Introduction
Circular RNAs (circRNAs) are a type of endogenous
non-coding RNA that have been a research hotspot in the field of
RNA. To date, thousands of endogenous circRNAs have been identified
and characterized. circRNAs exist ubiquitously in eukaryotes,
predominantly in the cytoplasm, exhibiting a tissue- and
developmental stage-specific pattern, serving as gene regulators in
mammals (1–3). Furthermore, circRNAs may serve roles in
neurological disorders, atherosclerotic vascular disease risk and
diverse cancer types (4–6).
Gastrointestinal cancer is a class of cancer
affecting the organs of the digestive system. The most frequently
occurring types of gastrointestinal cancer are colorectal cancer
(CRC), gastric cancer (GC), pancreatic cancer, hepatocellular
carcinoma (HCC) and esophageal carcinoma. Although there are
several options with regards to diagnostic and therapeutic methods,
the 5-year survival rate of patients with gastrointestinal cancer
remains poor due to a lack of effective tools for early diagnosis
and therapy (7). Recently, mounting
evidence has demonstrated that a number of circRNAs exhibit
potential biological functions in gastrointestinal cancer, and
consequently, may be prognostically and diagnostically relevant
biomarkers (8–10).
The present review first briefly describes the
features, biogenesis and biological functions of circRNAs across
eukaryotes. Secondly, potential functions and implications of
circRNA in gastrointestinal cancer are discussed, and their roles
in gastrointestinal tumor occurrence and development are explored.
Finally, the future of research progress and direction, in respect
to circRNA in gastrointestinal cancer diagnosis and targeted
therapy, are discussed.
Features of circRNA
Structure
circRNAs are characterized by covalently closed
continuous loop structures with neither 5′ to 3′ polarity nor a
polyadenylated tail, as well as the resistance to exonucleolytic
degradation (2,3). As a result, circRNAs are highly stable
in vivo compared with their linear counterparts.
Biogenesis
Studies have revealed that circRNAs may be generated
from exons or introns (11–15). Two models of circRNA biogenesis have
been proposed, namely lariat splicing and direct back-splicing
(11,12). In lariat splicing, a looped
intermediate containing exons is formed and the introns in the
lariat are removed, generating exonic circRNA. In direct
back-splicing, exons are spliced in non-canonical order, whereby
the downstream 5′ splice site is spliced to an upstream 3′ splice
site, generating a circular transcript. The principle distinction
between the two models is in the first step of circRNA generation.
In the two models, the canonical spliceosome has been implicated in
the generation of circRNA (13,14).
Additionally, Zhang et al (15) proposed a model of alternative
circularization; the study demonstrated that the alternative
formation of inverted repeated Alu pairs and the competition
between them led to alternative circularization, resulting in
multiple circRNA transcripts produced from a single gene.
Notably, RNA-binding proteins (RBPs) may also
regulate the biogenesis of circRNAs in certain conditions.
Previously, studies have revealed that the RBPs, quaking and
muscleblind (MBL), may serve as factors involved in circRNA
biogenesis (13,16). The RNA-editing enzyme adenosine
deaminase, RNA-specific is able to bind to double-stranded RNA to
antagonize circRNA biogenesis (17).
Conservation
circRNA expression appears to be conserved across
eukaryotic species. Thus far, circRNAs have been detected in
yeasts, protists, plants and numerous animals, ranging from fly to
human, which suggests that circRNAs are an ancient feature of gene
expression that have been conserved over the course of eukaryotic
evolution (2,3,18). To
further support this view, circRNAs exhibit sequence conservation.
A previous study revealed that 457 out of the 2,121 circRNAs in
humans were identified in mice as circular orthologues (2). Another study demonstrated that between
15 and 20% of the circRNAs produced in a mouse brain utilize splice
sites that are orthologous to those used in a pig brain (19). In addition, it has also been estimated
that 23.6% of the circRNAs identified in murine neutrophils are
also expressed in rat neutrophils (20).
