Introduction
Based on the 2014 World Health Organization report,
breast cancer is the second most life-threatening tumor (following
lung cancer) for women in China (1).
Numerous genes were identified to be abnormal in breast cancer,
with different biological significance (2). It is well known that the Erb-B2 receptor
tyrosine kinase 2 gene [also known as human epidermal growth factor
receptor (HER)2], which encodes a member of the ErbB family, serves
essential roles in breast cancer carcinogenesis, invasion and
metastasis (3,4). Additionally, HER2 amplification
is a well-established biomarker for the treatment of breast and
gastric carcinomas with trastuzumab (5,6).
Epidermal growth factor receptor (EGFR),
which also encodes a family member of the ErbB family, serves
essential roles in breast cancer. EGFR is a well-established
treatment target for colorectal cancer, non-small cell lung cancer,
and squamous cell carcinoma of the head and neck (7). Furthermore, a high EGFR gene copy
number was significantly associated with poor clinical outcome
(8–10). EGFR overexpression was reported
to be significantly correlated with poor clinical outcome in breast
cancer (11). EGFR is also a
target for EGFR-tyrosine kinase inhibitor therapy for EGFR
mutation and EGFR amplification of cancer patients (5,6).
Since both EGFR and HER2 belong to the
same family and share a high degree of structural and functional
homology (12), the present study
evaluated the gene amplification status and clinical significance
in breast cancer of other members, including HER3 and
HER4. It has been reported that EGFR, HER2,
HER3 and HER4 constitute a complex network, coupling
various extracellular ligands to intracellular signal transduction
pathways, resulting in receptor interaction and cross-activation
(12). Members of the ErbB family are
critically involved in the development and progression of breast
cancer. Amplification of the four members of the ErbB family has
been detected by droplet digital polymerase chain reaction (ddPCR)
(13), fluorescence in situ
hybridization (FISH) (12) and
next-generation sequencing (NGS) (2,6) at
different rates, with no clinical outcome implications. Since these
molecules belong to the same family and share certain homologous
domains, the present study aimed to assess whether there are
invasive ductal carcinoma (IDC) patients with amplification of ≥2
ErbB family members. Additionally, the current study sought to
determine the clinical significance of the amplification of
multiple gene (such as, tumor genesis, invasion and metastasis), as
well as their prognostic values and therapeutic responses.
Thus, the quantification of all four ErbB family
member receptors as a whole panel in IDC may shed light on their
amplification status. Therefore, the amplification status of the
four ErbB family members and their clinical implications was
detected in 119 breast carcinoma patients with an average follow-up
of 27.0 months in the present study.
Materials and methods
Patients and sample preparation
The samples were human breast neoplasm tissue
specimens removed during surgery. Patients anonymity was preserved
in all cases. Approval for the study was granted by the Ethics
Committee of West China Hospital (Chengdu, China; approval no.
2013-191), who also waived the requirement for patient consent.
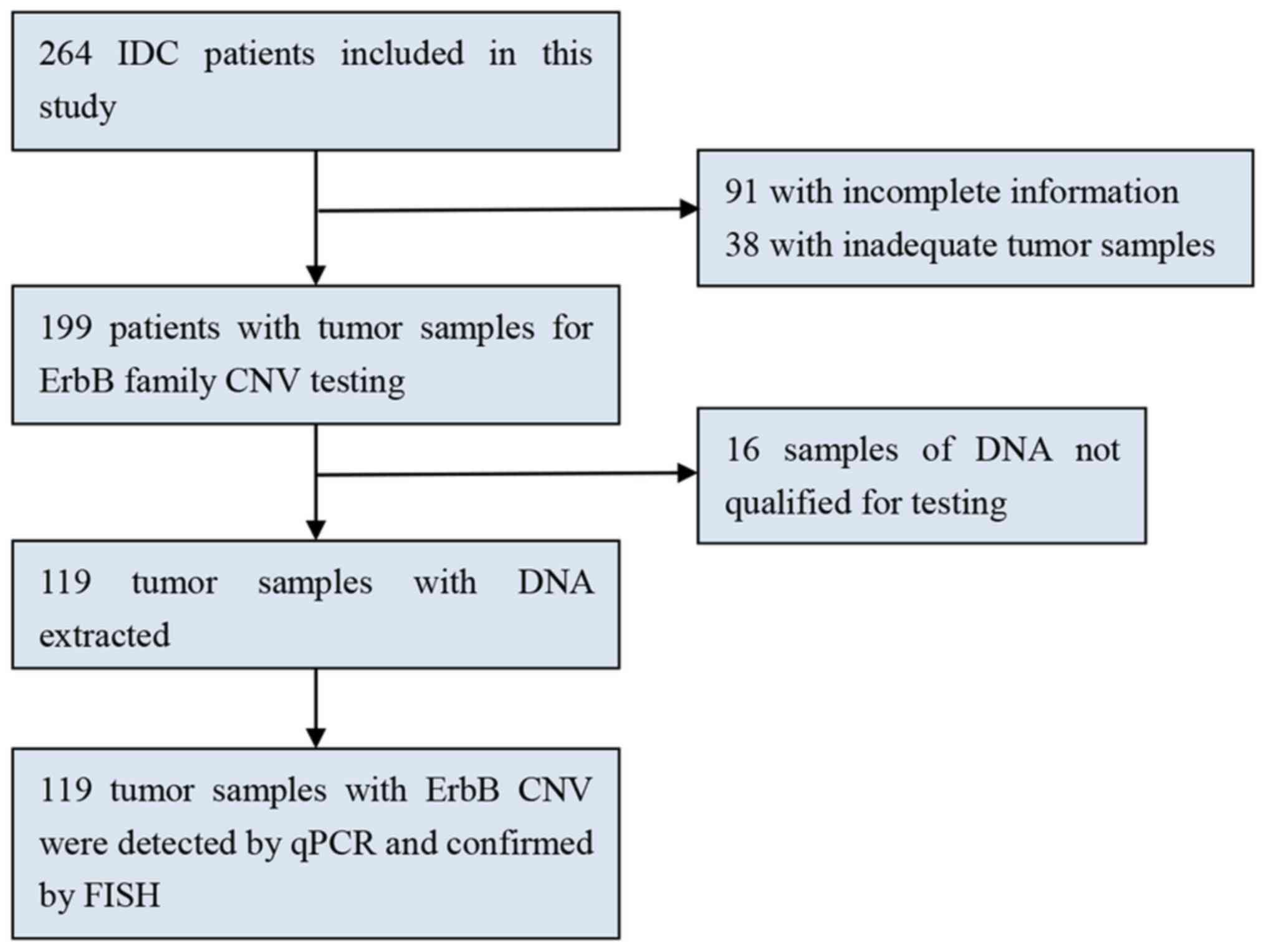
Formalin-fixed paraffin-embedded (FFPE) samples from 119 patients
with breast cancer who underwent breast mastectomy between January
2010 and December 2012 at West China Hospital were analyzed in the
present study (Fig. 1). Surgical
specimens were obtained prior to systemic treatment, and paraffin
embedding was performed within the framework of diagnostic
procedures. Disease-free survival (DFS) and overall survival (OS)
were defined as the time between the initial surgery and local or
distant metastatic relapse, and the time between surgery and
mortality, respectively.
