Introduction
Breast cancer is the most commonly diagnosed type of
cancer and the second leading cause of cancer-associated mortality
in females worldwide, accounting for 29% of the total new cancer
cases and 14% of the total cancer-associated mortalities in 2016
(1). A previous study demonstrated
that various proteins are dysregulated in primary tumors, and are
associated with breast cancer development and progression (2). Therefore, an improved understanding of
the functions and the underlying molecular mechanisms of these
proteins, particularly in epithelial-mesenchymal transition, may
provide novel insights into the breast tumorigenesis and
progression, and enable the development of effective anti-cancer
therapeutics.
The SRY-box 4 (SOX4) gene, which is located on
chromosome 6p22.3, encodes a 47-kDa protein that is a member of the
sex-determining region Y-related high-mobility group-box
transcription factor family and has functions in embryonic
development and cell differentiation (3,4). SOX4 has
been demonstrated to be upregulated in a number of types of cancer,
and increased SOX4 activity was found to contribute to cancer
development and progression (5–8). SOX4 has
been revealed to induce EMT, a key process during organ development
and the progression of epithelial tumors to metastatic cancers
(9), via its activation of the Wnt
and transforming growth factor β signaling pathways in cancer cells
(10,11). Previously, it has been demonstrated
that certain transcriptional targets of SOX4 are associated with
cancer development and progression and the processing of microRNAs
(miRNAs/miRs) (12).
miRNAs are small non-coding RNAs (~22 nucleotides in
length) that regulate the expression of their target genes by
translational repression or mRNA degradation (13,14).
miRNAs participate in crucial biological processes, including
development, differentiation and proliferation (15,16).
Previous studies have suggested that miRNAs are involved in a
number of types of cancer (17,18).
Differential expression of miRNAs has been widely described in
breast cancers, and suggests that certain miRNAs, including miR-206
(19), miR-129 (20), miR-200 (21) and miR-34 (22), may function as oncogenes or tumor
suppressors in breast cancer. Therefore, miRNAs are now considered
to be important in the development of biomarkers, and may be
targets for the diagnosis and treatment of breast cancer patients
(23).
To understand the molecular mechanism of SOX4 in
breast cancer development and progression, the present study aimed
to identify miRNAs that regulate the expression of SOX4, which
revealed miR-320 as a potential candidate. It has been previously
identified that miR-320 is significantly downregulated, and can
inhibit proliferation, metastasis and angiogenesis, in a number of
types of cancer, including cervical cancer, colon cancer, glioma
and prostate cancer; this evidence suggests that miR-320 functions
as a tumor suppressor (24–29). However, the function and molecular
mechanism of miR-320 remain unknown.
In the present study, miR-320 was demonstrated to be
frequently downregulated in breast cancer tissues compared with
adjacent normal tissues. Furthermore, SOX4 was confirmed as a
direct target of miR-320, and miR-320 was shown to suppress breast
cancer development and progression via the downregulation of SOX4.
The results of the present study indicate that miR-320
overexpression may represent a novel therapeutic strategy for
patients with breast cancer.
Materials and methods
Cell culture and tissue specimens
The human breast cancer cell lines MCF7, T47D,
MDA-MB-231, MDA-MB-468 and 293FT were purchased from the American
Type Culture Collection (Manassas, VA, USA). MDA-MB-231 and
MDA-MB-468 cells were maintained in Leibovitz's L15 medium (Gibco;
Thermo Fisher Scientific, Inc., Waltham, MA, USA), and 293FT, MCF7
and T47D cells were maintained in Dulbecco's modified Eagle's
medium (Gibco; Thermo Fisher Scientific, Inc.). All cell lines were
supplemented with 10% fetal bovine serum (FBS), as well as
penicillin and streptomycin, and were incubated at 37°C in a
humidified atmosphere containing 5% CO2.
In addition, primary breast cancer tissues and
paired adjacent normal breast tissues were obtained from 15 females
aged between 40 to 65 years (mean age, 53 years) who underwent
breast surgery in the Affiliated Hospital of Inner Mongolia Medical
University between April 2015 and December in 2015. All patients
were diagnosed with invasive ductal carcinoma according to the
morphologic criteria described in the World Health Organization
histologic classification of tumors of the breast (30). Of the 15 patients, 5 patients were
stage II and 10 patients were stage I. All samples were evaluated
and subject to histological diagnosis by pathologists. All
protocols in the present study were approved by the ethics
committee of the Affiliated Hospital of Inner Mongolia Medical
University, and all patients provided written informed consent.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from the breast cancer
tissues and cell lines using TRIzol reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) following the manufacturer's protocol. A
total of 2 µg RNA was used for RT-qPCR with the Qiagen OneStep
RT-PCR kit (Qiagen Benelux BV, Venlo, The Netherlands) in the
following conditions: 37°C for 30 min, 85°C for 5 sec and kept at
4°C until use. TaqMan microRNA assays (Applied Biosystems; Thermo
Fisher Scientific, Inc.) were performed to quantify the relative
expression of miR-320 using the following thermocycling conditions:
95°C for 3 min, 95°C for 15 sec and 60°C for 30 sec for a total of
40 cycles, as previously described (31). U6 was used as the normalization
control. The following primers were used: miR-320 forward,
5′-ACACTCCAGCTGGGAAAAGCTGGGTTGAGA-3′; miR-320 reverse,
5′-TGGTGTCGTGGAGTCG-3; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′; U6
reverse, 5′-AACGCTTCACGAATTTGCGT-3′; GAPDH forward,
5′-AATCCCATCACCATCTTCCA-3; GAPDH reverse,
5′-CCTGCTTCACCACCTTCTTG-3; SOX4 forward,
5′-ACCGGGACCTGGATTTTAAC-3′; and SOX4 reverse,
5′-AAACCAGGTTGGAGATGCTG-3′. The CT value of GAPDH or U6 was
subtracted from the CT value of target genes to obtain ΔCT. The
relative expression level of miR-320 and SOX4 was determined as
2−ΔCT as previously described (31). Three independent experiments were
conducted.