Biological functions across eukaryotes
To date, numerous studies have identified the
potential developmental functions of circRNAs in multiple
biological processes, including microRNA (miRNA) sponges,
alternative splicing, and transcriptional or post-transcriptional
gene regulation. The dysregulation of circRNAs leads to abnormal
cellular function and growth defects, and is involved in human
development and disease.
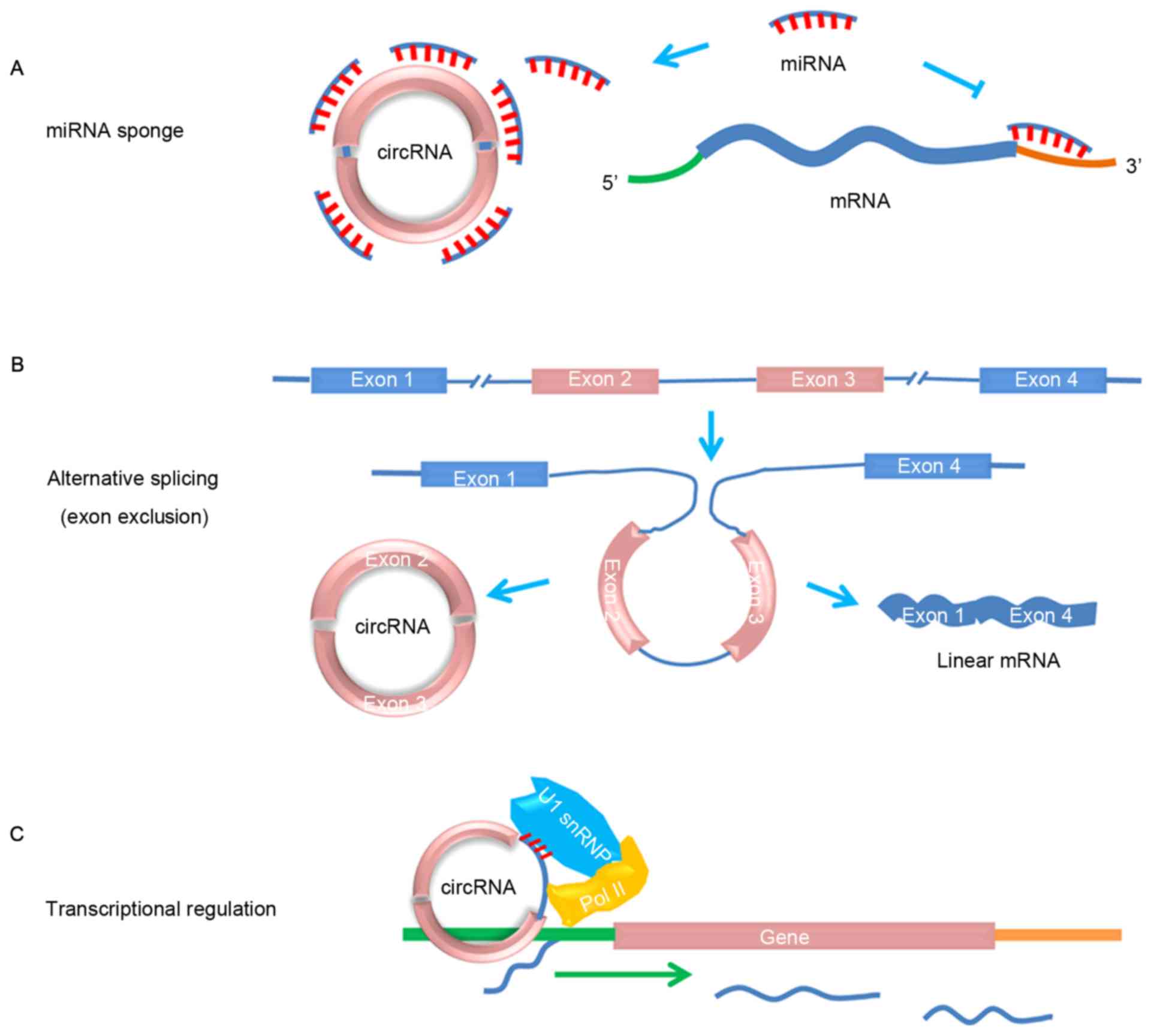
miRNA sponges
Sponge RNAs contain complementary binding sites to
miRNA, thereby serving as competitive inhibitors that suppress the
ability of miRNA to bind to its mRNA targets. Numerous studies have
provided evidence indicating that circRNAs are able to function as
miRNA sponges (Fig. 1A); for example,
cerebellar degeneration-related protein 1 (CDR1)-as, which is
derived from the Cdr1 antisense locus, targets miR-7 as an miRNA
sponge (1,2). The expression of CDR1-as, which contains
>70 miR-7 binding sites, results in increased levels of miR-7
targets. Additionally, murine sex-determining region Y circRNA, a
highly expressed circRNA in the testes, possesses 16 binding sites
for miR-138, thereby acting as a miR-138 sponge (1,21).
