,17); however, other studies concluded that
the pathogenesis and progression of cancer were associated with
reduced Beclin-1 expression, which is consistent with the present
study (6,9,18). These
data indicated that Beclin-1 may induce different effects in
different types of tumors, depending on the intrinsic
characteristics of the tumors themselves.
In the present study, Beclin-1 expression in gastric
carcinoma tissue samples was negatively associated with TNM stage
(Table II; P=0.008), although the
association between Beclin-1 expression and other clinical
parameters of the patients, including tumor size, histological
differentiation and metastatic status were not statistically
significant. A previous study identified that Beclin-1 expression
in stage 1 and 2 gastric cancer tissue samples was higher, whereas
Beclin-1 expression levels in stage 3 gastric cancer tissue samples
was significantly lower, than in normal adjacent tissues (4). These data were consistent with the data
of the present study, which revealed that Beclin-1 expression was
associated with gastric carcinoma TNM stage, implying that the
detection of high Beclin-1 expression levels in patients with
gastric cancer may be an effective strategy for predicting the
tumor invasion and stage phenotype.
Autophagy and apoptosis serve a central role in
maintaining homeostasis and disease progression (19), two processes that are regulated by
complex biological processes including interactions between the
autophagy-associated protein Beclin-1 and the anti-apoptotic
protein B-cell lymphoma 2 (Bcl-2) (20,21).
Previous studies identified that Beclin-1 possesses a BH3 domain
near its N-terminus, which may combine with the BH3 domains of
apoptosis-associated proteins including Bcl-2 and Bcl-xL (22,23). The
binding of Bcl-2 family members to Beclin-1 may suppress the
formation of the pro-autophagy Beclin-1/hVps34 complex formation
and reduce Beclin-1-associated PI3 K activity, therefore inhibiting
Beclin-1-induced autophagy (24). In
addition, previous studies demonstrated that higher Bcl-2/Bcl-xL
expression levels were significantly correlated with lower Beclin-1
expression levels in liver and lung cancer, and other types of
tumor tissue (25,26).
However, other BH3-binding proteins, including
Bcl-2-associated agonist of cell death, may also combine with the
Beclin-1 BH3 domain, interrupting the interaction between Beclin-1
and Bcl-2/Bcl-xL, therefore increasing autophagy activity and
inhibiting tumor growth (24,27). The present study identified that the
upregulation of Beclin-1 promoted apoptosis in gastric cancer
cells. A potential mechanism is that Beclin-1 overexpression
inhibited the expression of the anti-apoptotic factor Bcl-xL,
triggering the apoptosis pathway. These results suggest that the
mutual regulation of autophagy and apoptosis serve a pivotal role
in gastric tumorigenesis.
A previous study demonstrated that Beclin-1
lentivirus transfection may inhibit cell migration in tongue
squamous cell carcinoma cell (28).
Vascular endothelial growth factor, and matrix metalloproteinase-2
and −9 were identified as associated with the Beclin-1-mediated
inhibition of migration and invasion (28). Another study suggested that increased
Beclin-1 expression, as a tumor suppressor, contributed to the
inhibition of tumor growth and metastasis in gastric adenocarcinoma
through regulating the hedgehog signaling pathway (29). Therefore, it is reasonable to
hypothesize that Beclin-1 interaction with other associated
molecules regulated tumor cell migration. The data from the present
study demonstrated that the overexpression of Beclin-1 may inhibit
tumor cell migration, which also suggests that Beclin-1 expression
is associated with tumor invasion in gastric cancer. Novel
autophagy-based interventions, including Bcl-2 family regulation,
caspase-dependent cleavage of autophagy-related gene protein and
microRNA mimics to downregulate Beclin-1 have already been
clinically or experimentally applied, suggesting promising
approaches for novel clinical treatments (30–32).