DNA isolation and quantitative PCR
(qPCR)
Tumor areas (≥1 cm2) from 4.0 µm-thick
unstained FFPE sections were macrodissected. DNA was isolated from
two 4 µm-thick tissue sections using a QIAamp DNA FFPE Tissue kit
(Qiagen GmbH, Hilden, Germany) according to the manufacturer's
protocol. DNA quantitation was performed using a NanoDrop 2000
(Thermo Fisher Scientific, Inc., Wilmington, DE, USA). Finally, DNA
purity was confirmed by measuring the absorbance (A)260/A280 ratio.
Good-quality DNA was indicated by a ratio of A260/A280 nm =
1.70–1.95. Reactions were carried out using a CFX96 Touch™
Real-Time PCR Detection System (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA). The thermocycling conditions for qPCR were as
follows: 98°C for 2 min, followed by 39 amplification cycles at
98°C for 15 sec and 60°C for 15 sec. Each gene was measured in
triplicate and normalized relative to a set of two reference genes
[GAPDH and transferrin receptor (TFRC)] (Table I). Relative quantitation of ErbB gene
amplification in IDC was calculated by the 2−∆∆Cq method
(14) using the mean copy number in
50 normal control samples and reference genes (GAPDH and TFRC). A
sample was considered positive for EGFR, HER2, HER3
and HER4 gene amplification if the above ratio was >2,
whereas a ratio of <2 indicated that the sample was negative for
EGFR, HER2, HER3 and HER4 gene
amplification (15,16) (Table
I).
 | Table I.Quantitative polymerase chain
reaction primers of the ErbB family. |
Table I.
Quantitative polymerase chain
reaction primers of the ErbB family.
| Gene | GenBank no. | Oligo name | Oligo sequence | Target size
(bp) |
|---|
| TFRC | NC_000003.12 | TFRC-F |
5′-ACTTCCTCTCTCCCTACGTATC-3′ | 105 |
|
|
| TFRC-R |
5′-GCAGTTTCAAGTTCTCCAGTAAAG-3′ |
|
| GAPDH | NG_007073.2 | GAPDH-F |
5′-CCTCAAGATCATCAGCAATGCCTC-3′ | 100 |
|
|
| GAPDH-R |
5′-GTGGTCATGAGTCCTTCCACGATA-3′ |
|
| EGFR | NG_007726.3 | EGFR-F |
5′-CGGGACGTTTCGTTCTTCGG-3′ | 130 |
|
|
| EGFR-R |
5′-GAAAGTTGGGAGCGGTTCGG-3′ |
|
| HER2 | NG_007503.1 | HER2-F |
5′-ATGAGCTACCTGGAGGATGT-3′ | 103 |
|
|
| HER2-R |
5′-CCAGCCCGAAGTCTGTAATTT-3′ |
|
| HER3 | NG_011529.1 | HER3-F |
5′-CCTCAACCTGCTCCTCTTTATT-3′ | 168 |
|
|
| HER3-R |
5′-GGCTACAACAGTGAGACCATAG-3′ |
|
| HER4 | NG_011805.1 | HER4-F |
5′-TTGCACGACTTTCTCACGGC-3′ | 130 |
|
|
| HER4-R |
5′-GCTGCTGACCTGAAGGCACT-3′ |
|
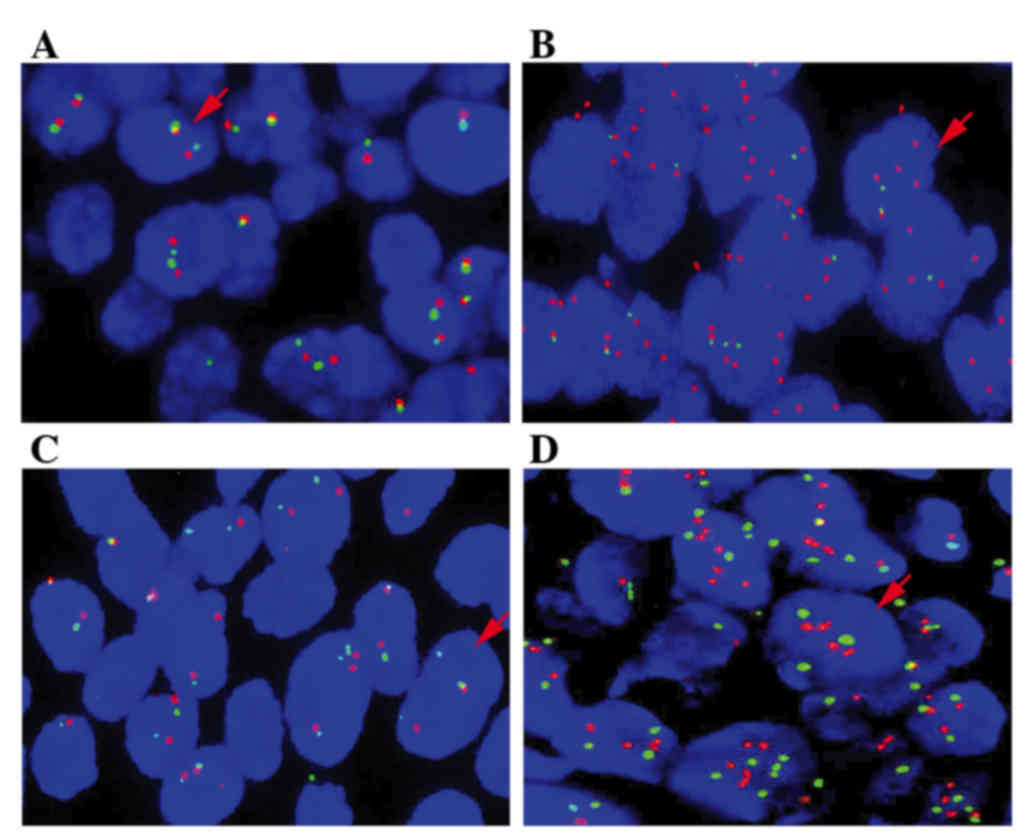
FISH
To confirm the EGFR and HER2 copy
number, FISH was conducted using EGFR and HER-2 DNA Probe kits (LBP
China, Inc., Guangzhou, China). FFPE tissues were prepared in
serial 4-µm sections on microscope slides. A set of tissue was used
for two-color FISH. SpectrumOrange-labeled gene-specific probes
were used together with SpectrumGreen-labeled probes (LBP China,
Inc.) for the respective centromere region as references. The probe
combinations were as follows: HER2, LBP EGFR
SpectrumOrange/centromere (CEP) 17 SpectrumGreen; and EGFR,
LBP EGFR SpectrumOrange/CEP7 SpectrumGreen. Prior to hybridization,
the tissues were deparaffinized, air dried, and dehydrated in 70,
85 and 100% ethanol, followed by denaturation for 5 min at 85°C.
