Introduction
MicroRNAs (miRNAs) are a class of endogenous
non-coding small RNAs molecules that trigger transcriptional or
post-transcriptional regulation by binding to the 3′-untranslated
region (UTR) of target genes (1–3). In recent
studies, miRNAs were revealed to function as either oncogenes or
tumor suppressor genes in biological processes, including cell
proliferation, migration, apoptosis and differentiation and in
tumorigenesis of a number of types of human cancer (3–7). In
females worldwide, ovarian cancer is a common gynecological
malignancy and remains a major problem, with high mortality due to
diagnosis at an advanced stage and distant metastases (8,9). Various
miRNAs are post-transcriptionally deregulated in ovarian cancer
(10–13). Although evidence has supported miRNAs
as a potential target in cancer diagnosis, therapy and prognosis,
the role of miRNAs in ovarian cancer remains unclear. miR-124, a
tumor suppressor miRNA, is significantly downregulated in a number
of human malignant tumors (14–17),
including hepatocellular carcinoma and gastric carcinoma; however,
the possible effect of miR-124 in ovarian cancer has not been fully
explored.
Programmed cell death 6 (PDCD6) is a calcium-binding
protein that regulates cell proliferation and death and belongs to
the penta-EF-hand protein family (18,19).
Previous studies have indicated that PDCD6 is critical in T-cell
receptor-, Fas- and glucocorticoid-induced programmed cell death
(19,20). Furthermore, PDCD6 was demonstrated to
be a pro-apoptotic protein involved in cell viability functions
(19,21); however, its role in regulating
migration and invasion in ovarian cancer is unclear.
The present study aimed to determine whether miR-124
and PDCD6 are involved in the apoptosis, migration and invasion of
ovarian cancer. It was revealed that miR-124 was downregulated in
ovarian cancer cell lines and revealed an association between
miR-124 expression level and ovarian cancer cell metastasis. The
present study further identified PDCD6 as a novel, direct target of
miR-124 that suppressed apoptosis in ovarian cancer progression,
and miR-124 rescue experiments restored PDCD6 expression level. The
results of the present study demonstrated that miR-124 suppressed
cell motility, migration and invasion and induced cell apoptosis by
downregulating PDCD6. Thus, miR-124 may provide a novel target for
effective treatment of ovarian cancer.
Materials and methods
Patients and tissue samples
Human ovarian tumor tissues were collected from the
30 patients who had ovarian epithelial carcinoma and non-neoplastic
ovarian tissues from 30 healthy individuals under 60 years of age
who received surgical resection between January 2009 and December
2012 at the Department of Gynecologic Oncology at Chongqing Cancer
Hospital (Chongqing Cancer Institute, Chongqing, China). The
present study was approved by the Ethics Committee of the Chongqing
Cancer Hospital (Chongqing Cancer Institute, Chongqing, China).
Written informed consent was provided by all patients prior to
enrollment in the present study.
Cell lines and cell culture
SKOV3 and OVCAR3 human ovarian cells were purchased
from the Cell Bank of the Chinese Academy of Sciences (Shanghai,
China). All cells were cultured in RPMI-1640 medium (Gibco; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10%
fetal bovine serum (FBS; Hyclone, Logan, UT, USA) in a humidified
atmosphere of 5% CO2 at 37°C.
Transient transfection of miR-124
The miR-124 mimics and the miR-124 mimics scramble
were synthesized by Shanghai GeneChem Co., Ltd. (Shanghai, China).
The sequences (5′-3′) were as follows: hsa-miR-124-5p mimics
forward, 5′-CGUGUUCACAGCGGACCUUGAU-3′ and reverse,
5′-AUCAAGGUCCGCUGUGAACACG-3′; miR-124 mimics scramble forward,
5′-UUCUCCGAACGUGUCAGU-3′ and reverse, 5′-ACGUGACACGUUCGGAGAA-3′.
The pcDNA3.1/PDCD6 and pcDNA3.1/control were synthesized by
Guangzhou RiboBio Co., Ltd. (Guangzhou, China). Cells were
harvested and plated in six-well plates at a density of
5×105 cells/ml. All cell transfections were performed
using Lipofectamine 2000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's instructions. In
the rescue experiment, cells were co-transfected with miR-124
mimics and plasmids.
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted using TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) from clinical
tumor samples and transfected cells according to the manufacturer's
instructions. RNA concentration and quality were evaluated and 0.5
µg RNA was used for RT. MMLV reverse transcriptase (Takara Bio.,
Inc., Otsu, Japan) was used to synthesize complementary DNA (cDNA).
For synthesis of miR-124 cDNA, a miR-124 RT primer was used and U6
small nuclear RNA was used as the internal control. cDNA for other
target genes was amplified using the primer oligo(dT) with
β-tubulin as the internal control. Target genes and controls were
analyzed by RT-qPCR using SYBR Premix Ex Taq (Promega Corporation,
Madison, WI, USA). The following primers were used for qPCR:
miR-124 forward, 5′-TGCGGTAAGGCACGCGGTG-3′ and reverse,
5′-CCAGTGCAGGGTCCGAGGT-3′; U6 forward, 5′-TGCGGGTGCTCGCTTCGGCAGC-3′
and reverse, 5′-CCAGTGCAGGGTCCGAGGT-3′; PDCD6 forward,
5′-TCCAGAGGGTCGATAAAGACA-3′ and reverse, 5′-TTCTGCCAGTCCGTGATGT-3′;
β-tubulin forward, 5′-TTGGCCAGATCTTTAGACCAGACAAC-3′; and reverse,
5′-CCGTACCACATCCAGGACAGAATC−3′. PCR was conducted at 94°C for 4
min, followed by 40 cycles of 94°C for 30 sec, 58°C for 30 sec and
72°C for 30 sec in an iQ5 Real-time PCR system (Bio-Rad
Laboratories, Inc., Hercules, CA, USA). All the samples were
assessed by relative quantification (2−ΔΔCq method)
(22).