In summary, the present study investigated the
association between Beclin-1 expression, clinically relevant
parameters and gastric carcinoma progression. Beclin-1 was
associated with the invasion status and TNM stage of gastric
carcinoma tissue samples, two major factors affecting the prognosis
of patients with gastric cancer. Increased apoptosis and reduced
cell migration were observed in gastric cancer cells overexpressing
Beclin-1, indicating a potential mechanism for the effect of
Beclin-1 in suppressing gastric cancer progression.
Acknowledgements
The present study was supported by grants from the
Natural Science Foundation of Shanghai Health and Family Planning
Commission (grant no. 201540088) and the China National Natural
Science Foundation of China (grant no. 81370961).
References
|
1
|
Chen N and Karantza-Wadsworth V: Role and
regulation of autophagy in cancer. Biochim Biophys Acta.
1793:1516–1523. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sakakura K, Takahashi H, Kaira K, Toyoda
M, Oyama T and Chikamatsu K: Immunological significance of the
accumulation of autophagy components in oral squamous cell
carcinoma. Cancer Sci. 106:1–8. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen Y, Li X, Wu X, He C, Guo L, Zhang S,
Xiao Y, Guo W and Tan B: Autophagy-related proteins LC3 and
Beclin-1 impact the efficacy of chemoradiation on esophageal
squamous cell carcinoma. Pathol Res Pract. 209:562–567. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fei B, Ji F, Chen X, Liu Z, Li S, Mo Z and
Fang X: Expression and clinical significance of Beclin-1 in gastric
cancer tissues of various clinical stages. Oncol Lett.
11:2271–2277. 2016.PubMed/NCBI
|
|
5
|
Zhao Y, Chen S, Gou WF, Xiao LJ, Takano Y
and Zheng HC: Aberrant Beclin 1 expression is closely linked to
carcinogenesis, differentiation, progression, and prognosis of
ovarian epithelial carcinoma. Tumour Biol. 35:1955–1964. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Qiu DM, Wang GL, Chen L, Xu YY, He S, Cao
XL, Qin J, Zhou JM, Zhang YX and E Q: The expression of beclin-1,
an autophagic gene, in hepatocellular carcinoma associated with
clinical pathological and prognostic significance. BMC Cancer.
14:3272014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Miracco C, Cevenini G, Franchi A, Luzi P,
Cosci E, Mourmouras V, Monciatti I, Mannucci S, Biagioli M and
Toscano M: Beclin 1 and LC3 autophagic gene expression in cutaneous
melanocytic lesions. Hum Pathol. 41:503–512. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zaanan A, Park JM, Tougeron D, Huang S, Wu
TT, Foster NR and Sinicrope FA: Association of beclin 1 expression
with response to neoadjuvant chemoradiation therapy in patients
with locally advanced rectal carcinoma. Int J Cancer.
137:1498–1502. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang Y, Wang C, Tang H, Wang M, Weng J,
Liu X, Zhang R, Huang H and Hou J: Decrease of autophagy activity
promotes malignant progression of tongue squamous cell carcinoma. J
Oral Pathol Med. 42:557–564. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liang XH, Jackson S, Seaman M, Brown K,
Kempkes B, Hibshoosh H and Levine B: Induction of autophagy and
inhibition of tumorigenesis by beclin 1. Nature. 402:672–676. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lozy F and Karantza V: Autophagy and
cancer cell metabolism. Semin Cell Dev Biol. 23:395–401. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kongara S, Kravchuk O, Teplova I, Lozy F,
Schulte J, Moore D, Barnard N, Neumann CA, White E and Karantza V:
Autophagy regulates keratin 8 homeostasis in mammary epithelial
cells and in breast tumors. Mol Cancer Res. 8:873–884. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gallagher LE, Williamson LE and Chan EY:
Advances in autophagy regulatory mechanisms. Cells. 5:pii: E242016.
