Introduction
Osteosarcoma (OS) is one of the most common human
bone malignancies and a leading cause of tumor-associated
mortalities in children and adolescents, which comprises ~2.4% of
all tumor types in pediatric patients (1,2). Emerging
evidence suggests that osteosarcoma is a type of differentiation
disease, which initiates in the regions where bone growth and
repair is activated (3). The clinical
treatment for osteosarcoma is limited given that the mortality rate
in amputated patients is high due to pulmonary metastases (4). Since the use of surgery and neoadjuvant
chemotherapy, the 5-year survival rate of osteosarcoma patients has
dramatically improved to ~60% (5).
However, there remains a significant proportion of patients who may
relapse, and distant metastasis may occur due to poor responses to
chemotherapy. The acquisition of chemoresistance is a major reason
for poor prognosis (6). Therefore,
the identification of the underlying mechanisms that are
responsible for chemoresistance is critical for improvements in
prognosis and therapeutic strategies.
Tripartite motif containing 37 (TRIM37), belongs to
the tripartite motif family and contains zinc-binding, RING finger
region, B-box motif and coiled-coil domains (7). The encoded TRIM37 gene is located at
17q22-23, and a number of novel mutations have been reported
(8,9).
During tumorigenesis and development, TRIM37 has essential roles in
regulating the expression of oncogenes due to its E3 ligase
activity in the ubiquitin-proteasome degradation system (10,11).
Upregulated TRIM37 expression has been reported in a number of
tumor types, including hepatocellular and pancreatic carcinoma, and
breast cancer (11,12). However, the expression pattern and the
function of TRIM37 in OS remain unclear.
The role of Wnt/β-catenin signaling pathway as a
major oncogenic pathway in a number of cancer types, including OS,
has been well established (13,14).
β-catenin is a key molecule in the Wnt/β-catenin signaling pathway
and is re-localized to the nucleus and forms a complex with T-cell
factor (TCF) to regulate gene expression upon Wnt ligand
stimulation (15). Studies have
demonstrated that Wnt/β-catenin is able to regulate a number of
downstream targets including cyclin D1, Myc proto-oncogene protein
and mitogen-activated protein kinase 8, which regulates cell
proliferation, migration and stemness (16–18).
Notably, previous studies have demonstrated that TRIM37 may
interact with β-catenin and be involved in aberrant activation of
the Wnt/β-catenin signaling pathway (19). However, the precise mechanisms
underlying the association between TRIM37 and Wnt/β-catenin
signaling pathway activation and the potential effect on
chemoresistance in OS cells remain to be elucidated.
The present study demonstrated that TRIM37
expression is induced by treatment with chemotherapy drugs,
potentially promoting chemoresistance in patients with pediatric
osteosarcoma. TRIM37-induced activation of the Wnt/β-catenin
signaling pathway may be responsible for the chemoresistance.
Therefore the current study hypothesized that decreasing TRIM37
levels combined with a Wnt/β-catenin signaling pathway inhibitor
may be a optimal therapeutic strategy in pediatric OS.
Materials and methods
Patient samples
The present study was approved by the Jining Medical
University Affiliated Hospital Research Ethics Committee (Jining,
China) and written informed consent was obtained from all patients.
A total of 41 OS tissues were collected from patients (<21 years
old; median, 17; range, 14–21) and the clinicopathological features
of patients are displayed in Table I.
Tissue samples were stored at −80°C.
 | Table I.Clinicopathological features of 41
patients with pediatric osteosarcoma. |
Table I.
Clinicopathological features of 41
patients with pediatric osteosarcoma.
| Clinicopathological
feature | n |
|---|
| Age, years |
|
| ≤13 | 24 |
|
>13 | 17 |
| Sex |
|
| Male | 22 |
|
Female | 19 |
| Tumor size, cm |
|
|
<5 | 30 |
| ≥5 | 11 |
| Tumor location |
|
|
Femur | 20 |
|
Tibia | 16 |
|
Humerus | 3 |
|
Other | 2 |
| Metastasis |
|
|
Present | 15 |
|
Absent | 26 |
Cell culture and transfection
Human OS cell lines (MG-63, SaOS-2, U-2 OS and
SOSP-9901) and osteoblasts (hFOB1.19) were obtained from the
American Type Culture Collection (Manassas, VA, USA). These cells
were maintained in Dulbecco's modified Eagle's medium (DMEM; Gibco;
Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with
10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.) at
37°C with 5% CO2 in a humidified incubator. The small
interfering RNAs (siRNAs) targeting TRIM37 were designed and
synthesized by Chang Jing Bio-Tech, Ltd. (Changsha, China; siRNA
sequences, 5′-ATTTGTATGGAGAAATTGC-3′ and
5′-CATTGCTCCAAACTGTGTTGTT-3′). The sequence of the control siRNA
was 5′-TTCTTCGAACGTGTCACGTT-3′ (Chang Jing Bio-Tech, Ltd.,
Changsha, China). Human TRIM37 cDNA was subcloned into the pcDNA3.1
expression vector with FLAG-tag. MG132 (cat. no. M8699; 10 mM) and
cycloheximide (CHX; cat. no. C4859; 10 µg/ml) were obtained from
Sigma (Merck KGaA, Darmstadt, Germany). Cell transfections with
pools of siRNA or overexpression vector were performed using
Lipofectamine 2000 reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was isolated from tissues samples or
cultured cells using Trizol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) and subsequently converted to single stranded
cDNA using the PrimeScript RT Master kit (Takara Biotechnology Co.,
Ltd., Dalian, China) according to the manufacturer's instructions.
The RT-qPCR was performed on a 7500 Real-time PCR System (Applied
Biosystems; Thermo Fisher Scientific, Inc.) with SYBR-Green PCR mix
(Takara Biotechnology Co., Ltd.). The PCR conditions were as
follows: 95°C 30 sec; 95°C 30 sec; 60°C 30 sec (40 cycles). The
gene expression was normalized to GAPDH by 2−ΔΔCq method
as previously described (20). The
primer sequences used were as follows: TRIM37 forward,
5′-TCAGCTGTATTAGGCGCTGG-3′ and reverse, 5′-ACTTCTTCTGCCCAACGACA-3′;
and housekeeping gene GAPDH forward, 5′-CATGAGAAGTATGACAACAGCCT-3′
and reverse, 5′-AGTCCTTCCACGATACCAAAGT-3′.