Target prediction
The prediction of the PDCD6 3′-UTR as a miR-124
binding target was determined using TargetScan (www.targetscan.org) and PicTar (www.pictar.org).
Dual-luciferase reporter assay
A luciferase reporter assay was used to monitor the
direct interaction between the miRNA and its target mRNA. The
binding site of miR-124 in the 3′-UTR of the target mRNA was cloned
into the pmirGLO Dual-Luciferase miRNA Target Expression Vector
(Promega Corporation), according to the manufacturer's
instructions. Cells were subsequently co-transfected with miRNA and
the wild type or mutant 3′-UTR for the luciferase assay. Following
transfection for 48 h at room temperature, the cells were harvested
for protein extraction. Luciferase intensity was examined using the
Dual Luciferase Reporter Gene Assay kit (Beyotime Institute of
Biotechnology, Haimen, China), according the manufacturer's
protocol. Renilla luciferase intensity was used an internal
control.
Cell migration and invasion
assays
Cell migration was evaluated using a wound-healing
assay. At 24 h after transfection, a wound was created by 20 µl
pipette tips. in all culture wells. Cultures were imaged at 0, 24
and 48 h to assess cell migration. Light microscopy was undertaken
at ×100 magnification at 0, 24 and 48 h to assess cell
migration.
The present study used a Transwell assay with a
Matrigel-coated membrane matrix (BD Biosciences, Franklin Lakes,
NJ, USA) to assess cell invasion. After transfection, cells
(5×104) were seeded in the upper chamber of a Millicell
Transwell chamber (EMD Millipore, Billerica, MA, USA) with 200 µl
RPMI-1640 medium without FBS. Cells that migrated through the
membrane were fixed with 4% methanol for 10 min at room
temperature. and stained with 0.1% crystal violet for 15 min at
room temperature. Cells were then imaged at ×100 magnification and
counted using an inverted microscope.
Cell survival assay
SKOV3 and OVCAR3 cells were plated onto 96-well
plates 48 h after transfection at a density of 5×104
cells/ml. A total of three replicate wells were examined for each
group. When cells reached 60–70% confluency, 10 µl 5 mg/ml MTT was
added to each well. After 4 h, the culture medium was discarded,
100 µl dimethyl sulfoxide was added to each well and the plates
were agitated at room temperature for 1 h. Absorbance (optical
density) was evaluated using a microplate reader at a wavelength of
490 nm. This experiment was repeated three times.
Western blot analysis
At the indicated time, SKOV3 and OVCAR3 whole-cell
lysates were extracted using radioimmunoprecipitation assay buffer
and incubated for 30 min on ice. Cell lysates were prepared from
cells seeded on 6-well plates at 106 cells per well
using radioimmunoprecipitation assay buffer (10 mM Tris-HCl, pH
7.4, 1% Triton X-100, 0.1% SDS, 1% NP- 40, 1 mM MgCl2) containing
protease inhibitors at 48 h post-transfection.
Proteins were quantified using the BCA method,
according to the manufacturer's instructions (Thermo Fisher
Scientific, Inc.). Proteins (10 µg) were separated by 10% SDS-PAGE
and then transferred to polyvinylidene difluoride membranes (EMD
Millipore). The membranes were blocked in 5% non-fat milk/TBST for
1 h at room temperature. Thereafter, they were incubated with
rabbit monoclonal anti-PDCD6 antibody (1:1,000; ab133326) and
anti-β-tubulin antibody (1:1,000; ab6046) (Abcam, Cambridge, UK)
overnight at 4°C. After adding peroxidase-conjugated goat
anti-rabbit antibody (1:5,000; BA1055; Boster Biological
Technology, Pleasanton, CA, USA) for 1 h at room temperature, the
membranes were visualized by Western Lightning®-ECL,
Enhanced Chemiluminescence Substrate (PerkinElmer, Inc., Waltham,
MA, USA, and X-ray film was applied to analyze the image and
intensity of band.
Cell cycle and apoptosis assays
At 48 h after transfection, 1×106 cells
were seeded onto 75-mm dishes. To analyze the cell cycle stage, the
cells were harvested, by trypsin and washed twice with cold PBS,
fixed in ice-cold 70% ethanol and stained with 100 µg/ml propidium
iodide (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) supplemented
with 1 mg/ml RNase A for 30 min at room temperature in the dark.
Subsequently, the cell cycle stages were analyzed using a flow
cytometer and Cell Quest software version 3.3 (BD Biosciences). To
analyze apoptosis, the cells were harvested from the culture dishes
by trypsinization. After washing twice with cold PBS, cells were
resuspended in 200 µl binding buffer and incubated with 10 µl
Annexin V-R-PE and 10 µl 7-ADD (SouthernBiotech, Birmingham, AL,
USA) in the dark at 4°C for 30 min. Thereafter, 380 µl binding
buffer was added to each tube and the samples were analyzed by flow
cytometry.