View Article : Google Scholar
|
|
14
|
Ozpolat B and Benbrook DM: Targeting
autophagy in cancer management-strategies and developments. Cancer
Manag Res. 7:291–299. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Washington K: 7th edition of the AJCC
cancer staging manual: Stomach. Ann Surg Oncol. 17:3077–3079. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Guo GF, Jiang WQ, Zhang B, Cai YC, Xu RH,
Chen XX, Wang F and Xia LP: Autophagy-related proteins Beclin-1 and
LC3 predict cetuximab efficacy in advanced colorectal cancer. World
J Gastroenterol. 17:4779–4786. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tang JY, Hsi E, Huang YC, Hsu NC, Chu PY
and Chai CY: High LC3 expression correlates with poor survival in
patients with oral squamous cell carcinoma. Hum Pathol.
44:2558–2562. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Burada F, Nicoli ER, Ciurea ME, Uscatu DC,
Ioana M and Gheonea DI: Autophagy in colorectal cancer: An
important switch from physiology to pathology. World J Gastrointest
Oncol. 7:271–284. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Nikoletopoulou V, Markaki M, Palikaras K
and Tavernarakis N: Crosstalk between apoptosis, necrosis and
autophagy. Biochim Biophys Acta. 1833:3448–3459. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yue Z, Jin S, Yang C, Levine AJ and Heintz
N: Beclin 1, an autophagy gene essential for early embryonic
development, is a haploinsufficient tumor suppressor. Proc Natl
Acad Sci USA. 100:pp. 15077–15082. 2003; View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhao GX, Pan H, Ouyang DY and He XH: The
critical molecular interconnections in regulating apoptosis and
autophagy. Ann Med. 47:305–315. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Pattingre S, Tassa A, Qu X, Garuti R,
Liang XH, Mizushima N, Packer M, Schneider MD and Levine B: Bcl-2
antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell.
122:927–939. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Maiuri MC, Le Toumelin G, Criollo A, Rain
JC, Gautier F, Juin P, Tasdemir E, Pierron G, Troulinaki K,
Tavernarakis N, et al: Functional and physical interaction between
Bcl-X(L) and a BH3-like domain in Beclin-1. EMBO J. 26:2527–2539.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Levine B, Sinha SC and Kroemer G: Bcl-2
family members: Dual regulators of apoptosis and autophagy.
Autophagy. 4:600–606. 2008. View Article : Google Scholar
|
|
25
|
Luo S and Rubinsztein DC: Apoptosis blocks
Beclin 1-dependent autophagosome synthesis: An effect rescued by
Bcl-xL. Cell Death Differ. 17:268–277. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kang R, Zeh HJ, Lotze MT and Tang D: The
Beclin 1 network regulates autophagy and apoptosis. Cell Death
Differ. 18:571–580. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lian J, Wu X, He F, Karnak D, Tang W, Meng
Y, Xiang D, Ji M, Lawrence TS and Xu L: A natural BH3 mimetic
induces autophagy in apoptosis-resistant prostate cancer via
modulating Bcl-2-Beclin1 interaction at endoplasmic reticulum. Cell
Death Differ. 18:60–71. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Weng J, Wang C, Wang Y, Tang H, Liang J,
Liu X, Huang H and Hou J: Beclin1 inhibits proliferation, migration
and invasion in tongue squamous cell carcinoma cell lines. Oral
Oncol. 50:983–990. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Won KY, Kim GY, Lim SJ, Sung JY, Kim YW,
Park YK, Lee J and Choi HS: Autophagy is related to the hedgehog
signaling pathway in human gastric adenocarcinoma: Prognostic
significance of Beclin-1 and Gli2 expression in human gastric
adenocarcinoma. Pathol Res Pract. 211:308–315. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mathew R, Kongara S, Beaudoin B, Karp CM,
Bray K, Degenhardt K, Chen G, Jin S and White E: Autophagy
suppresses tumor progression by limiting chromosomal instability.
Genes Dev. 21:1367–1381. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hasima N and Ozpolat B: Regulation of
autophagy by polyphenolic compounds as a potential therapeutic
strategy for cancer. Cell Death Dis. 5:e15092014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Geng Z, Xu F and Zhang Y:
MiR-129-5p-mediated Beclin-1 suppression inhibits endothelial cell
autophagy in atherosclerosis. Am J Transl Res. 8:1886–1894.
2016.PubMed/NCBI
|