Western blotting
The cells were harvested and lysed in
radioimmunoprecipitation assay buffer (Thermo Fisher Scientific,
Inc.) supplemented with phosphatase inhibitor and protease
inhibitor. Total protein concentration was measured by
bicinchoninic acid assay. Protein (60 µg) was separated by 10%
SDS-PAGE and subsequently transferred onto polyvinylidene
difluoride membranes. Following blocking with 5% bovine serum
albumin (Sangon Biotech Co., Ltd., Shanghai, China), the membrane
was incubated with the appropriate primary antibody at 4°C
overnight, followed by washing and incubation with horseradish
peroxidase goat-anti-rabbit Immunoglobulin G secondary antibody
(dilution, 1:2,000; no. ab6721; Abcam, Cambridge, MA, USA) for 1 h
at room temperature and detection using an enhanced
chemiluminescence kit (Thermo Fisher Scientific, Inc.). The primary
antibodies used were as follows: Anti-TRIM37 (dilution, 1:1,000;
cat. no. sc-49548; Santa Cruz Biotechnology, Inc., Dallas, TX,
USA), anti-Tubulin (dilution, 1:1,000; cat. no. sc-9104; Santa Cruz
Biotechnology, Inc.) and anti-cleaved poly(ADP-ribose) polymerase
(PARP; dilution, 1:1,000; anti-cleaved PARP; cat. no. 5625; Cell
Signaling Technology, Inc., Danvers, MA, USA).
Immunohistochemical staining
The osteosarcoma tissues were fixed in 4%
formaldehyde for 30 min at room temperature, embedded in paraffin
and sectioned (4 µm). The slides were deparaffinized with xylene
and rehydrated in graded ethanol, and immersed in 3%
H2O2 to block endogenous peroxidase activity.
Following antigen retrieval, the slides were blocked by 10% goat
serum (Sangon Biotech Co., Ltd.) and incubated with primary
anti-TRIM37 antibody overnight at 4°C. Visualization was performed
by adding biotinylated goat-anti-rabbit secondary antibody
(dilution, 1:2,000; no. SV0002; Wuhan Boster Biological Technology,
Ltd., Wuhan, China) for 30 min at room temperature, followed by the
3,3′-diaminobenzidine tetrahydrochloride and counterstained with
hematoxylin.
Colony formation and cell viability
assay
For colony formation and cell viability assay, cells
were seeded in 6-well plates (1,000 cells/well) and cultured at
37°C with 5% CO2 for 7 days. Following washing, the
plates were stained with crystal violet. For cell viability assay,
cells were seeded in 96-well plates (2,000 cells/well). Following
treatment with chemotherapy drugs [10 ng/µl doxorubicin (Dox;
Sigma-Aldrich; Merck KGaA), 20 µM cisplatin (Cis; Sigma-Aldrich;
Merck KGaA), 10 ng/µl methotrexate (Mtx; Sigma-Aldrich; Merck
KGaA), 10 µM XAV-939 (Selleck Chemicals, Shanghai, China) or
dimethylsufoxide]. The cells were incubated with 5 mg/ml MTT for 3
h. The medium was subsequently removed and dimethylsulfoxide was
added to solubilize the crystals. Absorbance was detected using a
spectrometer at a wavelength of 590 nm. Clone number (%) was
calculated by randomly selecting 5 fields in which the cells were
counted. The control group was 100% and test groups were calculated
according to this control.
Bioinformatics analysis
Microarray analysis was used to analyze the genes
where expression was altered following the knockdown of TRIM37 in
OS cells. SaOS-2 cells were treated with siRNA to knockdown levels
of TRIM37. Total RNA was extracted from these cells, as previously
stated, and sent to Shanghai Biomedical Laboratory, Co., Ltd.,
(Shanghai, China) to be analyzed using the Affymetrix Human U133
Plus 2.0 array. Gene Set Enrichment Analysis (GSEA) was performed
using the GSEA program (Broad Institute, Boston, MA; http://www.broadinstitute.org/gsea/index.jsp). GSEA
was used to analyze signaling pathways in which TRIM37-regulated
genes were enriched.
Statistical analysis
Statistical analysis was performed using SPSS
version 21.0 (IBM SPSS, Armonk, NY, USA) and GraphPad Prism 5
(GraphPad Software, Inc., La Jolla, CA, USA). Differences between
the mRNA levels in tumors and non-cancerous tissue were determined
using the Wilcoxon signed-rank test. Two-tailed Student's t-test
was used to analyze the significance between the experimental group
and the control. Data are presented as the mean ± standard
deviation in ≥3 independent experiments. P<0.05 was considered
to indicate a statistically significant difference.
Results
Upregulation of TRIM37 is induced by
chemical drug treatment in OS cells
To investigate the possible role of TRIM37, the
expression of TRIM37 in OS cells following treatment with
chemotherapy drugs (doxorubicin, cisplatin and methotrexate) was
examined. Notably, all three chemotherapy drugs markedly increased
TRIM37 mRNA and protein expression in MG-63 and SaOS-2 cells
(Fig. 1A and B). Furthermore,
transcription inhibitor cycloheximide was able to reduce
chemotherapy drug-induced TRIM37 expression in SaOS-2 cells
(Fig. 1C, left panel). MG132, which
is able to inhibit proteasome activity, was employed to further
analyze TRIM37 expression following chemotherapy drug treatment.