Transfection
MDA-MB-231 or MCF7 cells were seeded in 6-well
plates at a density of 5×106 cells/well. The cells in
each well were transfected with a solution of 3 µg pcDNA3.1-SOX4
(GenePharma Co., Ltd., Shanghai, China) or 20 nM RNA [miRNA-320
mimics, 5′-AAAAGCUGGGUUGAGAGGGCGA-3′; inhibitors,
5′-CCUCUCAACCCAGCUUUU-3′; negative control (NC)/mimic control,
5′-UUCUCCGAACGUGUCACGUTT-3′; and inhibitor control,
5′-CAGUACUUUUGUGUAGUACAA-3′; GenePharma Co., Ltd.] using
Lipofectamine 3000 (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. Subsequent experiments
were conducted 24 h after transfection.
Cell proliferation assays
MTT and colony formation assays were used to
evaluate cell proliferation. In brief, for the MTT assay
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany), a total of
5×103 MDA-MB-231 or MCF7 cells/well were seeded in
96-well plates, and MTT was added to a final concentration of 0.5
mg/ml, prior to the incubation of cells at 37°C for 4 h. Following
the removal of the culture medium, 150 µl dimethyl sulfoxide was
added and the absorbance at 570 nm was measured using a microplate
reader. The mimic or inhibitor control transfected cells were as
the negative control group.
For colony formation assay, 1×103/well
MDA-MB-231 or MCF7 cells were seeded into 6 cm dish after
transfection. After 2 weeks of culture, the cells were stained with
1% crystal violet for 15 min at room temperature, after fixation
with 10% formaldehyde for 30 min at room temperature.
Transwell invasion assay
Transwell Matrigel-coated chambers (BD Biosciences,
Franklin Lakes, NJ, USA) were used to determine cell invasion. A
total of 1×105/well cells transfected with miR-320
mimics, inhibitors or NC were suspended in 200 µl medium containing
1% FBS, and seeded on the upper chamber. A total of 600 µl medium
containing 10% FBS was added to the lower chamber. Following
incubation for 24 h, cells that remained in the upper chamber were
removed and the migrated cells were fixed in methanol, and
subsequently stained with crystal violet for 30 min at room
temperature. Cells were counted in five randomly selected fields
using a light microscope. The mimic or inhibitor control
transfected cells were as the negative control group. The cells
were counted in 5 randomly selected microscopic fields
(magnification, ×400) from each chamber.
Cell cycle and apoptosis assays
To determine cell cycle distribution,
1×106/well MDA-MB-231 or MCF7 cells were collected by
trypsinization and fixed in ice-cold 70% ethanol overnight.
Subsequently, cells were washed with PBS and incubated with
propidium iodide (PI; Sigma-Aldrich; Merck KGaA) containing RNase
for 30 min at room temperature. The cell cycle distribution was
analyzed using the FACScan flow cytometer (BD Biosciences). For the
apoptosis assay, 1×106/well MDA-MB-231 cells were
harvested and double-stained with annexin V-fluorescein
isothiocyanate (FITC) and PI by using the annexin V-FITC Apoptosis
Detection kit (BD Biosciences) following the manufacturer's
protocol. Subsequently, each sample was analyzed using the FACScan
flow cytometer. The data were processed using the ModFit LT 3.2
software (Verity Software House, Inc., Topsham, ME, USA).
Western blot analysis
Cells were harvested using cell scrapers, washed
with PBS and lysed using RIPA lysis buffer (Beyotime Institute of
Biotechnology, Haimen, China) with a protease inhibitor cocktail
(Pierce; Thermo Fisher Scientific, Inc.) and centrifuged at 10,000
× g for 15 min at 4°C to remove cellular debris. Protein samples
(50 µg) were separated using SDS-PAGE (10% gels) and subsequently
transferred to a polyvinylidene fluoride membrane (Millipore; Merck
KGaA). The membranes were blocked in 5% non-fat milk for 1 h and
incubated at 4°C overnight with the following primary antibodies:
Anti-GAPDH (catalog no. 2118; dilution, 1:2,000; Cell Signaling
Technology, Inc., Danvers, MA, USA), anti-SOX4 (catalog no.
ab86809; dilution, 1:1,000; Abcam, Cambridge, MA, USA),
anti-epithelial (E-)cadherin (catalog no. sc-59780; dilution,
1:1,000; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) and
anti-vimentin (catalog no. sc-66002; dilution, 1:2,000; Santa Cruz
Biotechnology, Inc.). Membranes were washed three times with
Tris-buffered Saline-Tween 20 and incubated for 1 h with an
anti-rabbit horseradish peroxidase (HRP)-conjugated secondary
antibody (catalog no. 7074; dilution, 1:2,500; Cell Signaling
Technology, Inc.) or an anti-mouse HRP-conjugated secondary
antibody (catalog no. 7076; dilution, 1:2,500; Cell Signaling
Technology, Inc.) at room temperature. Specific proteins were
detected using an ECL kit (Millipore; Merck KGaA).
Bioinformatic prediction of miR-320
potential targets
The microRNA.org
targets and expression (www.microrna.org) database was used to predict
potential targets for miR-320, as previously described (32).
Luciferase reporter assay
The SOX4 3′-untranslated region (UTR) containing the
wild-type (wt) or mutated form of the miR-320 binding site were
cloned into the psiCHECK2 vector (Promega Corporation, Madison, WI,
USA). A total of 2×105/well 293FT cells, cultured in
12-well plates, were co-transfected using Lipofectamine 3000 with
psiCHECK2 vectors containing either the wt or mutated SOX4 3′-UTR
fragments and the control vector, as well as the miR-320 mimics and
NC. Luciferase assays were performed 48 h after transfection, using
the Dual-Luciferase Reporter Assay kit (Promega Corporation),
according to the manufacturer's protocol. Firefly luciferase
activity was normalized to Renilla luciferase activity.
Statistical analysis
Statistical analysis was performed using SPSS
software (version 22.0; IBM Corp., Armonk, NY, USA). The Student's
t-test was used to perform comparisons between two groups of data.
Multiple comparisons between data were performed using a one-way
analysis of variance, followed by Dunnett's test. The results were
expressed as the mean ± standard error. P<0.05 was considered to
indicate a statistically significant difference. Three independent
experiments were conducted.