Upon overnight hybridization at 37°C in a humidified chamber, the
slides were washed and counterstained with 0.1% NP-40 in an
antifade solution (LBP China, Inc.), and viewed under a
fluorescence microscope. For each tumor, the predominant gene and
centromere copy numbers were estimated. Under a fluorescence
microscope, signals of the EGFR probe appear red, while
signals of the centromere 7 probe appear green. Red and green
signals were counted in 40 tumor cells, and the ratio of red:green
signals was calculated. EGFR and HER2 were considered
amplified if the oncogene/centromere ratio was >2 (17,18).
Statistical analysis
Statistical analyses were conducted using SPSS
version 16.0 software (SPSS, Inc., Chicago, IL, USA), and
two-tailed P<0.05 was considered to indicate a statistically
significant difference. Associations between the prevalence of
EGFR and HER2 amplification and clinical parameters
were evaluated using the χ2 test. Univariate survival
analysis was conducted using the Kaplan-Meier method, and
multivariate survival analysis was carried out using the Cox
proportional hazards model.
Results
Baseline clinical characteristics
All the patients included in the present study were
females, ranging in age from 29 to 74 years (mean, 49.3 years). The
mean DFS was 25.6 months, and the mean OS was 27.0 months. The DFS
and OS of the 119 patients are listed in Table II with respect to histopathological
characteristics and prognostic factors, including age, histological
grading, tumor size, nodal status, metastasis, clinical stage, and
estrogen receptor (ER), progesterone receptor (PR) and HER2/neu
status. As expected, nodal metastasis status, clinical state
(P=0.025) and distant metastasis status (when diagnosed)
(P<0.001) were observed to be significantly correlated with DFS.
Larger tumor size, positive-node status, higher clinical state and
metastasis (when diagnosed) were associated with DFS. However, none
of the histopathological characteristics was significantly
associated with OS (Table II).
 | Table II.Baseline clinical characteristics of
the study subjects (n=119). |
Table II.
Baseline clinical characteristics of
the study subjects (n=119).
|
|
| Disease-free
survival | Overall
survival |
|---|
|
|
|
|
|
|---|
| Characteristic | No. (%) | Log-rank | P-value | Log-rank | P-value |
|---|
| Age, years | 49.3
(29–74)b | 0.658 | 0.417 | 0.756 | 0.385 |
|
≤50 | 70 (58.8) |
|
|
|
|
|
>50 | 49 (41.2) |
|
|
|
|
| Grade |
| 2.245 | 0.134 | 2.633 | 0.105 |
|
G1-G2 | 40 (33.6) |
|
|
|
|
| G3 | 79 (66.4) |
|
|
|
|
| Tumor
sizea |
| 4.696 | 0.032c | 2.491 | 0.114 |
|
T0-2 | 111 (93.7) |
|
|
|
|
|
T3-4 | 7 (5.9) |
|
|
|
|
| Nodal
statusa |
| 5.065 | 0.024c | 1.567 | 0.211 |
| N0 | 54 (45.8) |
|
|
|
|
|
N1-N3 | 64 (54.2) |
|
|
|
|
| Metastasis |
| 118.000 |
<0.001c | 0.026 | 0.871 |
| M0 | 118 (98.3) |
|
|
|
|
| M1 | 1 (0.8) |
|
|
|
|
| Clinical
stagea |
| 5.020 | 0.025c | 0.725 | 0.394 |
|
I–II | 90 (76.3) |
|
|
|
|
|
III–IV | 28 (23.7) |
|
|
|
|
| ER status |
| 0.156 | 0.692 | 1.619 | 0.203 |
|
ER+ | 40 (33.6) |
|
|
|
|
|
ER− | 79 (66.4) |
|
|
|
|
| PR
statusa |
| 1.685 | 0.194 | 0.290 | 0.590 |
|
PR+ | 43 (36.8) |
|
|
|
|
|
PR− | 74 (63.2) |
|
|
|
|
| HER2a |
| 1.975 | 0.372 | 0.046 | 0.977 |
|
0-1+ | 65 (54.6) |
|
|
|
|
| 2+ | 25 (21.0) |
|
|
|
|
| 3+ | 28 (23.5) |
|
|
|
|
Gene amplification of ErbB family
members by qPCR and FISH
The gene amplification of 119 patients was detected
using qPCR (Table III) and was
confirmed by FISH (Fig. 2). The
relative quantitation of ErbB amplification in IDC was calculated
by the 2−∆∆Cq method using the mean copy number in 50
normal control samples and reference genes (GAPDH and TFRC). A
sample was considered positive for EGFR, HER2,
HER3 and HER4 gene amplification if the ratio was
>2, whereas a ratio <2 indicated that the sample was negative
for EGFR, HER2, HER3 and HER4 gene
amplification. EGFR amplification was detected in 30
patients (25.2%), while HER2 amplification was detected in
44 (36.9%) patients. However, in the present study, only one
patient was detected to have HER4 amplification but no
amplification of HER3. Furthermore, a group of 17 patients
(14.2%) with both EGFR and HER2 gene amplification
was identified. A total of 62 patients (52.1%) were identified to
have neither EGFR nor HER2 genes amplified. In one
patient with HER4 gene amplification, the EGFR,
HER2 and HER3 genes were not observed to be
amplified.
 | Table III.EGFR and HER2 gene
amplification in the present cohort. |
Table III.