TRIM37 expression was increased following MG132 treatment (Fig. 1C, right panel). These results
indicated that the upregulation of TRIM37 may be dependent on
transcriptional activation and protein stability. In addition, it
was observed that the upregulation of TRIM37 in OS cells (MG-63 and
SaOS-2) was time dependent in the case of doxorubicin (Fig. 1D), whilst this effect was not observed
in cells with cisplatin and methotrexate treatment. Collectively,
the present findings indicated that TRIM37 was upregulated when OS
cells were treated with chemotherapy drugs.
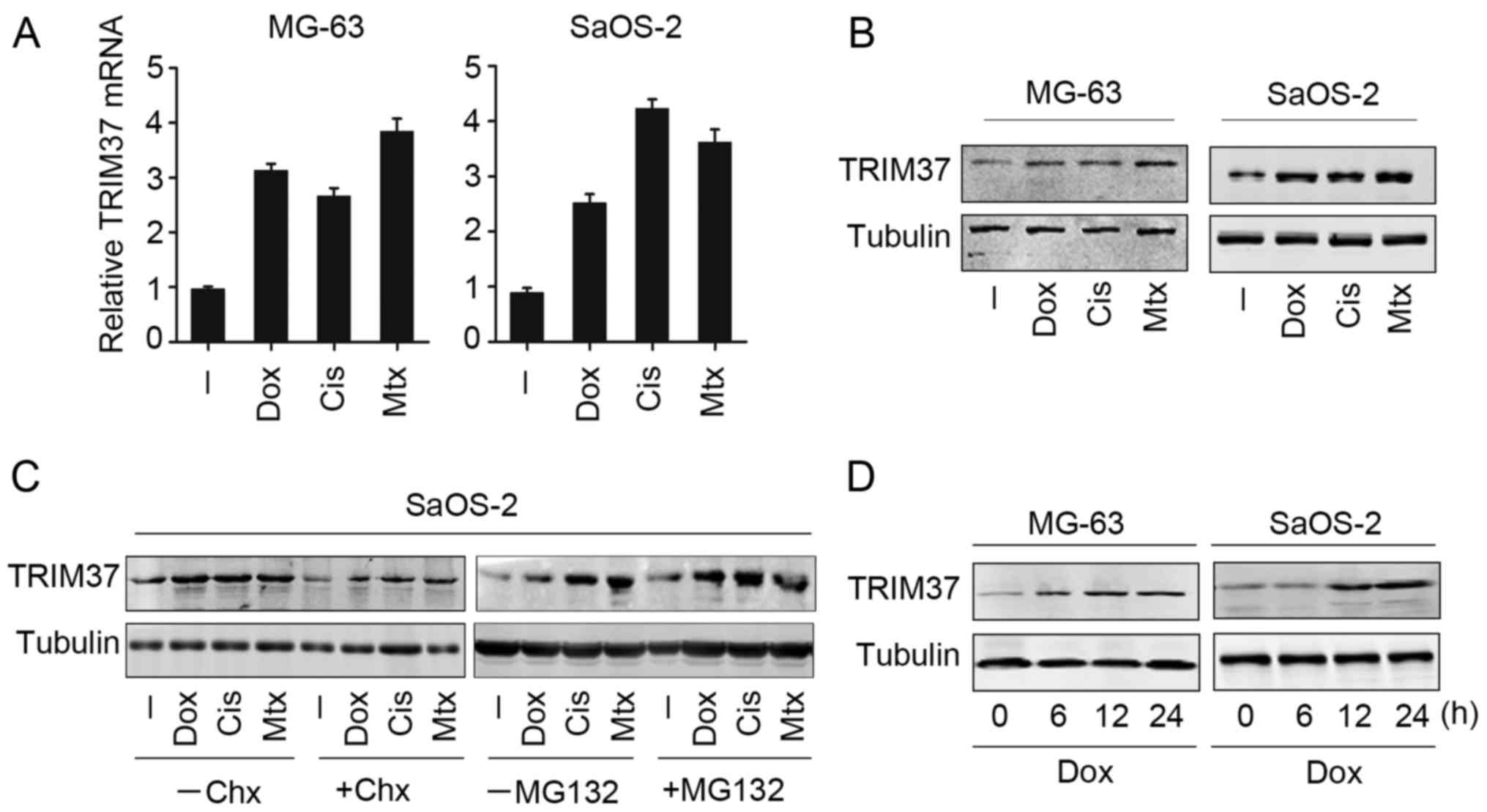 | Figure 1.TRIM37 expression may be induced by
chemotherapy drugs in osteosarcoma cells. MG-63 and SaOS-2 cell
lines were treated with doxorubicin, cisplatin and methotrexate for
24 h, and TRIM37 expression was analyzed by (A) reverse
transcription-quantitative polymerase chain reaction and (B)
western blotting. (C) SaOS-2 cells were treated with doxorubicin,
cisplatin and methotrexate in the presence and absence of
transcription inhibitor cycloheximide or MG132 treatment. TRIM37
expression was examined by western blotting. (D) MG-63 and SaOS-2
cells were treated with doxorubicin at 0, 6, 12 and 24 h, and
TRIM37 expression was examined by western blotting. TRIM37,
tripartite motif containing 37; chx, cycloheximide; cis, cisplatin;
dox, doxorubicin; mtx, methotrexate. |
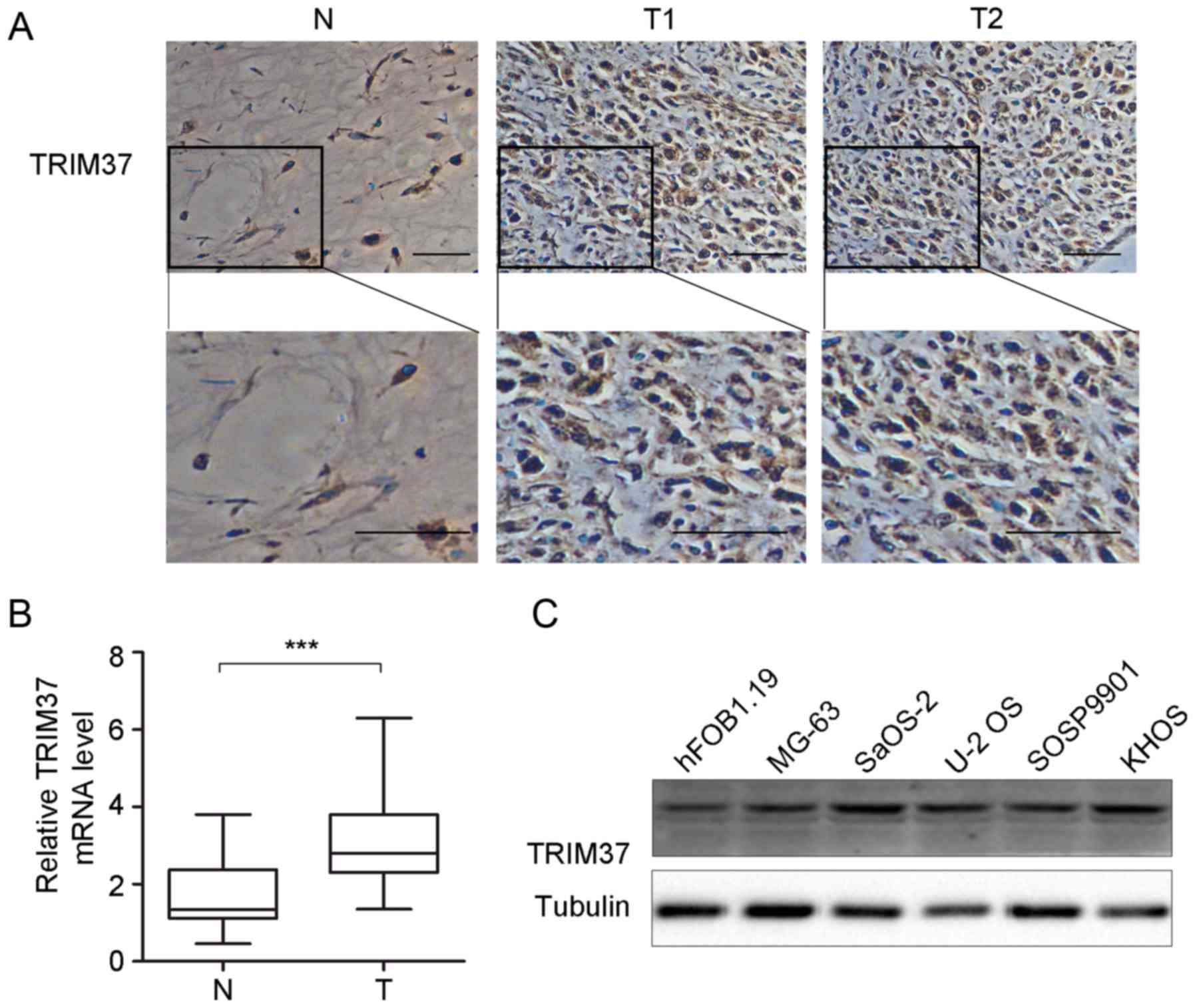
TRIM37 is upregulated in clinical OS
samples and OS cell lines
Expression of TRIM37 in pediatric OS was detected by
RT-qPCR and immunohistochemical analysis in patient tissue
specimens (n=41; Fig. 2). The results
indicated that TRIM37 expression was significantly increased at the
mRNA level and markedly increased at the protein level (Fig. 2B and C). Immunohistochemical staining
demonstrated relatively high TRIM37 expression in the cytoplasm and