Statistical analysis
All data were presented as the mean ± standard
deviation. Error bars represent standard error. One-way analysis of
variance, followed by Fisher's least significance test or
two-tailed Student's t-test were used to analyzed the statistical
significance. P<0.05 was considered to indicate a statistically
significant difference.
Results
miR-124 is downregulated in ovarian
cancer
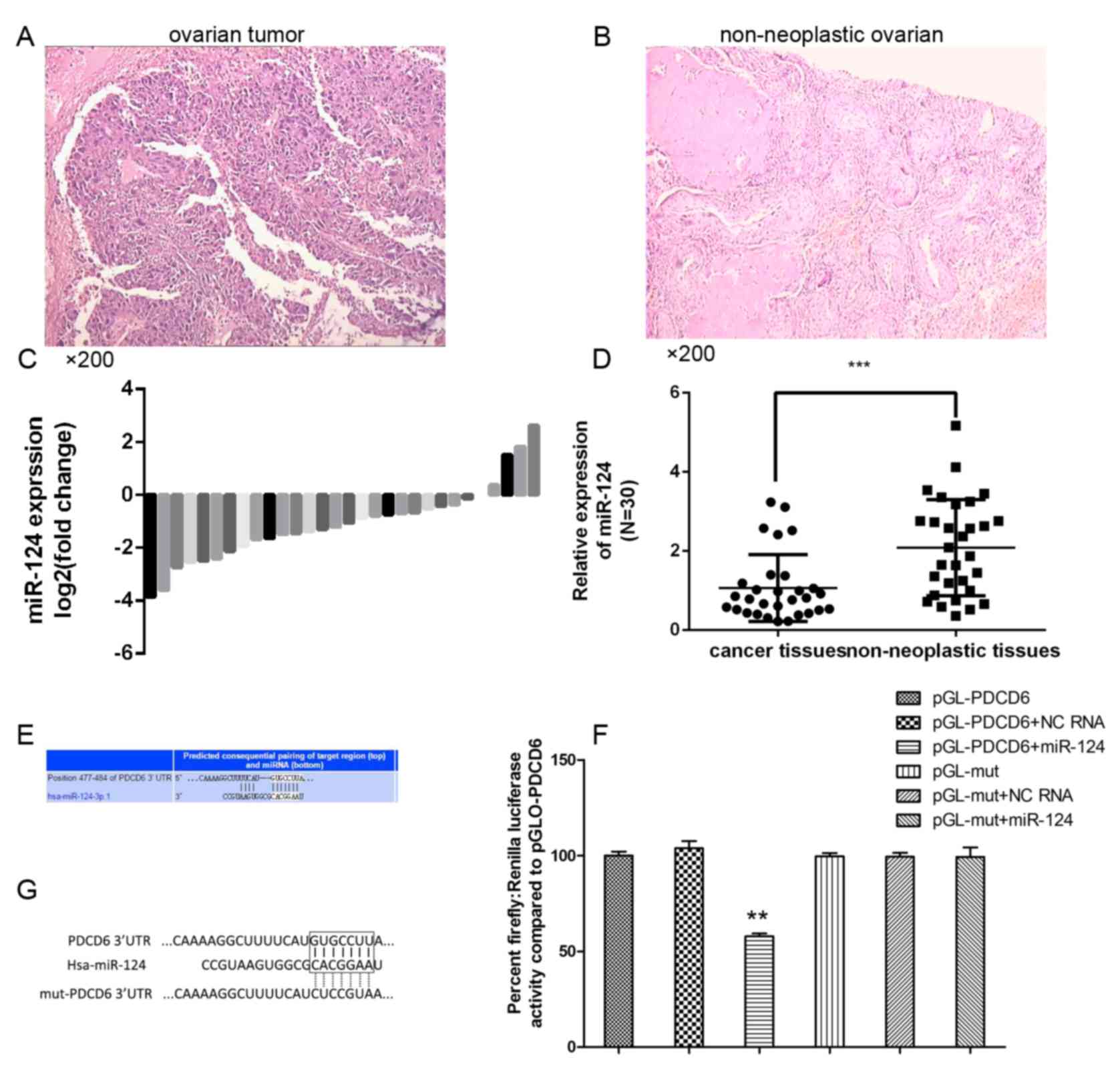
To analyze the expression level of miR-124 in
ovarian cancer progression, RT-qPCR was performed using TaqMan
probes. Ovarian cancer tissue and normal ovarian tissue have
distinct pathological patterns (Fig.
1). The present study compared the expression levels of 30
clinical tumor tissue samples and 30 non-neoplastic ovarian tissues
(Fig. 1C) and revealed that the
miR-124 expression level was significantly decreased in the
majority of ovarian cancer tissues compared with in the
non-neoplastic control tissues (Fig.
1D).