Similarly, circ-itchy E3 ubiquitin protein ligase (cir-ITCH)
harbors numerous miRNA binding sites that are able to bind to the
3′-untranslated region of ITCH, and may act as a sponge of
miR-7, miR-17 and miR-214 to increase the level of ITCH
(6).
Alternative splicing
Previous studies (13,22,23)
suggested that circRNAs contribute to alternative splicing
(Fig. 1B); general splicing factor
MBL may exhibit effects on alternative splicing that modulate the
balance between circRNA biogenesis and canonical splicing (13). circMBL and its flanking introns
contain conserved MBL binding sites, and are strongly and
specifically bound by MBL. This suggests that circRNAs are able to
function in gene regulation by competing with canonical pre-mRNA
splicing. Conversely, alternative splicing may also increase
circRNA diversity. Zhang et al (24) systematically annotated different types
and landscapes of alternative back-splicing and alternative
splicing events in circRNAs from various cell lines and provided a
valuable resource for studying the potential functions of circRNAs.
Collectively, the annotation of alternative splicing and circRNA
provides a means to investigate the molecular mechanism of circRNA
biogenesis and potential function in the future.
Transcriptional or
post-transcriptional gene regulation
Current evidence suggests that circRNAs may also
serve a role in transcriptional or post-transcriptional gene
regulation. For example, expression of the CDR1-as circRNA has been
revealed to promote the expression of CDR1-sense mRNA (25). Exon-intron circRNAs were recently
demonstrated to enhance the expression of their parental genes in
cis through interaction with U1 small nuclear
ribonucleoprotein and RNA polymerase II, which are components of
splicing and transcriptional machinery, respectively (Fig. 1C) (26).
The interplay between circRNAs and the transcriptional machinery
assists with the characterization of gene expression, although the
precise molecular mechanism by which this is achieved is unknown
and requires further investigation.
Techniques for investigating circRNA
Since the initial discovery of circRNAs, various
biochemical, molecular biology, high-throughput sequencing and
bioinformatic technologies have been developed to investigate the
properties, biogenesis and functions of circRNA. Among the various
tools to validate circRNAs, ribonuclease R, an exoribonuclease that
progressively degrades RNA from its 3′ to 5′ end, does not degrade
circRNAs, and as such, is often used to assess abundance of, as
well as enrich, circRNAs in combination with reverse
transcription-quantitative polymerase chain reaction, in sequencing
libraries (27). In addition to
exoribonuclease treatment, ribosomal RNA- and
polyadenylation-depletion are also used to enrich and quantify
circRNA expression via modified RNA sequencing approaches. For
in silico analyses, the online databases Circ2Traits
(28), CircBase (29), CircNet (30), deepBase v2.0 (31) and CircInteractome (32) allow researchers to study circRNA
expression profiles and circRNA-associated molecular interactions
that associate with diseases. Furthermore, several circRNA-specific
prediction or detection tools, including UROBORUS (33), find_circ (2) and DCC-CircTest (34), have also been developed.