Results
miR-320 expression is frequently
downregulated in breast cancer
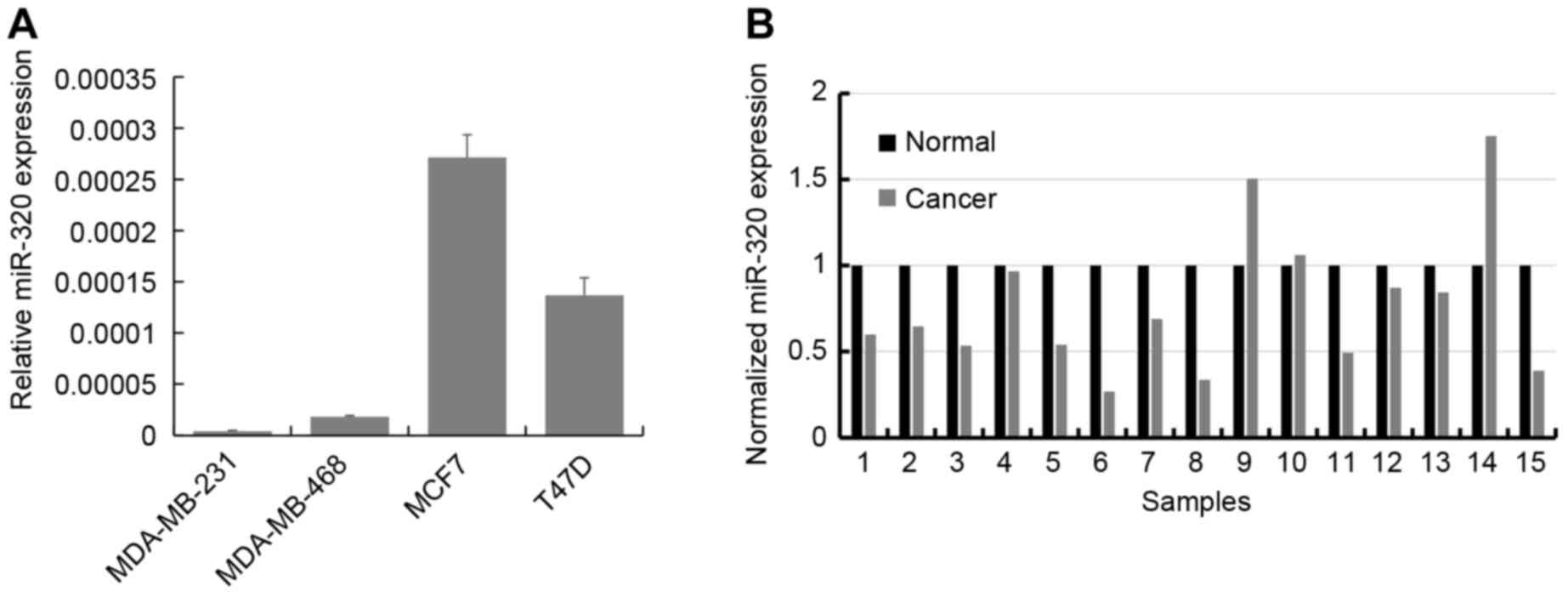
RT-qPCR was used to determine the expression levels
of miR-320 in the breast cancer cell lines MCF7, T47D, MDA-MB-231
and MDA-MB-468. The results demonstrated that miR-320 is expressed
mostly highly in MCF7 and T47D cells, and is expressed at a
relatively decreased level in MDA-MB-231 and MDA-MB-468 cells
(Fig. 1A). Subsequently, the
expression of miR-320 was analyzed using RT-qPCR in primary breast
cancer tissues and paired adjacent normal breast tissues from 15
patients. In 12 of the 15 cases (80%), the expression of miR-320
was downregulated in the breast cancer tissue compared with the
corresponding adjacent normal tissue (P<0.001; Fig. 1B).
miR-320 inhibits the proliferative and
invasive abilities of breast cancer cells
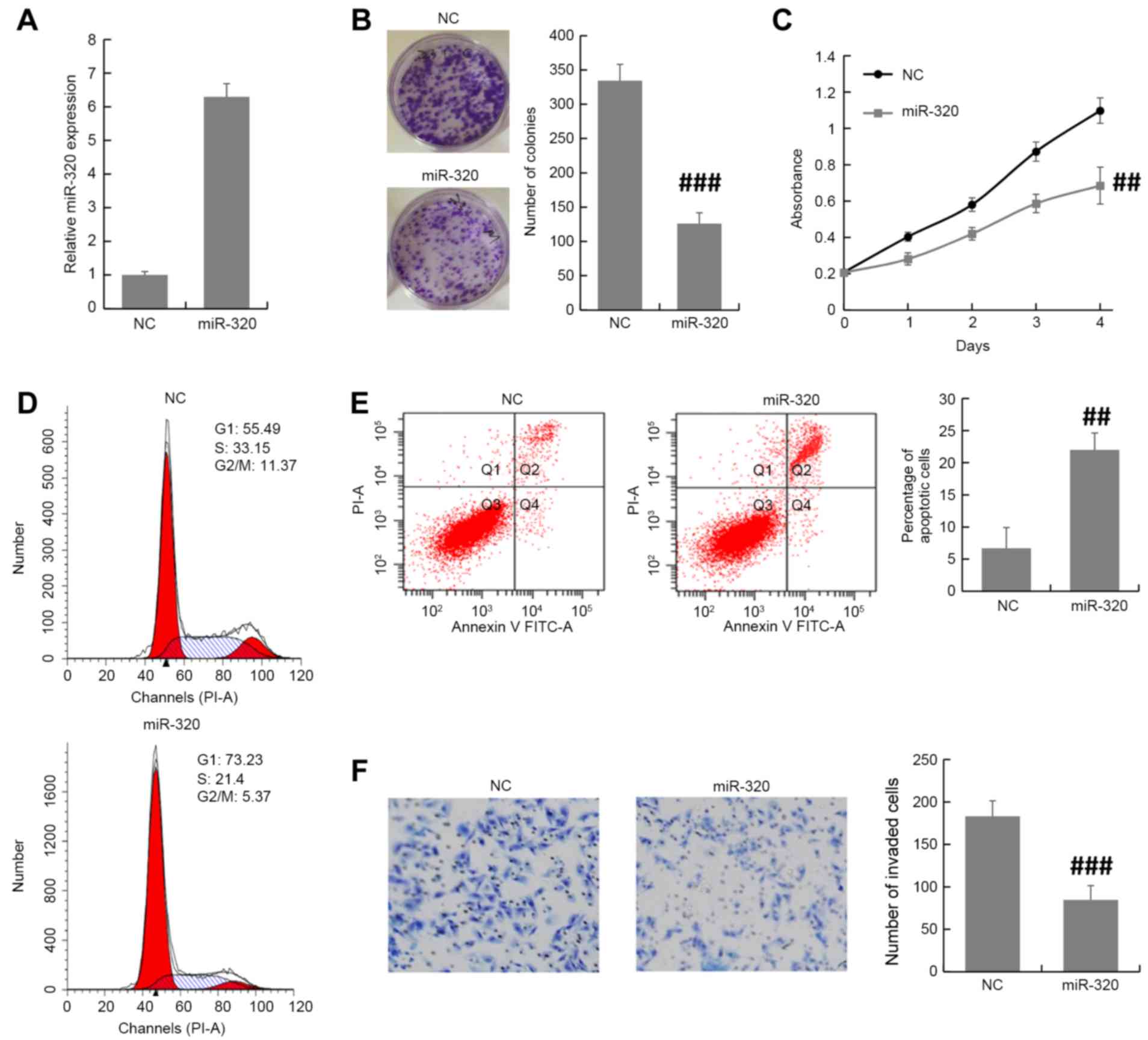
MDA-MB-231 cells were transfected with miR-320
mimics or NC oligonucleotides and the transfection efficiency was
examined by RT-qPCR. Using RT-qPCR, it was validated that miR-320
expression level was increased by miR-320 mimic in MDA-MB-231 cells
(Fig. 2A). Colony formation and MTT
assays revealed that overexpression of miR-320 inhibited the cell
proliferation of MDA-MB-231 cells (Fig.
2B and C). Furthermore, the cell cycle distribution assay
demonstrated that there was an accumulation in the G0/G1 phase