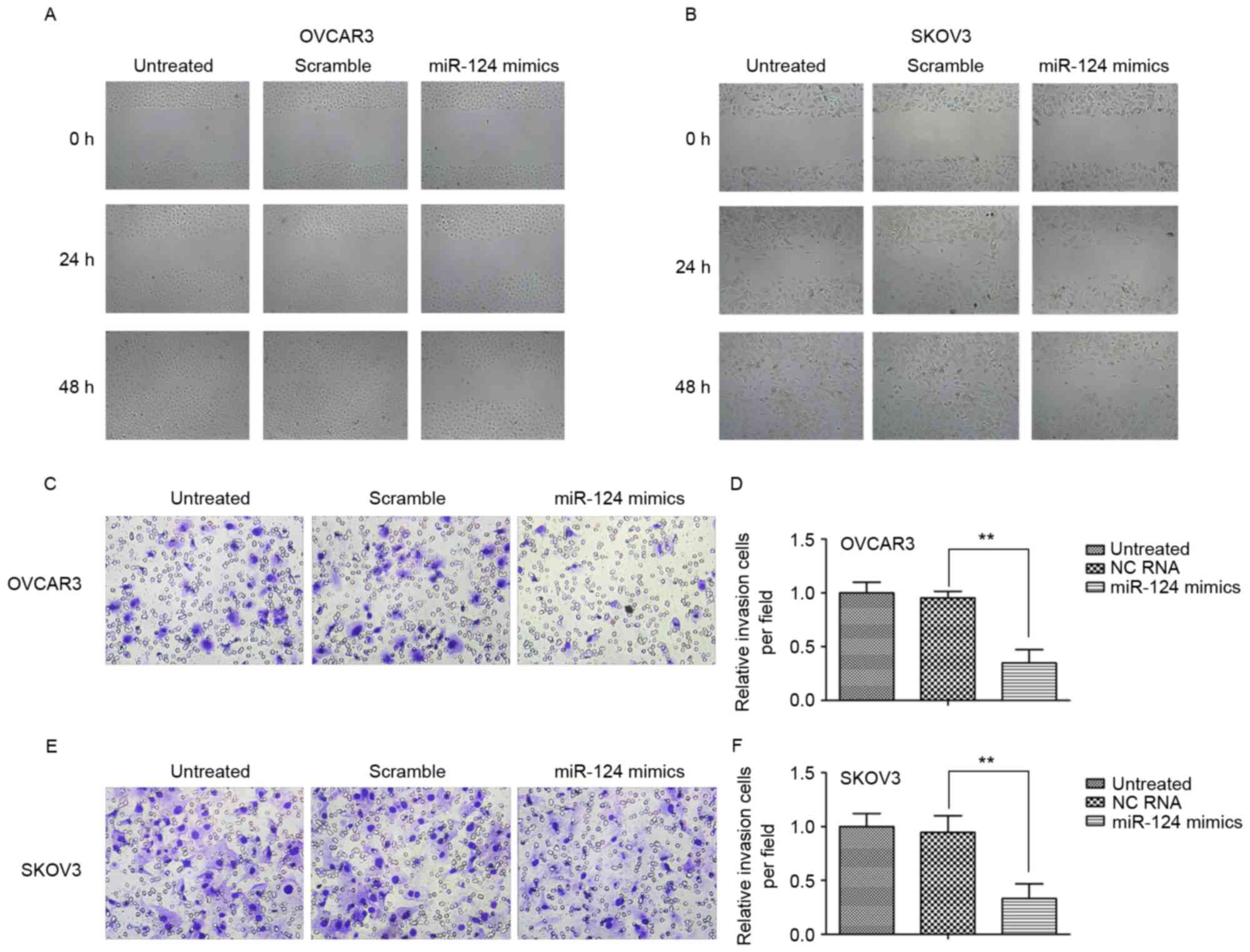
miR-124 suppresses the migration and
invasion of ovarian cancer cell lines
To investigate the biological role of miR-124 in
ovarian cancer, the present study observed its effect on the
migration and invasion of the ovarian cancer SKOV3 and OCVAR3 cell
lines by transient transfection with miR-124 mimics, miR-124 mimics
scramble or negative control (NC). The wound-healing assay
indicated that transfection with miR-124 mimics significantly
inhibited migration in both cell lines relative to the scramble
control or NC at 24 and 48 h (Fig. 2A and
B). Furthermore, ectopic expression of miR-124 markedly
decreased invasion of the cell lines compared with the scramble
control or NC (Fig. 2C-F).
PDCD6 is a direct target of miR-124 in
ovarian cancer
To investigate the potential targets of miR-124, the
present study performed bioinformatics analysis using TargetScan
and Pictar (Fig. 1E), which predicted
that miR-124 targets the PDCD6 3′-UTR region. To determine whether
the 3′-UTR of PDCD6 mRNA is a direct target of miR-124 in ovarian
cancer cells, the wild-type (wt) full-length 3′-UTR of PDCD6 or a
mutant (mt) sequence was cloned into a luciferase reporter vector
(Fig. 1G). Cells were co-transfected
with wt 3′-UTR or mt 3′-UTR vectors and miR-124 mimics. The results
indicated that overexpression of miR-124 significantly decreased
the luciferase activity of reporter genes with wt 3′-UTR compared
with the controls. The luciferase activity for the mt 3′-UTR vector
was not affected by transfection with miR-124 (Fig. 1F).