cell nucleus in 80.5% (33/41) of OS specimens, whilst low
expression was observed in normal tissues (Fig. 2A). Furthermore, TRIM37 expression was
examined in OS cell lines (n=5) and hFOB1.19 cells. As shown in
Fig. 2C, expression of TRIM37 was
increased in OS cell lines (MG-3, SaOS-2, U-2 OS, SOSP9901 and
KHOS) compared with the osteoblast cell line hFOB1.19. Taken
together, the results of the present study indicate that TRIM37
expression was increased in OS tissues and cell lines and may
contribute to tumor progression.
TRIM37 promotes cell proliferation and
inhibits chemotherapy-induced cell apoptosis
To investigate whether TRIM37 expression affects
proliferation and survival of OS cells, gain and loss-of-function
analysis was performed. Endogenous TRIM37 was knocked down by
co-transfection with 2 siRNAs targeting TRIM37. As shown in
Fig. 3A, transfection with TRIM37
siRNAs in SaOS-2 cells led to a marked decrease in TRIM37 protein
levels. TRIM37 knockdown reduced the proliferation rate as
determined by colony formation assay; the number of colonies
significantly decreased by ~60–85%, compared with control cells
(Fig. 3B). Next, the effect of TRIM37
knockdown on chemotherapy-induced cell death in OS cells was
examined. As indicated by the cell viability assays, significant
increases in apoptotic rates were observed following TRIM37
knockdown (Fig. 3C). This finding
suggested that TRIM37 knockdown may increase the sensitivity of OS
cells to doxorubicin, cisplatin and methotrexate. Furthermore,
western blotting was used to analyze the expression of the
apoptosis marker cleaved PARP and it was demonstrated that the
knockdown of TRIM37 resulted in an increase in the expression of
cleaved PARP protein. Notably, cleaved PARP expression was markedly
increased when TRIM37 knocked down cells were treated with
doxorubicin, cisplatin and methotrexate, compared with no treatment
(Fig. 3D).