among miR-320-overexpressing MDA-MB-231 cells compared with NC
cells (Fig. 2D). The apoptosis assay
revealed that the miR-320-overexpressing MDA-MB-231 cells exhibited
an increase in the apoptotic index compared with that of control
cells (Fig. 2E). To investigate the
function of miR-320 in breast cancer progression, the Transwell
invasion assay was performed in transfected MDA-MB-231 cells. The
results demonstrated that the overexpression of miR-320 inhibited
breast cancer cell invasion compared with the NC (Fig. 2F). These results suggested that
miR-320 suppresses breast cancer progression.
Inhibition of miR-320 promotes breast
cancer proliferation and invasion
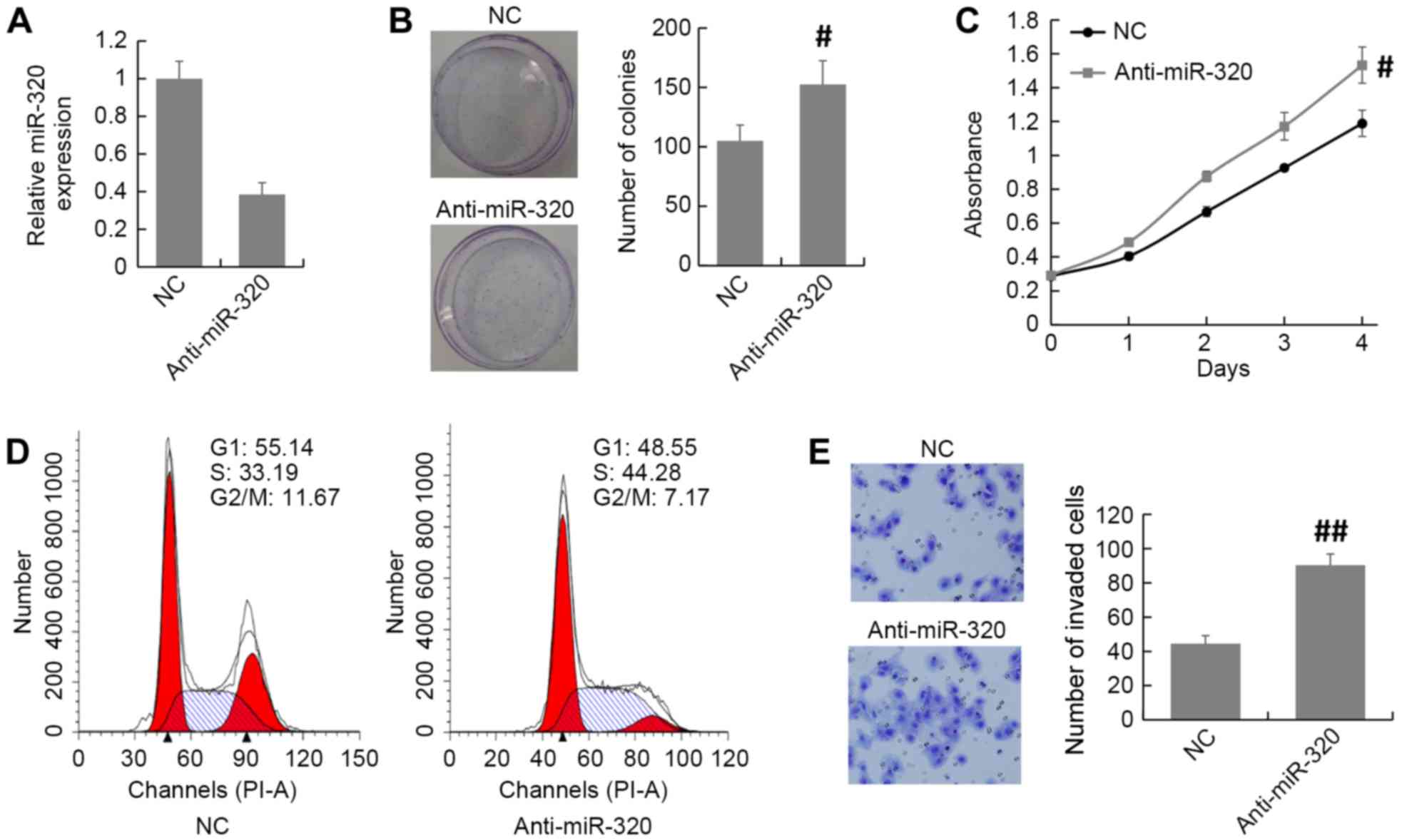
The expression of miR-320 in MCF7 cells was knocked
down by transfection of an miR-320 inhibitor or NC (Fig. 3A). The MTT and colony formation assays
revealed that knockdown of miR-320 expression promoted cell
proliferation in MCF7 cells compared with that in the NC group
(Fig. 3B and C). The cell cycle
distribution assay demonstrated that there was an accumulation of
S-phase cells among the miR-320-depleted MCF7 cells compared with
the NC cells (Fig. 3D). Furthermore,
the Transwell invasion assay demonstrated that the number of
invaded cells was increased in miR-320-depleted MCF7 cells compared
with the NC group (Fig. 3E). Thus,
these results indicate that inhibition of miR-320 promotes breast
cancer progression.