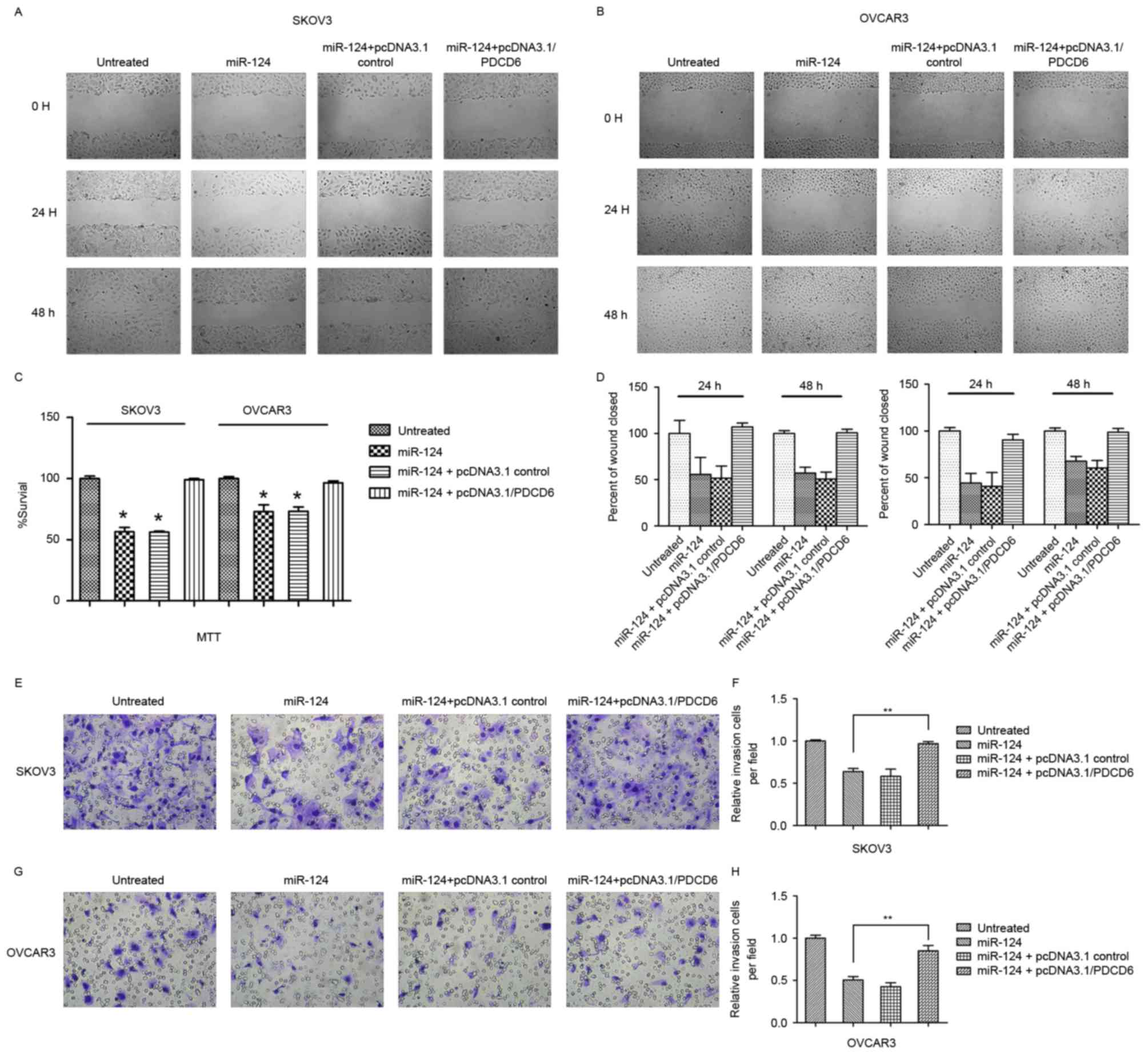
Expression of PDCD6 reversed the
miR-124-induced suppression of cellular migration and invasion, and
induction of cellular apoptosis
Transfection with miR-124 mimics or pcDNA3.1/vector
and miR-124 mimics significantly decreased ovarian cancer cell
proliferation and migration compared with the controls (Fig. 3). Conversely, co-transfection with
miR-124 mimics or pcDNA3.1/PDCD6 in SKOV3 or OCVAR3 cells induced
recovery of cell migration and invasion. To investigate whether
miR-124 affects cancer cell migration and invasion through PDCD6,
the present study performed rescue experiments by upregulating
PDCD6 expression in SKOV3 and OCVAR3 cells using pcDNA3.1/PDCD6.