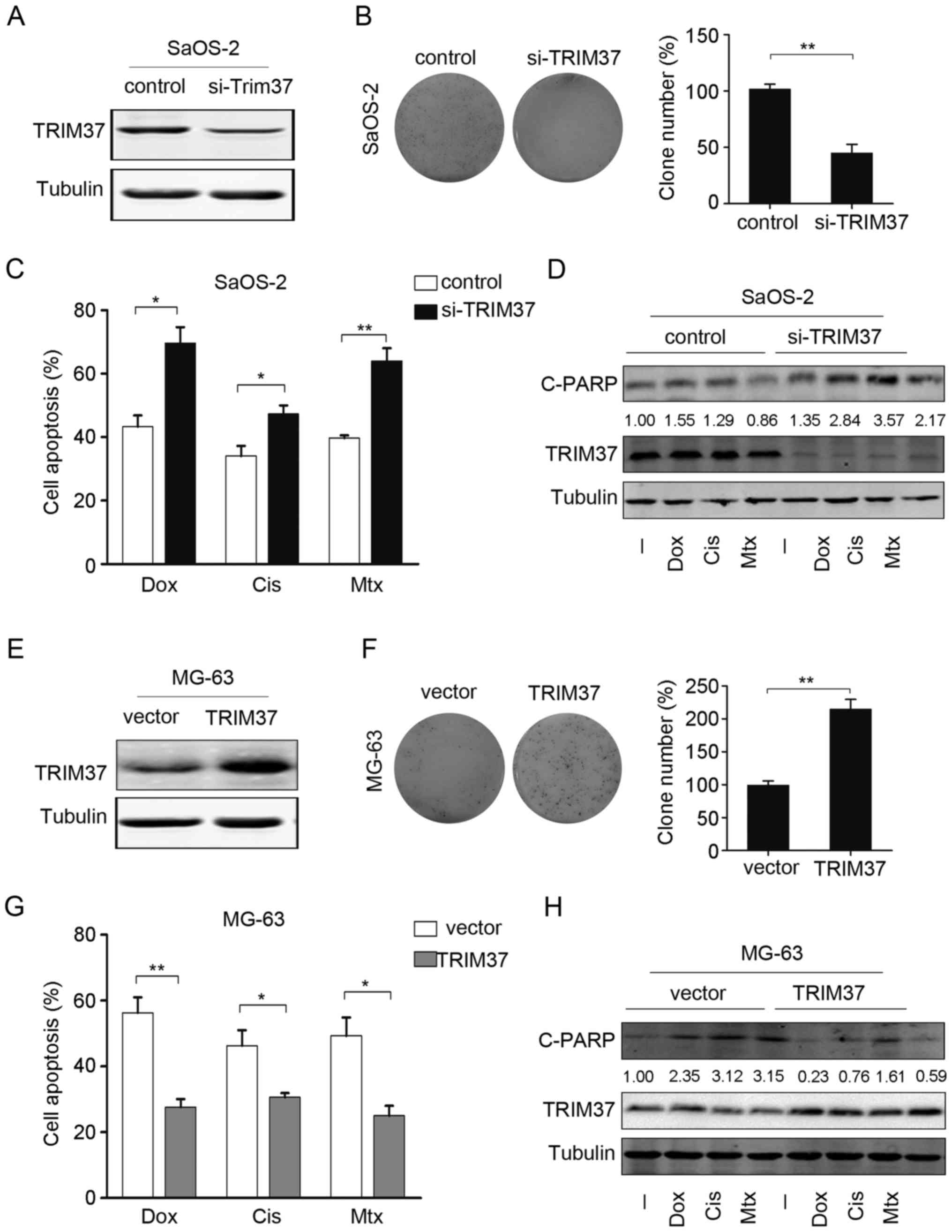 | Figure 3.TRIM37 promotes cell proliferation and
chemoresistance. (A) TRIM37 protein expression was assessed in
SaOS-2 cells that were transfected with si-TRIM37 and in the
control [negative control (NC) siRNA]. (B) Cell proliferation in
SaOS-2 cells transfected with si-TRIM37 was assessed by colony
formation assay (left panel) and the relative percentage of clones
was calculated (right panel). (C) Cell viability was assessed in
SaOS-2 cells transfected with si-TRIM37 and in the control (NC
siRNA) following treatment with doxorubicin, cisplatin and
methotrexate. (D) Cleaved poly(ADP-ribose) polymerase and TRIM37
protein expression was examined following treatment of SaOS-2 cells
si-TRIM37 transfected cells and the control (NC siRNA) with
doxorubicin, cisplatin and methotrexate. (E) TRIM37 expression was
examined in MG-63 cells overexpressing TRIM37 and in MG-63 cells
transfected with only vector. (F) Cell proliferation in MG-63 cells
transfected with si-TRIM37 was assessed by colony formation assay
(left panel), and the relative percentage of clones was calculated
(right panel). (G) Cell viability assay was performed in MG-63
cells overexpressing TRIM37 and in MG-63 cells transfected with
only vector following treatment with doxorubicin, cisplatin and
methotrexate. (H) C-PARP and TRIM37 expression was analyzed by
western blotting following treatment of MG-63 cells (cells
overexpressing TRIM37 and transfected with only vector) with
doxorubicin, cisplatin and methotrexate. Data are expressed as the
mean ± standard deviation of 3 independent experiments. *P<0.05,
**P<0.01. -, no treatment; si, small interfering; TRIM37,
tripartite motif containing 37; Dox, doxorubicin; Cis, cisplatin;
Mtx, methotrexate; C-PARP, cleaved poly(ADP-ribose) polymerase. |
The effect of TRIM37 overexpression was also
examined in an OS cell line (MG-63; Fig.
3E). Colony formation and cell viability assays indicated that
overexpression of TRIM37 significantly increased the cell
proliferation rate and significantly decreased the apoptotic rate
(Fig. 3F and G). The chemotherapy
drugs induced C-PARP expression, a marker of cell apoptosis, which
was attenuated by overexpression of TRIM37 (Fig. 3H). Collectively, these data suggested
that TRIM37 promoted cell proliferation and increased the
resistance of OS cells to chemotherapy drugs.
Wnt/β-catenin inhibitor abrogates
TRIM37-induced chemoresistance
To further investigate the mechanism by which TRIM37
affects OS proliferation and chemoresistance, genes regulated by
TRIM37 were identified by microarray analysis. GESA was
subsequently performed to investigate the potential downstream
signaling pathways that may be regulated by TRIM37. GESA indicated
that the Wnt/β-catenin signaling pathway was enriched, which was
verified in the present study by RT-qPCR detection for the
downstream genes (Fig. 4A and B).