EGFR and HER2 gene
amplification in the present cohort.
|
| HER2, no.
(%) |
|---|
|
|
|
|---|
| EGFR | Amp. | No amp. | Total |
|---|
| Amp. | 17 | 13 | 30 (25.2) |
| No amp. | 27 | 62 | 89 (74.8) |
| Total | 44 (36.9) | 75 (63.1) | 119 (100.0) |
Clinicohistopathological features of
EGFR and HER2 amplification in breast cancer
To identify any correlation between the gene
amplification status of the ErbB family and clinical
characteristics (Table IV), the
correlation between EGFR and HER2 amplification and
clinical features was analyzed. Patients with EGFR and
HER2 amplification, as well as those with EGFR and
HER2 co-amplification, were analyzed regarding age,
histological grading, tumor size, nodal status, metastasis,
clinical stage, ER, PR and HER2/neu status, local
recurrence, and distant metastasis. In the present study,
EGFR amplification was significantly associated with ER
expression (P=0.028), local recurrence (P=0.015) and distant
metastasis (following initial surgery) (P=0.011). Additionally,
EGFR amplification primarily occurred in tumors with a high
histological grade (Table IV).
HER2 amplification was associated with larger tumor size
(P=0.006), later clinical stage (P=0.003) and distant metastasis
(following initial surgery) (P=0.006). HER2 amplification,
as expected, was also significantly associated with HER2 expression
(P<0.001) and distant metastasis (following initial surgery)
(P=0.006) (Table IV).
 | Table IV.Prevalence of EGFR and
HER2 amplification in breast tumors stratified according to
clinical characteristics. |
Table IV.
Prevalence of EGFR and
HER2 amplification in breast tumors stratified according to
clinical characteristics.
|
| EGFR
amplification | HER2
amplification | EGFR and
HER2 co-amplification |
|---|
|
|
|
|
|
|---|
| Characteristic | P (n=30) No.
(%) | N (n=89) No.
(%) | P-value | P (n=44) No.
(%) | N (n=75) No.
(%) | P-value | P (n=17) No.
(%) | N (n=102) No.
(%) | P-value |
|---|
| Age, years |
|
| 0.781 |
|
| 0.468 |
|
| 0.595 |
|
≤50 | 17 (56.7) | 53 (59.6) |
| 24 (54.5) | 46 (61.3) |
| 11 (64.7) | 59 (57.8) |
|
|
>50 | 13 (43.3) | 36 (40.4) |
| 20 (45.5) | 29 (38.7) |
| 6 (35.3) | 43 (42.2) |
|
| Grading |
|
| 0.628 |
|
| 0.128 |
|
| 0.416 |
|
G1-G2 | 9 (30.0) | 31 (34.8) |
| 11 (25.0) | 29 (38.7) |
| 4 (23.5) | 36 (35.3) |
|
| G3 | 21 (70.0) | 58 (65.2) |
| 33 (75.0) | 46 (61.3) |
| 13 (76.5) | 66 (64.7) |
|
| Tumor
sizea |
|
| 0.844 |
|
| 0.006b |
|
| 0.271 |
|
T0-2 | 28 (93.3) | 83 (94.3) |
| 38 (86.4) | 73 (98.6) |
| 15 (88.2) | 96 (95.0) |
|
|
T3-4 | 2 (6.7) | 5 (5.7) |
| 6 (24.4) | 1 (1.4) |
| 2 (11.8) | 5 (5.0) |
|
| Nodal
statusa |
|
| 0.590 |
|
| 0.414 |
|
| 0.349 |
| N0 | 15 (50.0) | 39 (44.3) |
| 18 (40.9) | 36 (48.6) |
| 6 (35.3) | 48 (47.5) |
|
|
N1-N3 | 15 (50.0) | 49 (55.7) |
| 26 (59.1) | 38 (51.4) |
| 11 (64.7) | 53 (52.5) |
|
| Metastasis |
|
| 0.084 |
|
| 0.190 |
|
| 0.014b |
| M0 | 29 (96.7) | 89 (100.0) |
| 43 (97.7) | 75 (100.0) |
| 16 (94.1) | 102 (100.0) |
|
| M1 | 1 (3.3) | 0 (0.0) |
| 1 (2.3) | 0 (0.0) |
| 1 (5.9) | 0 (0.0) |
|
| Clinical
stagea |
|
| 0.661 |
|
| 0.003b |
|
| 0.068 |
|
I–II | 22 (73.3) | 68 (77.3) |
| 27 (61.4) | 63 (85.1) |
| 10 (58.8) | 80 (79.2) |
|
|
III–IV | 8 (26.7) | 20 (22.7) |
| 17 (38.6) | 11 (14.9) |
| 7 (41.2) | 21 (20.8) |
|
| ER status |
|
| 0.028b |
|
| 0.197 |
|
| 0.476 |
|
ER+ | 15 (50.0) | 25 (28.1) |
| 18 (40.9) | 22 (29.3) |
| 10 (58.8) | 69 (67.6) |
|
|
ER− | 15 (50.0) | 64 (71.9) |
| 26 (59.1) | 53 (70.7) |
| 7 (41.2) | 33 (32.4) |
|
| PR
statusa |
|
| 0.298 |
|
| 0.689 |
|
| 0.532 |
|
PR+ | 13 (44.8) | 30 (34.1) |
| 19 (45.2) | 24 (32.0) |
| 9 (56.2) | 65 (64.4) |
|
|
PR− | 16 (55.2) | 58 (65.9) |
| 23 (54.8) | 51 (68.0) |
| 7 (43.8) | 36 (35.6) |
|
| HER2a |
|
| 0.753 |
|
|
<0.001b |
|
|
|
|
0-1+ | 18 (60.0) | 47 (53.4) |
| 9 (20.5) | 56 (75.7) |
| 5 (29.4) | 60 (59.4) | 0.062 |
| 2+ | 5 (16.7) | 20 (22.7) |
| 8 (18.2) | 17 (23.0) |
| 5 (29.4) | 20 (19.8) |
|
| 3+ | 7 (25.4) | 21 (23.9) |
| 27 (61.4) | 1 (1.3) |
| 7 (41.2) | 21 (20.8) |
|
| Recurrence |
|
| 0.015b |
|
| 0.554 |
|
| 0.053 |
|
Yes | 3 (10.0) | 0 (0.0) |
| 2 (4.5) | 1 (1.3) |
| 2 (11.8) | 1 (1.0) |
|
| No | 27 (90.0) | 89 (100.0) |
| 42 (95.5) | 74 (98.7) |
| 15 (88.2) | 101 (99.0) |
|
| Distant
metastasis |
|
| 0.011b |
|
| 0.006b |
|
|
<0.001b |
|
Yes | 7 (23.3) | 5 (5.6) |
| 9 (20.5) | 3 (4.0) |
| 7 (41.2) | 5 (4.9) |
|
| No | 23 (76.7) | 85 (94.4) |
| 35 (79.5) | 72 (96.0) |
| 10 (58.8) | 97 (95.1) |
|
Furthermore, a subgroup of patients who harbored
EGFR and HER2 gene co-amplification was identified.