SOX4 is a target of miR-320
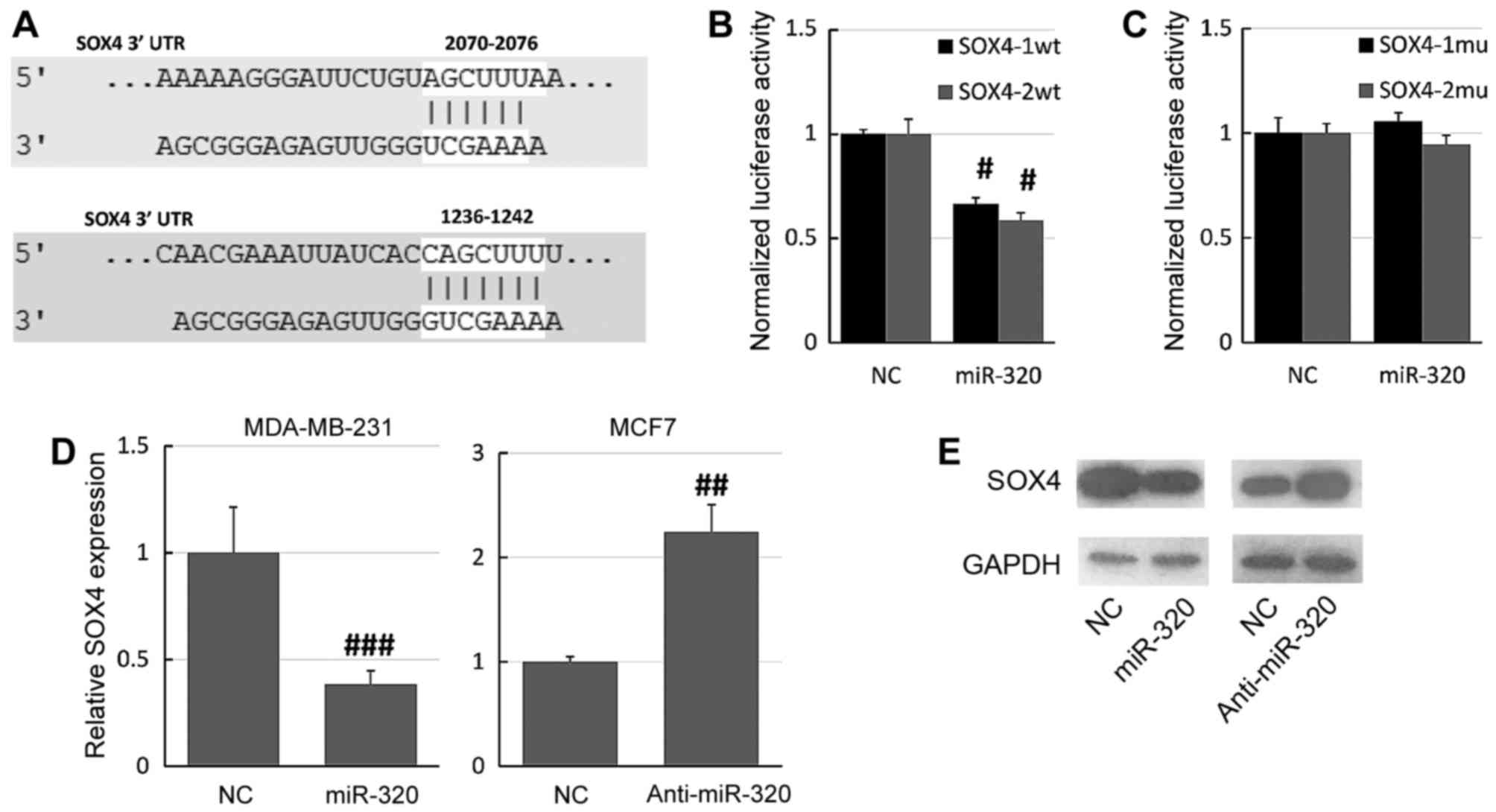
SOX4 was predicted as a potential target of miR-320
using the online database microRNA.org
(Fig. 4A). The online database
identified two potential miR-320 binding sites on the SOX4 3′-UTR,
so two different reporter plasmids were constructed (SOX4-1wt and
SOX4-2wt). The SOX4 3′-UTR was cloned into a luciferase reporter
vector and the putative miR-320 binding site was mutated using a
site-directed mutagenesis kit. The luciferase assay demonstrated
that overexpression of miR-320 significantly decreased the
luciferase activity of the SOX4-1wt and SOX4-2wt constructs
relative to NC-transfected cells (Fig.