This notable finding was consistent with the finding of a previous
study, which demonstrated that TRIM37 interacts with β-catenin and
induces transcriptional activity of β-catenin/TCF (12,19).
Notably, Wnt/β-catenin aberrant activation has been associated with
chemoresistance in different types of tumors, including
hepatocellular carcinoma, neuroblastoma and prostate cancer
(21–23). Therefore, the TRIM37-transfected OS
cells (MG-63 and SaoS-2) were treated with a specific Wnt/β-catenin
pathway inhibitor XAV-939 (10 µM), in combination with doxorubicin,
cisplatin and methotrexate. Western blotting was used to confirm
the inhibition of TRIM37-induced Wnt/β-catenin signaling (Fig. 4C, upper panel). As expected, 48 h
following transfection, XAV-939 significantly abrogated
TRIM37-induced chemoresistance in MG-63 and SaoS-2 cells (Fig. 4C, lower panel). These results further
indicated that the Wnt/β-catenin signaling pathway may be a major
target of TRIM37 and mediates TRIM37-induced chemoresistance.
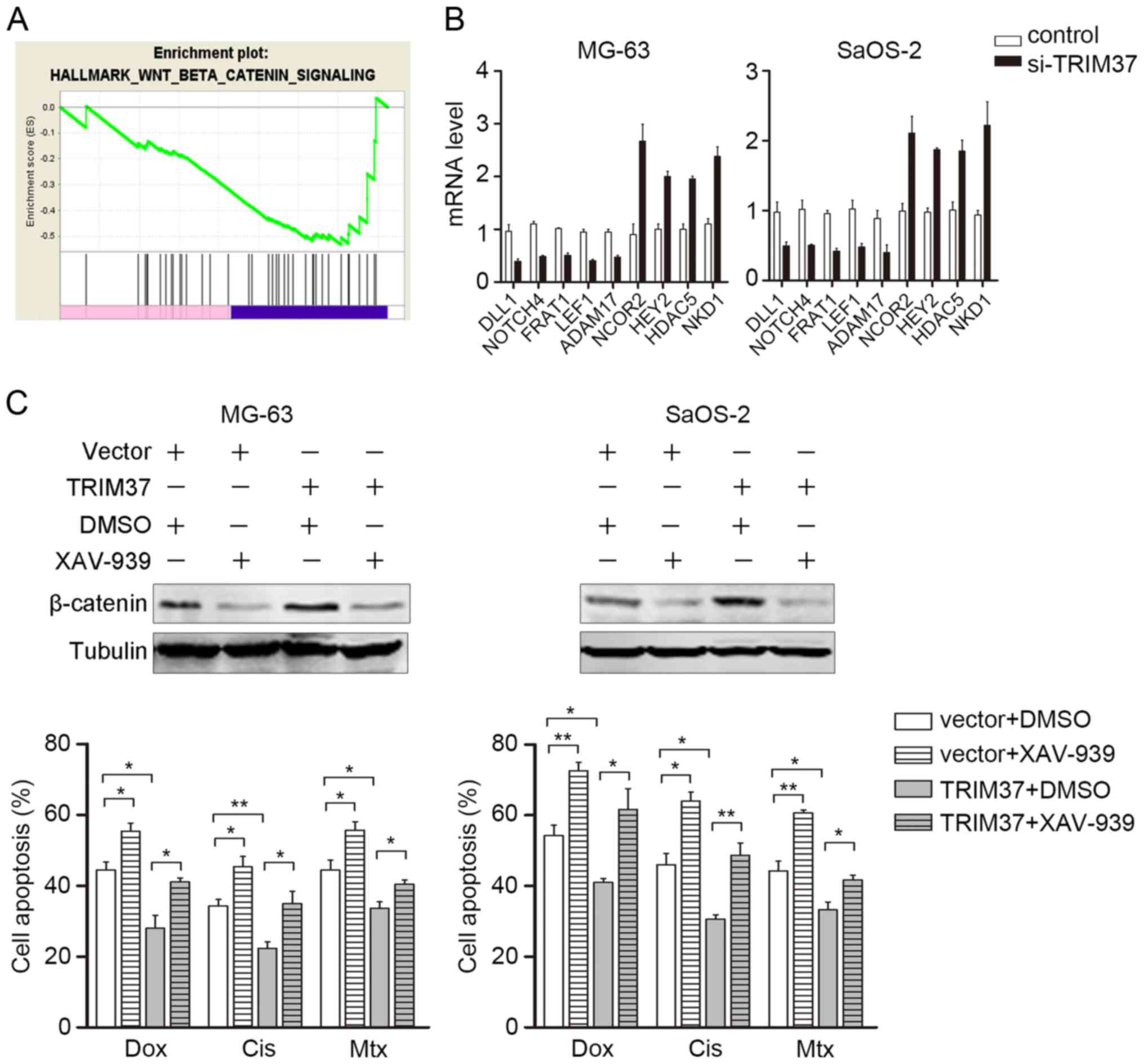 | Figure 4.Wnt/β-catenin inhibitor abrogates
TRIM37-induced chemoresistance. (A) Gene set enrichment analysis of
TRIM37-regulated genes. The Wnt/β-catenin pathway was enriched in
control group (blue) compared with the si-TRIM37 group (pink). (B)
mRNA transcripts of genes downstream of the Wnt/β-catenin signaling
pathway in MG-63 and SaOS-2 cells were examined by reverse
transcription-quantitative polymerase chain reaction. (C) Cell
viability assay was performed in MG-63 and SaOS-2 cells treated
with doxorubicin, cisplatin and methotrexate in the presence and
absence of XAV-939. Data are expressed as the mean ± standard
deviation of 3 independent experiments. *P<0.05, **P<0.01.