circRNA in gastrointestinal cancer
CRC
CRC was the third most commonly diagnosed cancer
worldwide in 2014 (35). A study by
Bachmayr-Heyda et al (36)
revealed that the ratio of circular to linear RNA isoforms was
consistently decreased in tumors compared with normal colon
samples, and demonstrated that this ratio correlated negatively
with the proliferation index; to the best of our knowledge, this
was the first negative correlation between global circRNA abundance
and proliferation in CRC to be identified. Specifically, the study
also demonstrated that the expression of Homo sapiens
(hsa)_circ_0006229 and hsa_circ_0007374 was dysregulated in CRC
compared with normal colon mucosa tissues. In addition, Huang et
al (9) identified that
cir-ITCH was significantly downregulated in CRC tissues, and
demonstrated that cir-ITCH functions as an miRNA sponge
serving an inhibitory role in CRC by regulating the Wnt/β-Catenin
pathway. Wang et al (37)
reported that the expression of hsa_circ_001988 was also decreased
in tumor tissues compared with normal mucosa, and that it was
significantly associated with perineural invasion and
differentiation. The aforementioned results suggest that specific
circRNAs may be potential novel biomarkers and targets for the
treatment of patients with CRC. Additionally, another previous
study demonstrated that certain circRNAs are enriched in exosomes
and that by assessing levels of exosomal circRNAs, patients with
CRC were able to be distinguished from healthy controls (38). Therefore, exosome-based circRNAs may
also be potential cancer biomarkers.
GC
GC was the second most common cause of
cancer-associated mortality worldwide in 2012 (39). According to bioinformatic analysis
using two circRNA databases (CircBase and circ2Traits), a series of
circRNAs may be associated with GC (28). In particular, hsa_circ_002059, a
commonly detected circRNA, was identified to be significantly
downregulated in GC tissues compared with paired adjacent non-tumor
tissues (10). This suggests that
certain circRNAs, including hsa_circ_002059, may be potential novel
and stable biomarkers for the diagnosis of gastric carcinoma.
Pancreatic cancer
Pancreatic ductal adenocarcinoma (PDAC) was the
fourth-leading cause of cancer-associated mortality worldwide in
2014, despite accounting for only 2.2% of all types of cancer
(40). Qu et al (41) used Arraystar Human Circular RNA
Microarray, which is able to test for circRNA gene expression, to
explore the expression profile of circRNAs in 4 PDAC samples
alongside paired adjacent normal tissues. The study revealed that
the circRNA expression signatures of PDAC were dysregulated; these
results indicate that circRNAs may be involved in the initiation
and progression of PDAC, and may serve a role as novel diagnostic
and treatment strategies for the disease. However, thus far,
specific circRNAs with the potential to serve as biomarkers have
not been identified in pancreatic cancer. Therefore, the
identification of novel differentially expressed circRNAs may be a
crucial step towards an improved understanding of PDAC in future
research and clinical application.
HCC
HCC ranked as the fifth most-common tumor, and the
third-leading cause for cancer-associated mortality, worldwide in
2010 (42). As in other
gastrointestinal cancers, there was a significant difference in the
global circRNA expression profile between HCC and adjacent normal
liver tissue; particularly, Yu et al (43) demonstrated that CDR1-as expression was
upregulated in HCC tissues compared with adjacent non-tumor
tissues. Furthermore, knockdown of CDR1-as suppressed HCC cell
proliferation and invasion, and suppressed the expression of miR-7,
which suggests that CDR1-as may act as an oncogene partly through
targeting miR-7. In addition, the aforementioned studies revealed
that hsa_circ_0005075 (8) and
hsa_circ_0001649 (44) exhibit a
significant difference in expression between HCC and normal
tissues, and identified them as potential HCC biomarkers.
Esophageal cancer
Esophageal cancer is the eighth most common type of
cancer and the sixth most common cause of cancer-associated
mortality worldwide in 2013 (45).