4B). Furthermore, mutation of the miR-320 binding sites
prevented this effect of miR-320 on luciferase activity: There were
no significant differences between the miR-320 mimic-transfected
and the NC cells (Fig. 4C).
Furthermore, RT-qPCR and western blot assays revealed that the
overexpression of miR-320 downregulated the expression of SOX4 in
MDA-MB-231 cells, whereas knockdown of miR-320 upregulated the
expression of SOX4 in MCF7 cells (Fig. 4D
and E). Together, these results suggested that SOX4 is a direct
target of miR-320.
miR-320 inhibits breast cancer
progression by downregulating SOX4
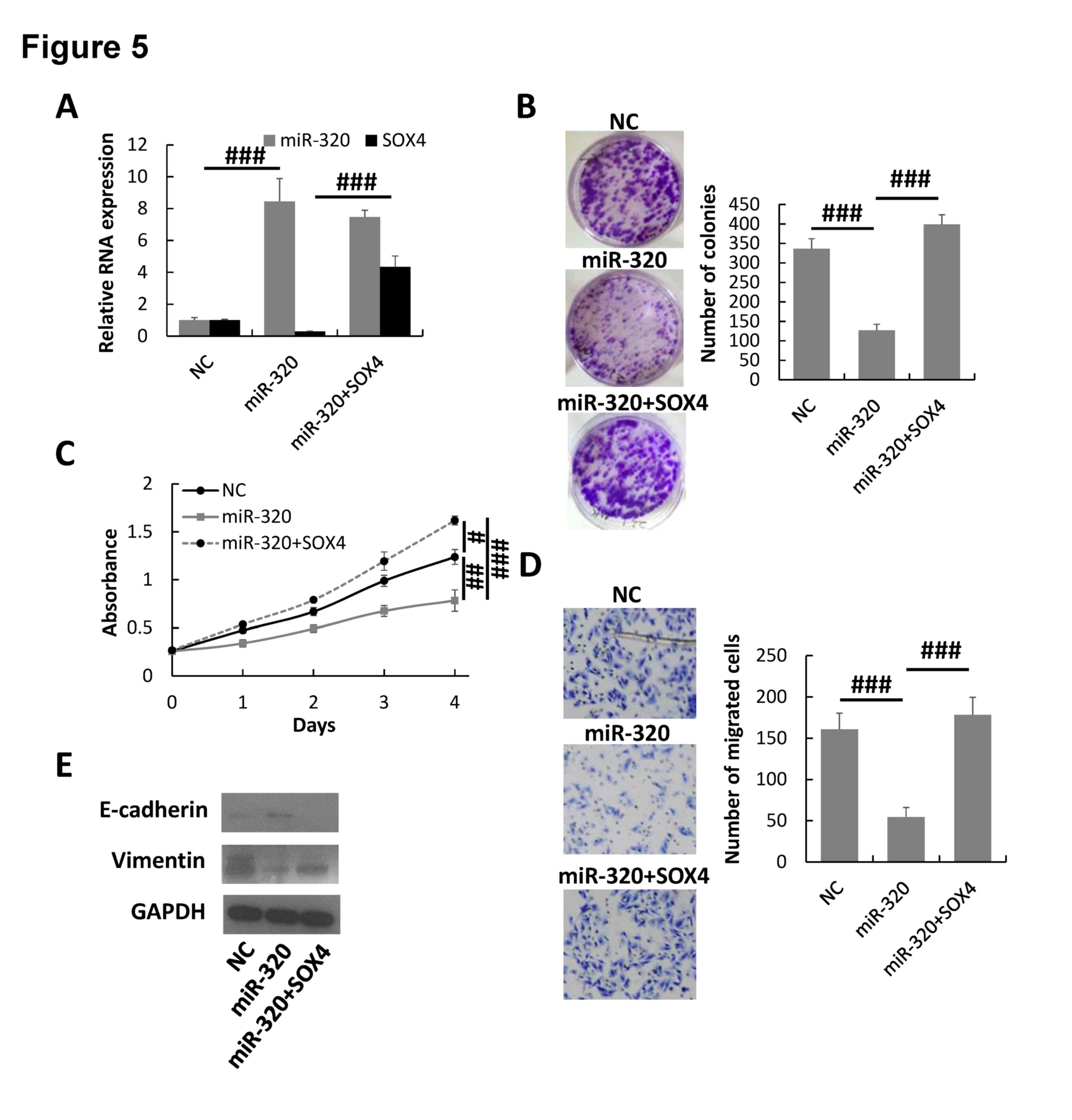
To additionally demonstrate the regulation of SOX4
by miR-320 in breast cancer cells, pcDNA3.1-SOX4 was transfected
into the miR-320-overexpressed MDA-MB-231 cells to rescue the SOX4
expression. The RT-qPCR assay indicated that the expression of SOX4
was significantly increased in miR-320/SOX4 co-transfected
MDA-MB-231 cells compared with that of NC cells and cells
transfected with miR-320 alone (Fig.
5A). The colony formation, MTT and Transwell invasion assays
demonstrated that overexpression of SOX4 could rescue the malignant
phenotype of miR-320-overexpressing MDA-MB-231 cells (Fig. 5B-D). Furthermore, the expression of
epithelial marker E-cadherin was downregulated and the mesenchymal
marker vimentin was upregulated in SOX4/miR-320-overexpressing
MDA-MB-231 cells compared with miR-320-overexpressing cells
(Fig. 5E). Therefore, the results
suggested that miR-320 inhibited breast cancer progression by
downregulating SOX4.
Discussion
miRNAs are critical regulators involved in a number
of biological processes, including proliferation, differentiation,
migration, metabolism and apoptosis (33). The abnormal expression of miRNAs has
been observed in a number types of cancer (34,35). Thus,
the identification and study of cancer-specific miRNAs and their
targets are critical for understanding their function and mechanism
in cancer development and progression. miR-320 has been identified
to be downregulated in a number of types of cancer, including
cervical cancer, colon cancer, oral cancer and osteosarcoma, and
was shown to be involved in tumorigenesis and progression as a
tumor suppressor (24,25,28,36). A
miRNA microarray analysis demonstrated that miR-321 was
downregulated in patients with breast cancer, suggesting that
miR-320 may serve an important function in breast cancer
development and progression (37).