DLL1, delta-like canonical notch ligand 1; NOTCH4, neurogenic locus
notch homolog protein 4, FRAT1, frequently rearranged in advanced
t-cell lymphomas 1; LEF1, ADAM17, ADAM metallopeptidase domain 17;
C-PARP, cleaved poly(ADP-ribose) polymerase; NCOR2, nuclear
receptor corepressor 2; HEY2, hairy-related transcription factor 2;
HDAC5, histone deacetylase 5; NKD1, naked cuticle homolog 1; si,
small interfering; Dox, doxorubicin; Cis, cisplatin; Mtx,
methotrexate; TRIM37, tripartite motif containing 37. |
Discussion
Increasing evidence indicates that TRIM37 may have a
role as an oncogene and be a potential molecular target for therapy
in human carcinoma. TRIM37 is highly upregulated in a number of
cancer types. Jiang et al (12) demonstrated that elevated expression of
TRIM37 promoted migration and metastasis of hepatocellular
carcinoma cells by upregulating the β-catenin signaling pathway.
Furthermore, a previous study indicated that TRIM37 interacts with
β-catenin and recruits the β-catenin/TCF complex to promote
pancreatic cancer progression. TRIM37 has been established to
mediate downstream target genes of β-catenin/TCF complex
transcriptional activation, which is independent of its E3 ligase
activity (19). Bhatnagar et
al (11) demonstrated the role of
TRIM37 as an E3 ubiquitin ligase for histone H2A, and the
association of TRIM37 with polycomb repressive complex 2 in breast
cancer. In addition, mutations in TRIM37 have been reported which
are associated with OS (24,25). Increasing evidence indicate that
TRIM37 may have key roles in tumor development and progression.
However, limited information has been reported regarding the impact
of TRIM37 on the Wnt/β-catenin-TCF axis in pediatric OS and
chemotherapy resistance.
In the present study, the role of TRIM37 in
pediatric OS tissues and cells was examined. Initially, it was
observed that TRIM37 expression at the mRNA and protein level was
induced by treatment with chemotherapy drugs (doxorubicin,
cisplatin and methotrexate). TRIM37 expression was significantly
enhanced in 41 pediatric osteosarcoma specimens and cell lines. The
data of the present study are also consistent with previous studies
that have reported that TRIM37 is upregulated in other types of
cancer, thus upregulation of TRIM37 is not tumor type-specific.
For assessing the function of TRIM37, siRNAs were
used to specifically knockdown TRIM37. It was observed in the
present study that TRIM37 regulates OS cell viability, apoptosis
and resistance to chemotherapy drugs. The results of the present
study suggested that TRIM37 downregulation increased cleaved-PARP
expression, which is an important mediator of cell apoptosis.
Furthermore, the pattern of gene expression of TRIM37 knockdown in
SaOS-2 OS cells was analyzed by microarray. The GSEA analysis
indicated that the genes with altered expression were enriched in
the Wnt/β-catenin signaling pathway, which suggest that the
inhibition of Wnt/β-catenin signaling caused by downregulation of
TRIM37 may be a potential mechanism for chemotherapy drug-induced
apoptosis in OS cells.
The involvement of Wnt/β signaling pathway in cell
survival and mobility is well recognized, and the activation of
Wnt/β catenin signaling increases resistance to drug induced
apoptosis. Wickström et al (26) reported that the inhibition of
β-catenin signaling is able to repress O6-methylguanine-DNA
methyltransferase activation and therefore prevents
chemoresistance. Hsieh et al (23) demonstrated that microRNA-320 is able
to inhibit β-catenin expression, and suppress chemoresistance and
tumorigenic abilities in prostate cancer. Furthermore, Ma et
al (27) observed that the
inhibition of Wnt/β-catenin and notch signaling sensitized OS cells
to chemotherapy. These results suggested that the Wnt/β-catenin
cascade is upregulated in OS cells. The GSEA demonstrated that the
expression of TRIM37-regulated genes is consistent with activation
of the Wnt/β-catenin signaling pathway, which is also in
concordance with the results of previous studies (28,29).
Furthermore, the reverse experiment using XAV-939 to specifically
inhibit Wnt/β-catenin signaling confirmed the hypothesis that
TRIM37 mediates Wnt/β-catenin signaling, which is involved in
chemoresistance of OS cells.
In summary, the present study demonstrated that the
effect of TRIM37 upregulation in OS on cellular proliferation and
resistance to chemotherapy drugs may be mediated by modulating the
expression of specific genes in the Wnt/β-catenin signaling
pathway. Thus, TRIM37 may be a potential therapeutic target for the
treatment of pediatric OS.
References
|
1
|
Ottaviani G and Jaffe N: The epidemiology
of osteosarcomaPediatric and Adolescent Osteosarcoma. Jaffe N,
Bruland OS and Bielack S: 152. Springer; US: pp. 3–13. 2010,
View Article : Google Scholar
|
|
2
|
Luetke A, Meyers PA, Lewis I and Juergens
H: Osteosarcoma treatment-where do we stand? A state of the art
review. Cancer Treat Rev. 40:523–532. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kansara M, Teng MW, Smyth MJ and Thomas
DM: Translational biology of osteosarcoma. Nat Rev Cancer.
14:722–735. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Salah S, Ahmad R, Sultan I, Yaser S and
Shehadeh A: Osteosarcoma with metastasis at initial diagnosis:
Current outcomes and prognostic factors in the context of a
comprehensive cancer center. Mol Clin Oncol. 2:811–816. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Subbiah V and Kurzrock R: Phase 1 clinical
trials for sarcomas: The cutting edge. Curr Opin Oncol. 23:352–360.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yang J and Zhang W: New molecular insights
into osteosarcoma targeted therapy. Curr Opin Oncol. 25:398–406.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kallijärvi J, Avela K, Lipsanen-Nyman M,
Ulmanen I and Lehesjoki AE: The TRIM37 gene encodes a peroxisomal
RING-B-box-coiled-coil protein: Classification of mulibrey nanism
as a new peroxisomal disorder. Am J Hum Genet. 70:1215–1228. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hämäläinen RH, Mowat D, Gabbett MT,
O'brien TA, Kallijärvi J and Lehesjoki AE: Wilms' tumor and novel
TRIM37 mutations in an Australian patient with mulibrey nanism.