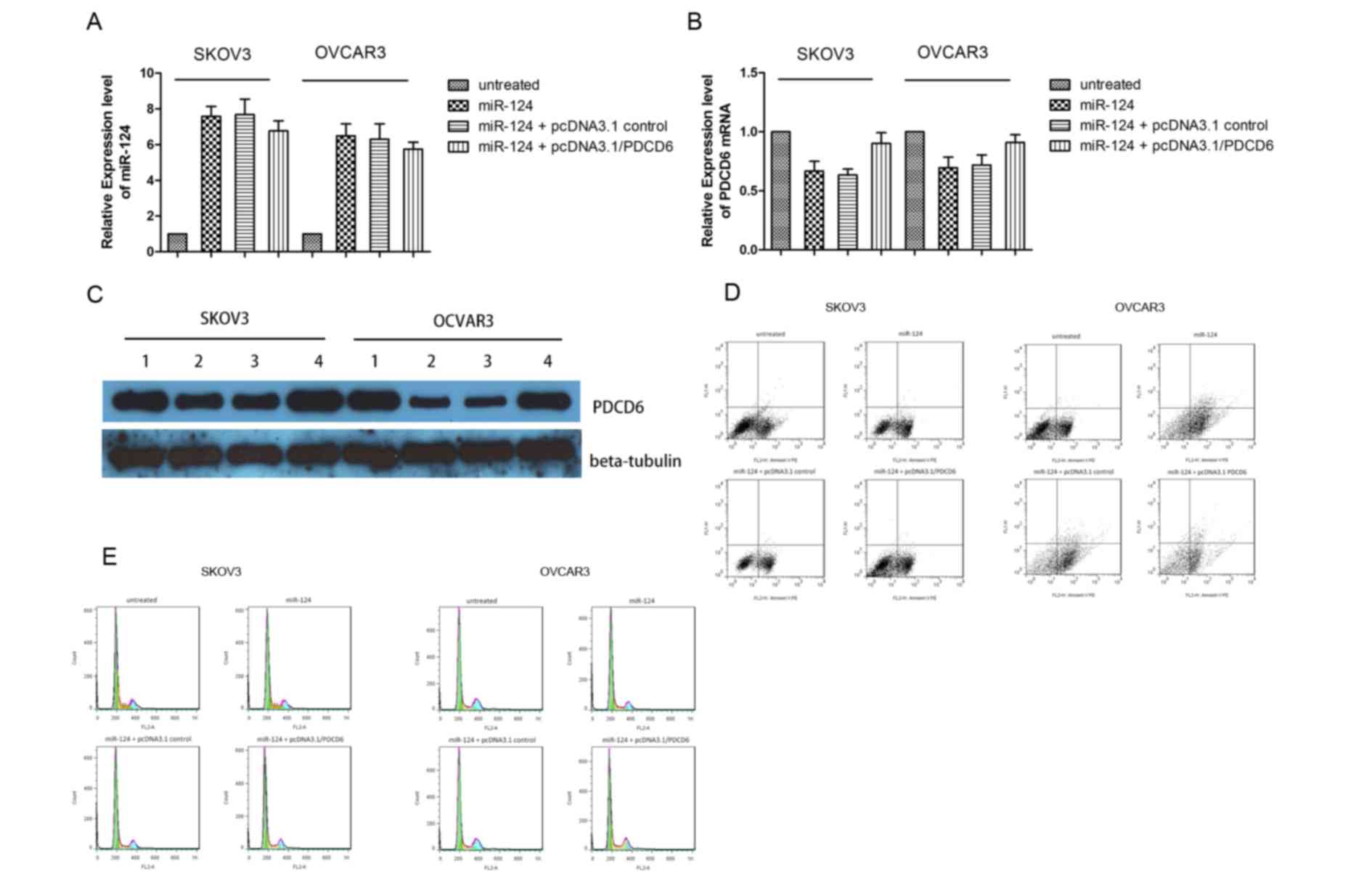
The expression level of PDCD6 mRNA was evaluated by qPCR (Fig. 4A and B). The expression level of
miR-124 significantly increased subsequent to transient
transfection with miR-124 mimics; however, the expression level of
PDCD6 mRNA markedly decreased, thus miR-124 downregulated PDCD6
mRNA expression level. Transient co-transfection of miR-124 mimics
together and pcDNA3.1/PDCD6 rescued the expression level of PDCD6
mRNA. Similar results were obtained by western blot analysis of
SKOV3 and OCVAR3 cells co-transfected with miR-124 mimics or NC and
pcDNA3.1/PDCD6 or pcDNA3.1/vector (Fig.
4C). The incidence of apoptosis and cell cycle arrest following
overexpression of miR-124 and overexpression of PDCD6 were
confirmed by flow cytometry. The results demonstrated that
transfection of miR-124 mimics markedly increased cell death
(Fig. 4D) and the proportion of SKOV3
and OVCAR3 cells in the G0/G1 cell cycle
phase, but decreased the proportion of cells in S phase (Fig. 4E). However, these effects were
reversed by co-transfection of miR-124 mimics and
pcDNA3.1/PDCD6.
Discussion
Currently, the majority of patients with ovarian
cancer are diagnosed with distant metastases at an advanced stage,
which challenges effective treatment (9,11,12,23).
Therefore, more research into potential therapeutic strategies for
ovarian cancer is required. Previous studies revealed that miRNAs
may regulate migration and invasion, acting as tumor suppressors or
oncogenes in human cancer types (16,17,24,25).
Further investigation of the roles of miRNAs in cancer development
is therefore important for understanding the molecular mechanisms
underlying cancer metastasis. Accumulating evidence has suggested
miR-124 may function as a tumor suppressor in ovarian cancer
(26). The present study revealed
that miR-124 was distinctly downregulated in clinical ovarian
cancer tissues samples compared with non-neoplastic ovarian tissues
from healthy individuals. Furthermore, miR-124 expression level was
lower in ovarian cancer lines, exhibiting greater migration and
invasion abilities, suggesting that it may be important in ovarian
cancer carcinogenesis. Thus, miR-124 appears to be gradually
downregulated during ovarian cancer progression, and its
upregulation may decrease proliferation and metastasis of ovarian
cancer. However, the mechanism underlying miR-124 regulation in
ovarian cancer has not been fully elucidated.