The expression of circRNAs is altered during the development of
esophageal squamous cell carcinoma (ESCC). One study revealed that
cir-ITCH expression was usually decreased in ESCC compared
with that in the peritumoral tissue (46). Li et al (6) suggested that cir-ITCH may exhibit
an inhibitory effect on ESCC by regulating the Wnt pathway. Studies
such as these further support the role of cir-ITCH as a
candidate for novel strategies for RNA-based esophageal cancer
diagnosis and therapy. Notably, a unique set of circRNAs and their
expression profiles were observed in radioresistant esophageal
cancer cells (47). The aberrant
expression of circRNAs may serve a role in the generation of
radiation-resistant esophageal cancer cells.
Conclusions and outlook
The studies explored in the present review
demonstrate that circRNAs are able to regulate various biological
processes. In particular, circRNAs may serve roles in
gastrointestinal tumor occurrence and development (Table I). These findings support the
hypothesis that circRNAs may be potential novel biomarkers for the
diagnosis of gastrointestinal cancer, and may be a tractable target
in the treatment of such cancer types.
 | Table I.Identified circRNAs associated with
gastrointestinal cancer. |
Table I.
Identified circRNAs associated with
gastrointestinal cancer.
| circRNA | Type of cancer | Expression | Potential
roles | (Refs.) |
|---|
|
cir-ITCH | Colorectal,
esophageal | ↓ | miRNA sponge,
oncogene, biomarker | (9) |
|
hsa_circ_001988 | Colorectal | ↓ | Biomarker,
invasion | (37) |
|
hsa_circ_0006229 | Colorectal | ↓ | Proliferation,
biomarker | (36) |
|
hsa_circ_0007374 | Colorectal | ↑ | Proliferation,
biomarker | (36) |
|
hsa_circ_002059 | Gastric | ↓ | Biomarker | (10) |
| CDR1-as | Hepatocellular | ↑ | miRNA sponge,
proliferation, invasion, oncogene | (43) |
|
hsa_circ_0005075 | Hepatocellular | ↑ | Biomarker | (8) |
|
hsa_circ_0001649 | Hepatocellular | ↓ | Metastasis,
biomarker | (44) |
Although the number of circRNAs with known functions
is expanding, the functions of thousands of circRNAs remain
unknown. Further studies are required to screen novel
differentially expressed circRNAs and confirm their roles in the
development of gastrointestinal cancer. More importantly, studying
the specific molecular mechanisms of the regulation of
gastrointestinal cancer occurrence and development by circRNA may
be a promising research field. If the regulation, mechanism and
function of circRNA are conserved across diseases, results may be
positively validated in, and applied to, other types of cancer and
diseases.
Acknowledgements
The present review was supported by the Research
Special Fund for Public Welfare Industry of Health (grant no.
201402001) and the Research Fund for the Doctoral Program of Higher
Education (grant nos. 20131106110008 and 20121106120048).
References
|
1
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Salzman J, Chen RE, Olsen MN, Wang PL and
Brown PO: Cell-type specific features of circular RNA expression.
PloS Genet. 9:e10037772013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z
and Sharpless NE: Expression of linear and novel circular forms of
an INK4/ARF-associated non-coding RNA correlates with
atherosclerosis risk. PLoS Genet. 6:e10012332010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hansen TB, Kjems J and Damgaard CK:
Circular RNA and miR-7 in cancer. Cancer Res. 73:5609–5612. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li F, Zhang L, Li W, Deng J, Zheng J, An
M, Lu J and Zhou Y: Circular RNA ITCH has inhibitory effect on ESCC
by suppressing the Wnt/β-catenin pathway. Oncotarget. 6:6001–6013.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shang X, Li G, Liu H, Li T, Liu J, Zhao Q
and Wang C: Comprehensive circular RNA profiling reveals that
hsa_circ_0005075, a new circular RNA biomarker, is involved in
hepatocellular crcinoma development. Medicine (Baltimore).