This group of patients was significantly correlated with metastasis
(at diagnosis) (P=0.014) and distant metastasis (subsequent to
initial surgery) (P<0.001). They were almost significantly
correlated with clinical stage (P=0.062), HER2 overexpression
(P=0.062) and local recurrence (P=0.053) (Table IV).
EGFR and HER2 amplification for IDC
prognosis
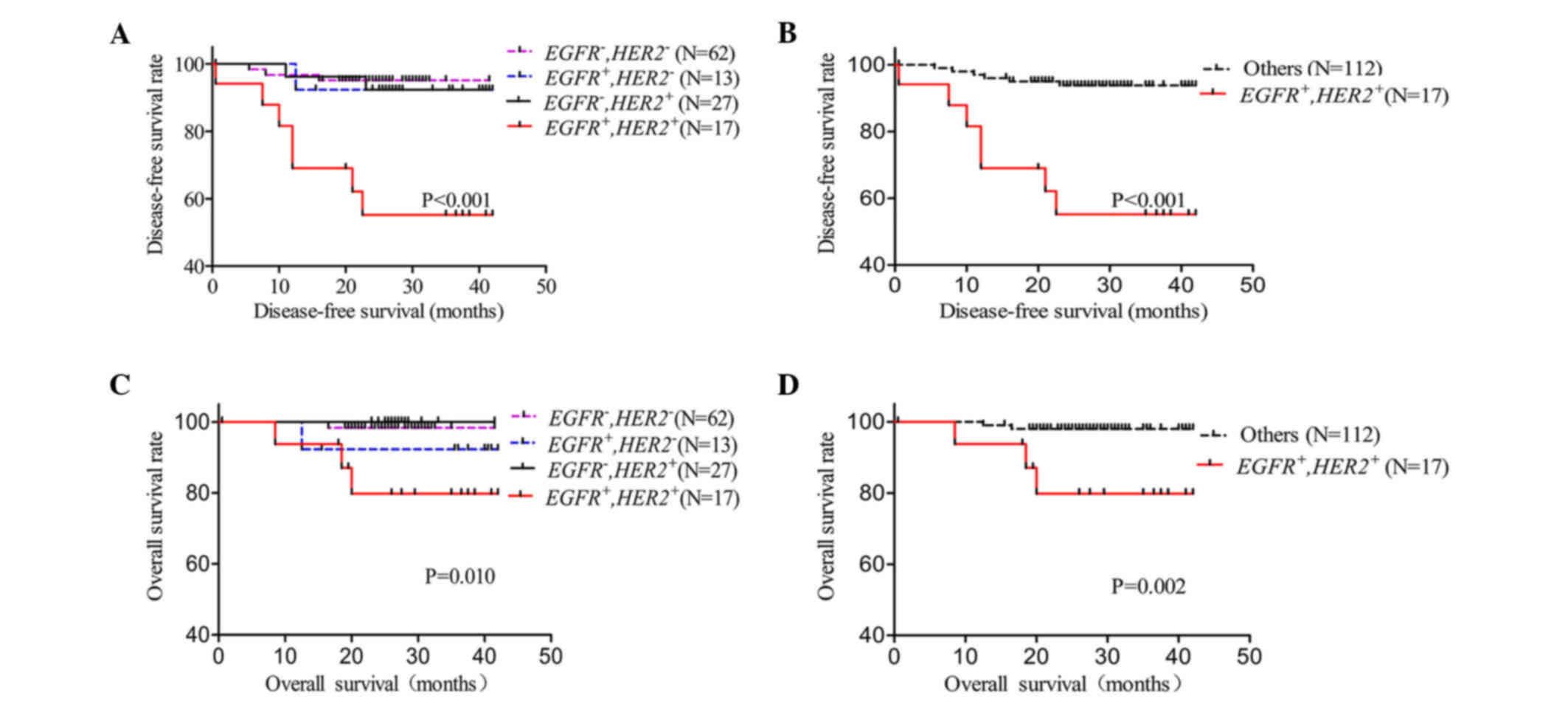
To further reveal the prognostic value of gene
amplification for EGFR or/and HER2 in IDC patients,
the EGFR and/or HER2 amplification status were
evaluated in association with DFS and OS by Kaplan-Meier analysis
(Fig. 3). The 119 patients were
divided into four groups: Nor EGFR or HER2
amplification; EGFR amplification but no HER2
amplification; no EGFR amplification but HER2
amplification; and EGFR and HER2 co-amplification.