However, the molecular mechanism by which miR-320 affects breast
cancer development and progression remains unknown. Therefore, the
present study aimed to elucidate the biological function and
mechanism of miR-320 in breast cancer.
Consistent with a previous study (38), the results of the present study
indicated that miR-320 was decreased in the majority of breast
cancer tissues compared with the corresponding adjacent normal
tissues. Overexpression of miR-320 inhibited proliferation and
invasion, and induced apoptosis, in breast cancer cells, suggesting
that miR-320 may be a tumor suppressor and a diagnostic biomarker
in breast cancer. In addition, the present study demonstrated a
potential underlying molecular mechanism of miR-320 in the
inhibition of breast cancer progression; the results indicated that
SOX4 is a direct target of miR-320 in breast cancer, which is
consistent with the mechanism of miR-320/SOX4 in colorectal cancer
(29). Furthermore, it was
demonstrated that the inhibitory effects of miR-320 overexpression
on breast cancer cell proliferation and invasion were partially
attenuated by the upregulation of SOX4 expression. Thus, it was
indicated that miR-320 inhibits breast cancer progression by
targeting SOX4.
EMT is a critical process in breast cancer
progression that causes epithelial cells to acquire fibroblast-like
properties, with reduced intercellular adhesion and increased
motility (20). SOX4 has been
observed to be overexpressed in a number of different types of
malignant tumor and is one of the master regulators in EMT-induced
cancer metastasis and chemoresistance (5,10,39,40). The
results of the present study demonstrated that miR-320 inhibits the
EMT in breast cancer cells, as demonstrated by the retention of the
epithelial phenotype, and the EMT-inhibiting effect of miR-320 may
be attenuated by the overexpression of SOX4. Thus, to the best of
our knowledge, the current study is the first to demonstrate that
miR-320 is a regulator of SOX4 in breast cancer cells, which
provides one possible mechanism underlying the function of miR-320
in breast cancer progression.
In summary, the results of the present study
indicated that miR-320 is frequently decreased in breast cancer
cells and that miR-320 inhibits breast cancer progression by
targeting SOX4. This novel miR-320/SOX4 axis may provide insight
into the molecular mechanisms underlying breast cancer progression,
and the upregulation of miR-320 expression may be a therapeutic
strategy for the treatment of breast cancer.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Karlsson MC, Gonzalez SF, Welin J and Fuxe
J: Epithelial-mesenchymal transition in cancer metastasis through
the lymphatic system. Mol Oncol. 11:781–791. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Poncy A, Antoniou A, Cordi S, Pierreux CE,
Jacquemin P and Lemaigre FP: Transcription factors SOX4 and SOX9
cooperatively control development of bile ducts. Dev Biol.
404:136–148. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jang SM, Kim JW, Kim CH, An JH, Johnson A,
Song PI, Rhee S and Choi KH: KAT5-mediated SOX4 acetylation
orchestrates chromatin remodeling during myoblast differentiation.
Cell Death Dis. 6:e18572015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tiwari N, Tiwari VK, Waldmeier L, Balwierz
PJ, Arnold P, Pachkov M, Meyer-Schaller N, Schübeler D, van
Nimwegen E and Christofori G: Sox4 is a master regulator of
epithelial-mesenchymal transition by controlling Ezh2 expression
and epigenetic reprogramming. Cancer Cell. 23:768–783. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang B, Li Y, Tan F and Xiao Z: Increased
expression of SOX4 is associated with colorectal cancer
progression. Tumour Biol. 37:9131–9137. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bilir B, Osunkoya AO, Wiles WG IV,
Sannigrahi S, Lefebvre V, Metzger D, Spyropoulos DD, Martin WD and
Moreno CS: SOX4 is essential for prostate tumorigenesis initiated
by PTEN ablation. Cancer Res. 76:1112–1121. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hasegawa S, Nagano H, Konno M, Eguchi H,
Tomokuni A, Tomimaru Y, Asaoka T, Wada H, Hama N, Kawamoto K, et
al: A crucial epithelial to mesenchymal transition regulator,
Sox4/Ezh2 axis is closely related to the clinical outcome in
pancreatic cancer patients. Int J Oncol. 48:145–152. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sung WJ, Kim H and Park KK: The biological
role of epithelial-mesenchymal transition in lung cancer (Review).
Oncol Rep. 36:1199–1206. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang J, Liang Q, Lei Y, Yao M, Li L, Gao
X, Feng J, Zhang Y, Gao H, Liu DX, et al: SOX4 induces
epithelial-mesenchymal transition and contributes to breast cancer
progression. Cancer Res. 72:4597–4608. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sinner D, Kordich JJ, Spence JR, Opoka R,
Rankin S, Lin SC, Jonatan D, Zorn AM and Wells JM: Sox17 and Sox4
differentially regulate beta-catenin/T-cell factor activity and
proliferation of colon carcinoma cells. Mol Cell Biol.
27:7802–7815. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jiao C, Song Z, Chen J, Zhong J, Cai W,
Tian S, Chen S, Yi Y and Xiao Y: lncRNA-UCA1 enhances cell
proliferation through functioning as a ceRNA of Sox4 in esophageal
cancer. Oncol Rep. 36:2960–2966. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang W, Zhang E and Lin C: MicroRNAs in
tumor angiogenesis. Life Sci. 136:28–35. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Huang J, Zhang SY, Gao YM, Liu YF, Liu YB,
Zhao ZG and Yang K: MicroRNAs as oncogenes or tumour suppressors in
oesophageal cancer: Potential biomarkers and therapeutic targets.