95:e38112016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Huang G, Zhu H, Shi Y, Wu W, Cai H and
Chen X: cir-ITCH plays an inhibitory role in colorectal cancer by
regulating the Wnt/beta-catenin pathway. PLoS One. 10:e01312252015.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li P, Chen S, Chen H, Mo X, Li T, Shao Y,
Xiao B and Guo J: Using circular RNA as a novel type of biomarker
in the screening of gastric cancer. Clin Chim Acta. 444:132–136.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jeck WR and Sharpless NE: Detecting and
characterizing circular RNAs. Nat Biotechnol. 32:453–461. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ashwal-Fluss R, Meyer M, Pamudurti NR,
Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N and
Kadener S: circRNA biogenesis competes with Pre-mRNA splicing. Mol
Cell. 56:55–66. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Starke S, Jost I, Rossbach O, Schneider T,
Schreiner S, Hung LH and Bindereif A: Exon circularization requires
canonical splice signals. Cell Rep. 10:103–111. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL
and Yang L: Complementary sequence-mediated exon circularization.
Cell. 159:134–147. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Conn SJ, Pillman KA, Toubia J, Conn VM,
Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA and
Goodall GJ: The RNA binding protein quaking regulates formation of
circRNAs. Cell. 160:1125–1134. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ivanov A, Memczak S, Wyler E, Torti F,
Porath HT, Orejuela MR, Piechotta M, Levanon EY, Landthaler M,
Dieterich C, et al: Analysis of intron sequences reveals hallmarks
of circular RNA biogenesis in animals. Cell Rep. 10:170–177. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang PL, Bao Y, Yee MC, Barrett SP, Hogan
GJ, Olsen MN, Dinneny JR, Brown PO and Salzman J: Circular RNA is
expressed across the eukaryotic tree of life. PloS One.
9:e908592014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Venø MT, Hansen TB, Venø ST, Clausen BH,
Grebing M, Finsen B, Holm IE and Kjems J: Spatio-temporal
regulation of circular RNA expression during porcine embryonic
brain development. Genome Biol. 16:2452015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
You X, Vlatkovic I, Babic A, Will T,
Epstein I, Tushev G, Akbalik G, Wang M, Glock C, Quedenau C, et al:
Neural circular RNAs are derived from synaptic genes and regulated
by development and plasticity. Nat Neurosci. 18:603–610. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Capel B, Swain A, Nicolis S, Hacker A,
Walter M, Koopman P, Goodfellow P and Lovell-Badge R: Circular
transcripts of the testis-determining gene Sry in adult mouse
testis. Cell. 73:1019–1030. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Salzman J, Chen RE, Olsen MN, Wang PL and
Brown PO: Cell-type specific features of circular RNA expression.
PLoS Genet. 9:e10037772013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang XO, Dong R, Zhang Y, Zhang JL, Luo
Z, Zhang J, Chen LL and Yang L: Diverse alternative back-splicing
and alternative splicing landscape of circular RNAs. Genome Res.
26:1277–1287. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hansen TB, Wiklund ED, Bramsen JB,
Villadsen SB, Statham AL, Clark SJ and Kjems J: miRNA-dependent
gene silencing involving Ago2-mediated cleavage of a circular
antisense RNA. EMBO J. 30:4414–4422. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li Z, Huang C, Bao C, Chen L, Lin M, Wang
X, Zhong G, Yu B, Hu W, Dai L, et al: Exon-intron circular RNAs
regulate transcription in the nucleus. Nat Struct Mol Biol.
22:256–264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Suzuki H, Zuo Y, Wang J, Zhang MQ,
Malhotra A and Mayeda A: Characterization of RNase R-digested
cellular RNA source that consists of lariat and circular RNAs from
pre-mRNA splicing. Nucleic Acids Res. 34:e632006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ghosal S, Das S, Sen R, Basak P and
Chakrabarti J: Circ2Traits: A comprehensive database for circular
RNA potentially associated with disease and traits. Front Genet.