The present study revealed that patients with EGFR and
HER2 co-amplification had a significantly shorter DFS
(P<0.001) and OS (P=0.010) than any other group (Fig. 3A and C). Next, differences in the
EGFR and HER2 co-amplification group vs. the no
co-amplification group were analyzed for DFS and OS. In the present
study, EGFR and HER2 co-amplification was observed to
be correlated with both DFS (P<0.001) and OS (P=0.002) (Fig. 3B and D). DFS and OS were also
calculated by Kaplan-Meier analysis for triple-negative breast
cancer (TNBC) (ER−, PR− and HER2−)
(19) in the present study as the
control. The TNBC group did not exhibit any significant difference
with the non-TNBC group for DFS (P=0.538) or OS (P=0.633) (data not
show).
Furthermore, multivariate analysis indicated that
EGFR and HER2 co-amplification was associated with
both DFS (co-amplification vs. no co-amplification; hazard ratio,
10.145; 95% confidence interval, 2.820–36.499; P<0.001) and OS
(co-amplification vs. no co-amplification; hazard ratio, 51.564;
95% confidence interval, 1.467–1,890.000; P=0.032) (data not
shown). Concerning the treatment regimen, EGFR and
HER2 co-amplification patients were significantly correlated
with poor DFS regarding chemotherapy (P<0.001), radiotherapy
(P<0.001) and hormonal therapy (P=0.001) (Table V).
 | Table V.Prevalence of epidermal growth factor
receptor and human epidermal growth factor receptor 2
co-amplification and treatment response. |
Table V.
Prevalence of epidermal growth factor
receptor and human epidermal growth factor receptor 2
co-amplification and treatment response.
|
|
| Disease-free
survival |
|---|
|
|
|
|
|---|
| Treatment | No. (%) | Log-rank | P-value |
|---|
| Chemotherapy | 117 |
|
|
|
Co-amplification | 17 (14.5) | 22.219 |
<0.001a |
| No
co-amplification | 100 (85.5) |
|
|
|
Radiotherapy | 40 | 15.694 |
<0.001a |
|
Co-amplification | 6 (15.0) |
|
|
| No
co-amplification | 34 (85.0) |
|
|
| Hormonal
therapy | 74 | 13.330 | 0.001a |
|
Co-amplification | 9 (12.2) |
|
|
| No
co-amplification | 65 (87.8) |
|
|
Discussion
The gene copy number of ErbB family members has been
determined in a group of 119 IDC patients with an average follow-up
of 27.0 months, and has been compared with clinicopathological
features. The reliability of all of the amplification-positive
tumors for EGFR and HER2 was confirmed by FISH. Of
the four detected ErbB family members of IDC in the present study,
14.2% (17/119) represented an EGFR and HER2
co-amplification subgroup. This subgroup was significantly
correlated with a higher possibility of metastasis (when diagnosed)
(P=0.014) and distant metastasis (following initial surgery)
(P<0.001), while they were almost significantly associated with
local recurrence (P=0.053). EGFR and HER2
co-amplification was noticed to be significantly associated with
DFS (P<0.001) and OS (P=0.002). Concerning the treatment
regimen, EGFR and HER2 co-amplification patients were
significantly correlated with poor DFS regarding chemotherapy
(P<0.001), radiotherapy (P<0.001) and hormonal therapy
(P=0.001). Thus, EGFR and HER2 co-amplification may
be an independent prognostic indicator of poor DFS and OS.
In the present study, the EGFR amplification
rate was 25.2%, a value similar to that reported in previous
studies (7.9–33.1%) (10,17,20). The
rate of HER2 amplification (36.9%) in the present study was
higher than that reported in previous studies (21–26),
suggesting that the frequency of HER2 may vary according to
the different detection methods (e.g. qPCR-based methods vs.
FISH-based assays). According to the FISH assay, the HER2
status can be classified as non-amplified (HER2/CEP17 ratio
<1.8), amplified (HER2/CEP17 ratio >2.2) or equivocal (1.8
<HER2/CEP17 <2.2). However, qPCR-based assays can only
identify certain patients as equivocally amplified cases (22). Thus, by combining qPCR-based and FISH
assays, more HER2-amplified cases were identified, which
revealed that certain FISH equivocal patients were actually
amplified cases (22). A rate of
EGFR and HER2 co-amplification of 14.2% (17/119) was
detected in the present study, but no HER3 amplification was
detected. Only one patient had HER4 amplification in the
present study. Previous reports had mentioned the frequency of
HER3 and HER4 in breast cancer. However, this
frequency varies depending on the cut-off value for FISH, ddPCR and
NGS-based assays. The cut-off value is difficult to determine. For
HER2 FISH assay, the cut-off value was not well defined, and
the cut-off values for HER3 and HER4 amplification
were not defined either. There were almost no patients in whom the
HER3 and HER4 FISH ratio was >2.0 (12).
It was previously shown that EGFR or
HER2 amplification was an independent poor clinical
prognostic indicator in breast cancer (10,23,24).
However, to date, no study has been reported concerning EGFR
and HER2 co-amplification in breast cancer. The present
study further investigated the association between EGFR and
HER2 co-amplification with the clinical prognosis of breast
cancer. The present study confirmed the association of HER2
amplification with HER2 overexpression (Table IV) (25). In the current study, HER2
amplification was also significantly associated with DFS and OS, as
has been reported previously (3,24). It was
also confirmed that a variable EGFR copy number can be
useful for predicting outcomes in patients (8,10). Certain
clinicopathological analyses of ErbB family receptors in breast
cancer were limited to single ErbB family members (8,27–32). The present study detected gene
amplification of four members of the ErbB family, and observed that
EGFR and HER2 co-amplification in the present study
was significantly associated with short DFS and OS (Fig. 3). When analyzing the co-amplification
subgroup with chemotherapy, radiotherapy and hormonal therapy, it
was observed that the co-amplification subgroup was significantly
correlated with DFS. However, due to the relatively short
follow-up, the association between the co-amplification subgroup
and OS regarding the treatment regimens could not be
determined.
To assess the prognostic value of EGFR and
HER2 co-amplification in breast cancer patients, the present
study analyzed DFS and OS for this subgroup of patients. Another
classification by expression profile, e.g. TNBC patients,
were also analyzed as a control (19). In the present study, EGFR and
HER2 co-amplification exhibited a significant difference
compared with the non-co-amplified group for both DFS and OS, but
the TNBC group did not show any significant difference compared
with the non-TNBC group for DFS in such a relatively short
follow-up period (26,33,34). This
result suggests that EGFR and HER2 co-amplification
can be considered to indicate a poor prognosis.