Clin Genet. 70:473–479. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jagiello P, Hammans C, Wieczorek S, Arning
L, Stefanski A, Strehl H, Epplen JT and Gencik M: A novel splice
site mutation in the TRIM37 gene causes mulibrey nanism in a
Turkish family with phenotypic heterogeneity. Hum Mutat.
21:630–635. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kallijärvi J, Lahtinen U, Hämäläinen R,
Lipsanen-Nyman M, Palvimo JJ and Lehesjoki AE: TRIM37 defective in
mulibrey nanism is a novel RING finger ubiquitin E3 ligase. Exp
Cell Res. 308:146–155. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bhatnagar S, Gazin C, Chamberlain L, Ou J,
Zhu X, Tushir JS, Virbasius CM, Lin L, Zhu LJ, Wajapeyee N and
Green MR: TRIM37 is a new histone H2A ubiquitin ligase and breast
cancer oncoprotein. Nature. 516:116–120. 2014.PubMed/NCBI
|
|
12
|
Jiang J, Yu C, Chen M, Tian S and Sun C:
Over-expression of TRIM37 promotes cell migration and metastasis in
hepatocellular carcinoma by activating Wnt/β-catenin signaling.
Biochem Biophys Res Commun. 464:1120–1127. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cai Y, Cai T and Chen Y: Wnt pathway in
osteosarcoma, from oncogenic to therapeutic. J Cell Diochem.
115:625–631. 2014. View Article : Google Scholar
|
|
14
|
Anastas JN and Moon RT: WNT signalling
pathways as therapeutic targets in cancer. Nat Rev Cancer.
13:11–26. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mora-Blanco EL, Mishina Y, Tillman EJ, Cho
YJ, Thom CS, Pomeroy SL, Shao W and Roberts CW: Activation of
β-catenin/TCF targets following loss of the tumor suppressor SNF5.
Oncogene. 33:933–938. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tetsu O and McCormick F: β-Catenin
regulates expression of cyclin D1 in colon carcinoma cells. Nature.
398:422–426. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li YJ, Wei ZM, Meng YX and Ji XR:
Beta-catenin up-regulates the expression of cyclinD1, c-myc and
MMP-7 in human pancreatic cancer: Relationships with carcinogenesis
and metastasis. World J Gastroenterol. 11:2117–2123. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Trierweiler C, Blum HE and Hasselblatt P:
The transcription factor c-Jun protects against liver damage
following activated β-catenin signaling. PLoS One. 7:e406382012.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jiang J, Tian S, Yu C, Chen M and Sun C:
TRIM37 promoted the growth and migration of the pancreatic cancer
cells. Tumor Biol. 37:2629–2634. 2016. View Article : Google Scholar
|
|
20
|
Wang Y, Guo Q, Zhao Y, Chen J, Wang S, Hu
J and Sun Y: BRAF-activated long non-coding RNA contributes to cell
proliferation and activates autophagy in papillary thyroid
carcinoma. Oncol Lett. 8:1947–1952. 2014.PubMed/NCBI
|
|
21
|
Flahaut M, Meier R, Coulon A, Nardou KA,
Niggli FK, Martinet D, Beckmann JS, Joseph JM, Mühlethaler-Mottet A
and Gross N: The Wnt receptor FZD1 mediates chemoresistance in
neuroblastoma through activation of the Wnt/beta-catenin pathway.
Oncogene. 28:2245–2256. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Noda T, Nagano H, Takemasa I, Yoshioka S,
Murakami M, Wada H, Kobayashi S, Marubashi S, Takeda Y, Dono K, et
al: Activation of Wnt/beta-catenin signalling pathway induces
chemoresistance to interferon-alpha/5-fluorouracil combination
therapy for hepatocellular carcinoma. Br J Cancer. 100:1647–1658.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hsieh IS, Chang KC, Tsai YT, Ke JY, Lu PJ,
Lee KH, Yeh SD, Hong TM and Chen YL: MicroRNA-320 suppresses the
stem cell-like characteristics of prostate cancer cells by
down-regulating the Wnt/beta-catenin signaling pathway.
Carcinogenesis. 34:530–538. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hämäläinen RH, Avela K, Lambert JA,
Kallijärvi J, Eyaid W, Gronau J, Ignaszewski AP, McFadden D, Sorge
G, Lipsanen-Nyman M and Lehesjoki AE: Novel mutations in the TRIM37
gene in Mulibrey Nanism. Hum Mutat. 23:5222004. View Article : Google Scholar
|
|
25
|
Kumpf M, Hämäläinen RH, Hofbeck M and
Baden W: Refractory congestive heart failure following delayed
pericardectomy in a 12-year-old child with Mulibrey nanism due to a
novel mutation in TRIM37. Eur J Pediatr. 172:1415–1418. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wickström M, Dyberg C, Milosevic J, Einvik
C, Calero R, Sveinbjörnsson B, Sandén E, Darabi A, Siesjö P, Kool
M, et al: Wnt/β-catenin pathway regulates MGMT gene expression in
cancer and inhibition of Wnt signalling prevents chemoresistance.
Nat Commun. 6:89042015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ma Y, Ren Y, Han EQ, Li H, Chen D, Jacobs
JJ, Gitelis S, O'Keefe RJ, Konttinen YT, Yin G and Li TF:
Inhibition of the Wnt-β-catenin and Notch signaling pathways
sensitizes osteosarcoma cells to chemotherapy. Biochem Biophys Res
Commun. 431:274–279. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:pp. 15545–15550. 2005;
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Mootha VK, Lindgren CM, Eriksson KF,
Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E,
Ridderstråle M, Laurila E, et al: PGC-1alpha-responsive genes
involved in oxidative phosphorylation are coordinately
downregulated in human diabetes. Nat Genet. 34:267–273. 2003.
View Article : Google Scholar : PubMed/NCBI
|