4:2832013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Glazar P, Papavasileiou P and Rajewsky N:
circBase: A database for circular RNAs. RNA. 20:1666–1670. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu YC, Li JR, Sun CH, Andrews E, Chao RF,
Lin FM, Weng SL, Hsu SD, Huang CC, Cheng C, et al: CircNet: A
database of circular RNAs derived from transcriptome sequencing
data. Nucleic Acids Res. 44:D209–D215. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zheng LL, Li JH, Wu J, Sun WJ, Liu S, Wang
ZL, Zhou H, Yang JH and Qu LH: deepBase v2.0: Identification,
expression, evolution and function of small RNAs, LncRNAs and
circular RNAs from deep-sequencing data. Nucleic Acids Res.
44:D196–D202. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Dudekula DB, Panda AC, Grammatikakis I, De
S, Abdelmohsen K and Gorospe M: CircInteractome: A web tool for
exploring circular RNAs and their interacting proteins and
microRNAs. RNA Biol. 13:34–42. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Song X, Zhang N, Han P, Moon BS, Lai RK,
Wang K and Lu W: Circular RNA profile in gliomas revealed by
identification tool UROBORUS. Nucleic Acids Res. 44:e872016.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cheng J, Metge F and Dieterich C: Specific
identification and quantification of circular RNAs from sequencing
data. Bioinformatics. 32:1094–1096. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Stewart BW and Wild CP: World Cancer
Report 2014World Health Organization. International Agency for
Research on Cancer; Lyon: 2014
|
|
36
|
Bachmayr-Heyda A, Reiner AT, Auer K,
Sukhbaatar N, Aust S, Bachleitner-Hofmann T, Mesteri I, Grunt TW,
Zeillinger R and Pils D: Correlation of circular RNA abundance with
proliferation-exemplified with colorectal and ovarian cancer,
idiopathic lung fibrosis, and normal human tissues. Sci Rep.
5:80572015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang X, Zhang Y, Huang L, Zhang J, Pan F,
Li B, Yan Y, Jia B, Liu H, Li S, et al: Decreased expression of
hsa_circ_001988 in colorectal cancer and its clinical
significances. Int J Clin Exp Pathol. 8:16020–16025.
2015.PubMed/NCBI
|
|
38
|
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J,
Chen D, Gu J, He X and Huang S: Circular RNA is enriched and stable
in exosomes: A promising biomarker for cancer diagnosis. Cell Res.
25:981–984. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Jones OP, Melling JD and Ghaneh P:
Adjuvant therapy in pancreatic cancer. World J Gastroenterol.
20:14733–14746. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Qu S, Song W, Yang X, Wang J, Zhang R,
Zhang Z, Zhang H and Li H: Microarray expression profile of
circular RNAs in human pancreatic ductal adenocarcinoma. Genom
Data. 5:385–387. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yu L, Gong X, Sun L, Yao H, Lu B and Zhu
L: miR-454 functions as an oncogene by inhibiting CHD5 in
hepatocellular carcinoma. Oncotarget. 6:39225–39234. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yu L, Gong X, Sun L, Zhou Q, Lu B and Zhu
L: The circular RNA Cdr1as act as an oncogene in hepatocellular
carcinoma through targeting miR-7 expression. PLoS One.
11:e01583472016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Qin M, Liu G, Huo X, Tao X, Sun X, Ge Z,
Yang J, Fan J, Liu L and Qin W: Hsa_circ_0001649: A circular RNA
and potential novel biomarker for hepatocellular carcinoma. Cancer
Biomark. 16:161–169. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Sakai NS, Samia-Aly E, Barbera M and
Fitzgerald RC: A review of the current understanding and clinical
utility of miRNAs in esophageal cancer. Semin Cancer Biol.
23:512–521. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wei W, Li M, Wang J, Nie F and Li L: The
E3 ubiquitin ligase ITCH negatively regulates canonical Wnt
signaling by targeting dishevelled protein. Mol Cell Biol.
32:3903–3912. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Su H, Lin F, Deng X, Shen L, Fang Y, Fei
Z, Zhao L, Zhang X, Pan H, Xie D, et al: Profiling and
bioinformatics analyses reveal differential circular RNA expression
in radioresistant esophageal cancer cells. J Transl Med.
14:2252016. View Article : Google Scholar : PubMed/NCBI
|