Cell Prolif. 47:277–286. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kim VN, Han J and Siomi MC: Biogenesis of
small RNAs in animals. Nat Rev Mol Cell Biol. 10:126–139. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Thomson DW, Bracken CP and Goodall GJ:
Experimental strategies for microRNA target identification. Nucleic
Acids Res. 39:6845–6853. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rupaimoole R, Calin GA, Lopez-Berestein G
and Sood AK: miRNA deregulation in cancer cells and the tumor
microenvironment. Cancer Discov. 6:235–246. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Magee P, Shi L and Garofalo M: Role of
microRNAs in chemoresistance. Ann Transl Med. 3:3322015.PubMed/NCBI
|
|
19
|
Yin K, Yin W, Wang Y, Zhou L, Liu Y, Yang
G, Wang J and Lu J: MiR-206 suppresses epithelial mesenchymal
transition by targeting TGF-β signaling in estrogen receptor
positive breast cancer cells. Oncotarget. 7:24537–24548. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yu Y, Zhao Y, Sun XH, Ge J, Zhang B, Wang
X and Cao XC: Down-regulation of miR-129-5p via the Twist1-Snail
feedback loop stimulates the epithelial-mesenchymal transition and
is associated with poor prognosis in breast cancer. Oncotarget.
6:34423–34436. 2015.PubMed/NCBI
|
|
21
|
Rhodes LV, Martin EC, Segar HC, Miller DF,
Buechlein A, Rusch DB, Nephew KP, Burow ME and Collins-Burow BM:
Dual regulation by microRNA-200b-3p and microRNA-200b-5p in the
inhibition of epithelial-to-mesenchymal transition in
triple-negative breast cancer. Oncotarget. 6:16638–16652. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yang S, Li Y, Gao J, Zhang T, Li S, Luo A,
Chen H, Ding F, Wang X and Liu Z: MicroRNA-34 suppresses breast
cancer invasion and metastasis by directly targeting Fra-1.
Oncogene. 32:4294–4303. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Pichler M and Calin GA: MicroRNAs in
cancer: From developmental genes in worms to their clinical
application in patients. Br J Cancer. 113:569–573. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wan LY, Deng J, Xiang XJ, Zhang L, Yu F,
Chen J, Sun Z, Feng M and Xiong JP: miR-320 enhances the
sensitivity of human colon cancer cells to chemoradiotherapy in
vitro by targeting FOXM1. Biochem Biophys Res Commun. 457:125–132.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang T, Zou P, Wang T, Xiang J, Cheng J,
Chen D and Zhou J: Down-regulation of miR-320 associated with
cancer progression and cell apoptosis via targeting Mcl-1 in
cervical cancer. Tumour Biol. 37:8931–8940. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sun JY, Xiao WZ, Wang F, Wang YQ, Zhu YH,
Wu YF, Miao ZL and Lin YC: MicroRNA-320 inhibits cell proliferation
in glioma by targeting E2F1. Mol Med Rep. 12:2355–2359. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hsieh IS, Chang KC, Tsai YT, Ke JY, Lu PJ,
Lee KH, Yeh SD, Hong TM and Chen YL: MicroRNA-320 suppresses the
stem cell-like characteristics of prostate cancer cells by
downregulating the Wnt/beta-catenin signaling pathway.
Carcinogenesis. 34:530–538. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wu YY, Chen YL, Jao YC, Hsieh IS, Chang KC
and Hong TM: miR-320 regulates tumor angiogenesis driven by
vascular endothelial cells in oral cancer by silencing neuropilin
1. Angiogenesis. 17:247–260. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Vishnubalaji R, Hamam R, Yue S, Al-Obeed
O, Kassem M, Liu FF, Aldahmash A and Alajez NM: MicroRNA-320
suppresses colorectal cancer by targeting SOX4, FOXM1, and FOXQ1.
Oncotarget. 7:35789–35802. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lakhani SR, Ellis IO, Schnitt SJ, Tan PH
and van de Vijver MJ: WHO Classification of Tumours of the Breast.
4th edition. Lyon: IARC Press; 2012
|
|
31
|
Zhang YF, Yu Y, Song WZ, Zhang RM, Jin S,
Bai JW, Kang HB, Wang X and Cao XC: miR-410-3p suppresses breast
cancer progression by targeting Snail. Oncol Rep. 36:480–486. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Betel D, Wilson M, Gabow A, Marks DS and
Sander C: The microRNA.org resource: Targets and expression.
Nucleic Acids Res. 36:(Database Issue). D149–D153. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Adlakha YK and Saini N: MicroRNA: A
connecting road between apoptosis and cholesterol metabolism.
Tumour Biol. 37:8529–8554. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chen Y, Yang X, Xu Y, Cao J and Chen L:
Genomic analysis of drug resistant small cell lung cancer cell
lines by combining mRNA and miRNA expression profiling. Oncol Lett.
13:4077–4084. 2017.PubMed/NCBI
|
|
35
|
Macedo T, Silva-Oliveira RJ, Silva VAO,
Vidal DO, Evangelista AF and Marques MMC: Overexpression of mir-183
and mir-494 promotes proliferation and migration in human breast
cancer cell lines. Oncol Lett. 14:1054–1060. 2017.PubMed/NCBI
|
|
36
|
Cheng C, Chen ZQ and Shi XT: MicroRNA-320
inhibits osteosarcoma cells proliferation by directly targeting
fatty acid synthase. Tumour Biol. 35:4177–4183. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yan LX, Huang XF, Shao Q, Huang MY, Deng
L, Wu QL, Zeng YX and Shao JY: MicroRNA miR-21 overexpression in
human breast cancer is associated with advanced clinical stage,
lymph node metastasis and patient poor prognosis. RNA.
14:2348–2360. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang B, Yang Z, Wang H, Cao Z, Zhao Y,
Gong C, Ma L, Wang X, Hu X and Chen S: MicroRNA-320a inhibits
proliferation and invasion of breast cancer cells by targeting
RAB11A. Am J Cancer Res. 5:2719–2729. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Jin Y, Zhao M, Xie Q, Zhang H, Wang Q and
Ma Q: MicroRNA-338-3p functions as tumor suppressor in breast
cancer by targeting SOX4. Int J Oncol. 47:1594–1602. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Liu Y, Li Y, Liu J, Wu Y and Zhu Q:
MicroRNA-132 inhibits cell growth and metastasis in osteosarcoma
cell lines possibly by targeting Sox4. Int J Oncol. 47:1672–1684.
2015. View Article : Google Scholar : PubMed/NCBI
|