Resistance to treatment regiments, including
chemotherapy, radiotherapy, hormonal therapy and target therapy, is
a nearly universal and ultimately lethal consequence for breast
cancer patients (35–37). Numerous theories have attempted to
explain drug resistance during treatment, including the cancer stem
cell theory, the epithelial-mesenchymal transition theory and
certain somatic tumor cell mutations (38–41). Since
EGFR and HER2 co-amplified tumor cells were abnormal
in the corresponding signaling pathway, the patients may respond
differently to treatment regimens (5,42). In the
present study, patients with EGFR and HER2
co-amplification exhibited poor clinical outcome for both DFS and
OS. Notably, this is also true for DFS with respect to treatment
regimen for chemotherapy (P<0.001), radiotherapy (P<0.001)
and hormonal therapy (P=0.001). However, the response to current
treatment of this group of patients requires further detailed
studies. In addition, the present study explored the response to
target therapy of this subgroup of patients, including Herceptin
treatment for HER2-amplified patients. Only 8 patients in
the present study received Herceptin treatment. Of these, 3
patients were EGFR and HER2 co-amplified. Although
all 3 patients exhibited distant metastasis following initial
surgery, there are not statistically significant data showing
resistance to Herceptin treatment in EGFR and HER2
co-amplified patients due to the limited number of patients
included in the present study. The other 5 patients who received
target therapy were not co-amplified patients, who did not show any
recurrence or distant metastasis in a mean of 26.2 months of
follow-up. All the 8 patients exhibited HER2 amplification
and HER2 overexpression (3+), but there were no other statistically
significant deferences between the two groups. Further studies on
the response to different treatment of this particular subgroup of
patients should be carried out, although the present data strongly
suggest that EGFR and HER2 co-amplified cancer cells
may be the cell source responsible for drug resistance.
In summary, the present study detected ErbB family
gene amplification using qPCR and FISH, and the results suggested
that EGFR and HER2 co-amplification has a
considerable prognostic relevance regarding clinical outcomes in
breast cancer. EGFR and HER2 co-amplification may be
a novel particular subgroup in IDC that can be considered
predictive of poor clinical outcomes. Regarding treatment regimen
analysis, the results of the present study indicate that patients
with EGFR and HER2 co-amplification exhibit
resistance to chemotherapy, radiotherapy and hormonal therapy.
Specific treatment regimens may be required for this particular
subgroup of patients.
Acknowledgements
The present study was funded by grants from the
National Natural Science Foundation of China (grant nos. 31000601
and 81200461) and the Young Investigator Scholarship in Sichuan
University (grant no. 2012SCU04A14).
References
|
1
|
World Health Organization: Cancer Country
Profiles. 2014, http://www.who.int/cancer/country-profiles/chn_en.pdf
|
|
2
|
Cancer Genome Atlas Network, .
Comprehensive molecular portraits of human breast tumours. Nature.
490:61–70. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Callahan R: Genetic alterations in primary
breast cancer. Breast Cancer Res Treat. 13:191–203. 1989.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Eccles SA: The role of c-erbB-2/HER2/neu
in breast cancer progression and metastasis. J Mammary Gland Biol
Neoplasia. 6:393–406. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lemmon MA: The EGF receptor family as
therapeutic targets in breast cancer. Breast Dis. 18:33–43. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Vasan N, Yelensky R, Wang K, Moulder S,
Dzimitrowicz H, Avritscher R, Wang B, Wu Y, Cronin MT, Palmer G, et
al: A targeted next-generation sequencing assay detects a high
frequency of therapeutically targetable alterations in primary and
metastatic breast cancers: Implications for clinical practice.
Oncologist. 19:453–458. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chong CR and Jänne PA: The quest to
overcome resistance to EGFR-targeted therapies in cancer. Nat Med.
19:1389–1400. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Park HS, Jang MH, Kim EJ, Kim HJ, Lee HJ,
Kim YJ, Kim JH, Kang E, Kim SW, Kim IA and Park SY: High EGFR gene
copy number predicts poor outcome in triple-negative breast cancer.
Mod Pathol. 27:1212–1222. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cho EY, Choi YL, Han JJ, Kim KM and Oh YL:
Expression and amplification of Her2, EGFR and cyclin D1 in breast
cancer: Immunohistochemistry and chromogenic in situ hybridization.
Pathol Int. 58:17–25. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lv N, Xie X, Ge Q, Lin S, Wang X, Kong Y,
Shi H, Xie X and Wei W: Epidermal growth factor receptor in breast
carcinoma: Association between gene copy number and mutations.
Diagn Pathol. 6:1182011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lee HJ, Seo AN, Kim EJ, Jang MH, Kim YJ,
Kim JH, Kim SW, Ryu HS, Park IA, Im SA, et al: Prognostic and
predictive values of EGFR overexpression and EGFR copy number
alteration in HER2-positive breast cancer. Br J Cancer.
112:103–111. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sassen A, Rochon J, Wild P, Hartmann A,
Hofstaedter F, Schwarz S and Brockhoff G: Cytogenetic analysis of
HER1/EGFR, HER2, HER3 and HER4 in 278 breast cancer patients.
Breast Cancer Res. 10:R22008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zaczek A, Wełnicka-Jaśkiewicz M, Bielawski
KP, Jaśkiewicz J, Badzio A, Olszewski W, Rhone P and Jassem J: Gene
copy numbers of HER family in breast cancer. J Cancer Res Clin
Oncol. 134:271–279. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak and Schmittgen: Analysis of relative
gene expression data using real-time quantitative PCR and the
2-ΔΔCt method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gjerdrum LM, Sorensen BS, Kjeldsen E,
Sorensen FB, Nexo E and Hamilton-Dutoit S: Real-time quantitative
PCR of microdissected paraffin-embedded breast carcinoma: An
alternative method for HER-2/neu analysis. J Mol Diagn. 6:42–51.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bernardi CC, Ede S Ribeiro, Cavalli IJ,
Chautard-Freire-Maia EA and Souza RL: Amplification and deletion of
the ACHE and BCHE cholinesterase genes in sporadic breast cancer.