PDCD6 is a calcium binding protein in the
penta-EF-hand family, and is involved in T-cell receptor-, Fas- and
glucocorticoid-induced apoptosis (20,27–30). PDCD6
was identified in rat liver hepatoma and human lung cancer tissues,
suggesting that it may be involved in survival pathways, in
addition to its pro-apoptotic function (19). Although numerous studies have
investigated the association between PDCD6 expression level and
disease risk, the results are controversial (19,21,31–34).
A previous study indicated that PDCD6 was involved in T-cell
receptor-induced apoptosis as a pro-apoptotic factor and that Alix
and PDCD6 interaction with pro-caspase-8 significantly induced cell
death via tumor necrosis factor receptor 1 (21). However, another study demonstrated
that PDCD6 expression was upregulated in 7,371 tumor tissue samples
from lung, breast or colon cancer, which indicated that PDCD6 may
be involved in maintenance of cellular viability (19,35,36).
Similarly, PDCD6 has been suggested to have a role in cancer cell
pathology and contribute to cancer cell viability.
In the present study, PDCD6 was identified as a
direct target of miR-124 via luciferase assay and bioinformatics
analysis. The present study observed that miR-124 targeted PDCD6 to
function as a tumor suppressor in ovarian cancer cells.
Furthermore, PDCD6 upregulation rescued the effects of miR-124
overexpression in ovarian cancer lines. These results indicated
that the effects of miR-124 on cell proliferation, cell cycle
progression, apoptosis, migration and invasion of ovarian cancer
cells is partly mediated by inhibiting the expression of PDCD6.
In future studies, the upstream or downstream
modulators of PDCD6 and the effect of miR-124 on signaling pathways
of tumor progression by targeting PDCD6 will be investigated. In
addition, as chemotherapy resistance is one of the main obstacles
in treatment of ovarian cancer, the regulation and function of
miR-124 and PDCD6 in chemotherapy resistance in ovarian cancer will
continue to be investigated.
In conclusion, the present study provides evidence
that the aberrant expression of miR-124 is important in cancer cell
migration and invasion. Furthermore, miR-124 induced apoptosis in
ovarian cancer cell by regulating PDCD6, a novel and direct target
of miR-124. The upregulation of miR-124 markedly inhibited ovarian
cancer cell growth, invasion and migration. These results provided
a foundation for future studies exploring a new approach to
suppressing cancer by targeting miR-124 and suggested that it may
provide a novel therapeutic target for ovarian cancer.
References
|
1
|
Farazi TA, Hoell JI, Morozov P and Tuschl
T: MicroRNAs in human cancer. Adv Exp Med Biol. 774:1–20. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Iorio MV and Croce CM: MicroRNAs in
cancer: Small molecules with a huge impact. J Clin Oncol.
27:5848–5856. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Iorio MV, Visone R, Di Leva G, Donati V,
Petrocca F, Casalini P, Taccioli C, Volinia S, Liu CG, Alder H, et
al: MicroRNA signatures in human ovarian cancer. Cancer Res.
67:8699–8707. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Abba M, Mudduluru G and Allgayer H:
MicroRNAs in cancer: Small molecules, big chances. Anticancer
Agents Med Chem. 12:733–743. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gandellini P, Giovannetti E and Nicassio
F: MicroRNAs in cancer management: Big challenges for small
molecules. Biomed Res Int. 2015:9821562015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Calin GA, Sevignani C, Dumitru CD, Hyslop
T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M
and Croce CM: Human microRNA genes are frequently located at
fragile sites and genomic regions involved in cancers. Proc Natl
Acad Sci USA. 101:pp. 2999–3004. 2004, View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kim K, Zang R, Choi SC, Ryu SY and Kim JW:
Current status of gynecological cancer in China. J Gynecol Oncol.
20:72–76. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lees B and Leath CA III: The impact of
diabetes on gynecologic cancer: Current status and future
directions. Curr Obstet Gynecol Rep. 4:234–239. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Pal MK, Jaiswar SP, Dwivedi VN, Tripathi
AK, Dwivedi A and Sankhwar P: MicroRNA: A new and promising
potential biomarker for diagnosis and prognosis of ovarian cancer.
Cancer Biol Med. 12:328–341. 2015.PubMed/NCBI
|
|
11
|
Langhe R: microRNA and ovarian cancer. Adv
Exp Med Biol. 889:119–151. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Maalouf SW, Liu WS and Pate JL: MicroRNA
in ovarian function. Cell Tissue Res. 363:7–18. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zaman MS, Maher DM, Khan S, Jaggi M and
Chauhan SC: Current status and implications of microRNAs in ovarian
cancer diagnosis and therapy. J Ovarian Res. 5:442012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang Y, Zheng L, Huang J, Gao F, Lin X,
He L, Li D, Li Z, Ding Y and Chen L: MiR-124 Radiosensitizes human
colorectal cancer cells by targeting PRRX1. PLoS One. 9:e939172014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang W, Mao YQ, Wang H, Yin WJ, Zhu SX
and Wang WC: MiR-124 suppresses cell motility and adhesion by
targeting talin 1 in prostate cancer cells. Cancer Cell Int.