Cancer Genet Cytogenet. 197:158–165. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Park K, Han S, Shin E, Kim HJ and Kim JY:
EGFR gene and protein expression in breast cancers. Eur J Surg
Oncol. 33:956–960. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Burkhardt L, Grob TJ, Hermann I, Burandt
E, Choschzick M, Jänicke F, Müller V, Bokemeyer C, Simon R, Sauter
G, et al: Gene amplification in ductal carcinoma in situ of the
breast. Breast Cancer Res Treat. 123:757–765. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bauer KR, Brown M, Cress RD, Parise CA and
Caggiano V: Descriptive analysis of estrogen receptor
(ER)-negative, progesterone receptor (PR)-negative, and
HER2-negative invasive breast cancer, the so-called triple-negative
phenotype: A population-based study from the California cancer
Registry. Cancer. 109:1721–1728. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Brandt B, Vogt U, Schlotter CM, Jackisch
C, Werkmeister R, Thomas M, von Eiff M, Bosse U, Assmann G and
Zänker KS: Prognostic relevance of aberrations in the erbB
oncogenes from breast, ovarian, oral and lung cancers:
Double-differential polymerase chain reaction (ddPCR) for clinical
diagnosis. Gene. 159:35–42. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nanda R: Targeting the human epidermal
growth factor receptor 2 (HER2) in the treatment of breast cancer:
Recent advances and future directions. Rev Recent Clin Trials.
2:111–116. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Belgrader P, Tanner SC, Regan JF, Koehler
R, Hindson BJ and Brown AS: Droplet digital PCR measurement of HER2
copy number alteration in formalin-fixed paraffin-embedded breast
carcinoma tissue. Clin Chem. 59:991–994. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Slamon DJ, Clark GM, Wong SG, Levin WJ,
Ullrich A and McGuire WL: Human breast cancer: Correlation of
relapse and survival with amplification of the HER-2/neu oncogene.
Science. 235:177–182. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Menard S, Fortis S, Castiglioni F, Agresti
R and Balsari A: HER2 as a prognostic factor in breast cancer.
Oncology. 61 Suppl 2:S67–S72. 2001. View Article : Google Scholar
|
|
25
|
Hoang MP, Sahin AA, Ordòñez NG and Sneige
N: HER-2/neu gene amplification compared with HER-2/neu protein
overexpression and interobserver reproducibility in invasive breast
carcinoma. Am J Clin Pathol. 113:852–859. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bhatti AB, Khan AI, Siddiqui N, Muzaffar
N, Syed AA, Shah MA and Jamshed A: Outcomes of triple-negative
versus non-triple-negative breast cancers managed with
breast-conserving therapy. Asian Pac J Cancer Prev. 15:2577–2581.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Stern DF: Tyrosine kinase signalling in
breast cancer: ErbB family receptor tyrosine kinases. Breast Cancer
Res. 2:176–183. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ge H, Gong X and Tang CK: Evidence of high
incidence of EGFRvIII expression and coexpression with EGFR in
human invasive breast cancer by laser capture microdissection and
immunohistochemical analysis. Int J Cancer. 98:357–361. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Olayioye MA: Update on HER-2 as a target
for cancer therapy: Intracellular signaling pathways of ErbB2/HER-2
and family members. Breast Cancer Res. 3:385–389. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ellis MJ: Neoadjuvant endocrine therapy
for breast cancer: Medical perspectives. Clin Cancer Res.
7:s4388–s4391. 2001.
|
|
31
|
Lemoine NR, Barnes DM, Hollywood DP,
Hughes CM, Smith P, Dublin E, Prigent SA, Gullick WJ and Hurst HC:
Expression of the ERBB3 gene product in breast cancer. Br J Cancer.
66:1116–1121. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kew TY, Bell JA, Pinder SE, Denley H,
Srinivasan R, Gullick WJ, Nicholson RI, Blamey RW and Ellis IO:
c-erbB-4 protein expression in human breast cancer. Br J Cancer.
82:1163–1170. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Nguyen PL, Taghian AG, Katz MS, Niemierko
A, Abi Raad RF, Boon WL, Bellon JR, Wong JS, Smith BL and Harris
JR: Breast cancer subtype approximated by estrogen receptor,
progesterone receptor and HER-2 is associated with local and
distant recurrence after breast-conserving therapy. J Clin Oncol.
26:2373–2378. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li CY, Zhang S, Zhang XB, Wang P, Hou GF
and Zhang J: Clinicopathological and prognostic characteristics of
triple-negative breast cancer (TNBC) in Chinese patients: A
retrospective study. Asian Pac J Cancer Prev. 14:3779–3784. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Diver EJ, Foster R, Rueda BR and Growdon
WB: The therapeutic challenge of targeting HER2 in endometrial
cancer. Oncologist. 20:1058–1068. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Feldinger K and Kong A: Profile of
neratinib and its potential in the treatment of breast cancer.
Breast Cancer (Dove Med Press). 7:147–162. 2015.PubMed/NCBI
|
|
37
|
Black JC, Atabakhsh E, Kim J, Biette KM,
Van Rechem C, Ladd B, Burrowes PD, Donado C, Mattoo H, Kleinstiver
BP, et al: Hypoxia drives transient site-specific copy gain and
drug-resistant gene expression. Genes Dev. 29:1018–1031. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang S, Mou Z, Ma Y, Li J, Li J, Ji X, Wu
K, Li L, Lu W and Zhou T: Dopamine enhances the response of
sunitinib in the treatment of drug-resistant breast cancer:
Involvement of eradicating cancer stem-like cells. Biochem
Pharmacol. 95:98–109. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Chiotaki R, Polioudaki H and
Theodoropoulos PA: Cancer stem cells in solid and liquid tissues of
breast cancer patients: Characterization and therapeutic
perspectives. Curr Cancer Drug Targets. 15:256–269. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhang P, Sun Y and Ma L: ZEB1: At the
crossroads of epithelial-mesenchymal transition, metastasis and
therapy resistance. Cell Cycle. 14:481–487. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Jaspers JE, Sol W, Kersbergen A, Schlicker
A, Guyader C, Xu G, Wessels L, Borst P, Jonkers J and Rottenberg S:
BRCA2-deficient sarcomatoid mammary tumors exhibit multidrug
resistance. Cancer Res. 75:732–741. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Montemurro F and Scaltriti M: Biomarkers
of drugs targeting HER-family signalling in cancer. J Pathol.
232:219–229. 2014. View Article : Google Scholar : PubMed/NCBI
|