15:492015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lu Y, Yue X, Cui Y, Zhang J and Wang K:
MicroRNA-124 suppresses growth of human hepatocellular carcinoma by
targeting STAT3. Biochem Biophys Res Commun. 441:873–879. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zheng YB, Xiao GC, Tong SL, Ding Y, Wang
QS, Li SB and Hao ZN: Paeoniflorin inhibits human gastric carcinoma
cell proliferation through up-regulation of microRNA-124 and
suppression of PI3K/Akt and STAT3 signaling. World J Gastroenterol.
21:7197–7207. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Vito P, Lacanà E and D'Adamio L:
Interfering with apoptosis: Ca(2+)-binding protein ALG-2 and
Alzheimer's disease gene ALG-3. Science. 271:521–525. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
la Cour JM, Høj BR, Mollerup J, Simon R,
Sauter G and Berchtold MW: The apoptosis linked gene ALG-2 is
dysregulated in tumors of various origin and contributes to cancer
cell viability. Mol Oncol. 1:431–439. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
D'Adamio L, Lacanà E and Vito P:
Functional cloning of genes involved in T-cell receptor-induced
programmed cell death. Semin Immunol. 9:17–23. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mahul-Mellier AL, Strappazzon F, Petiot A,
Chatellard-Causse C, Torch S, Blot B, Freeman K, Kuhn L, Garin J,
Verna JM, et al: Alix and ALG-2 are involved in tumor necrosis
factor receptor 1-induced cell death. J Biol Chem. 283:34954–34965.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Giulietti A, Overbergh L, Valckx D,
Decallonne B, Bouillon R and Mathieu C: An overview of real-time
quantitative PCR: Applications to quantify cytokine gene
expression. Methods. 25:386–401. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Dwivedi SK, Mustafi SB, Mangala LS, Jiang
D, Pradeep S, Rodriguez-Aguayo C, Ling H, Ivan C, Mukherjee P,
Calin GA, et al: Therapeutic evaluation of microRNA-15a and
microRNA-16 in ovarian cancer. Oncotarget. 7:15093–15104. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cheng CJ, Bahal R, Babar IA, Pincus Z,
Barrera F, Liu C, Svoronos A, Braddock DT, Glazer PM, Engelman DM,
et al: MicroRNA silencing for cancer therapy targeted to the tumour
microenvironment. Nature. 518:107–110. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Maltby S, Plank M, Tay HL, Collison A and
Foster PS: Targeting MicroRNA function in respiratory diseases:
Mini-review. Front Physiol. 7:212016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang H, Wang Q, Zhao Q and Di W: MiR-124
inhibits the migration and invasion of ovarian cancer cells by
targeting SphK1. J Ovarian Res. 6:842013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
la Cour JM, Mollerup J, Winding P,
Tarabykina S, Sehested M and Berchtold MW: Up-regulation of ALG-2
in hepatomas and lung cancer tissue. Am J Pathol. 163:81–89. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Su D, Xu H, Feng J, Gao Y, Gu L, Ying L,
Katsaros D, Yu H, Xu S and Qi M: PDCD6 is an independent predictor
of progression free survival in epithelial ovarian cancer. J Transl
Med. 10:312012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jung YS, Kim KS, Kim KD, Lim JS, Kim JW
and Kim E: Apoptosis-linked gene 2 binds to the death domain of Fas
and dissociates from Fas during Fas-mediated apoptosis in Jurkat
cells. Biochem Biophys Res Commun. 288:420–426. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhou B, Bai P, Xue H, Zhang Z, Shi S,
Zhang K, Wang Y, Wang K, Quan Y, Song Y and Zhang L: Single
nucleotide polymorphisms in PDCD6 gene are associated with the
development of cervical squamous cell carcinoma. Fam Cancer.
14:1–8. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Jang IK, Hu R, Lacaná E, D'Adamio L and Gu
H: Apoptosis-linked gene 2-deficient mice exhibit normal T-cell
development and function. Mol Cell Biol. 22:4094–4100. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Aviel-Ronen S, Coe BP, Lau SK, da Cunha
Santos G, Zhu CQ, Strumpf D, Jurisica I, Lam WL and Tsao MS:
Genomic markers for malignant progression in pulmonary
adenocarcinoma with bronchioloalveolar features. Proc Natl Acad Sci
USA. 105:pp. 10155–10160. 2008, View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Mahul-Mellier AL, Hemming FJ, Blot B,
Fraboulet S and Sadoul R: Alix, making a link between
apoptosis-linked gene-2, the endosomal sorting complexes required
for transport, and neuronal death in vivo. J Neurosci. 26:542–549.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Suzuki K, Dashzeveg N, Lu ZG, Taira N,
Miki Y and Yoshida K: Programmed cell death 6, a novel
p53-responsive gene, targets to the nucleus in the apoptotic
response to DNA damage. Cancer Sci. 103:1788–1794. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Høj BR, la Cour JM, Mollerup J and
Berchtold MW: ALG-2 knockdown in HeLa cells results in G2/M cell
cycle phase accumulation and cell death. Biochem Biophys Res
Commun. 378:145–148. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
la Cour JM, Mollerup J, Winding P,
Tarabykina S, Sehested M and Berchtold MW: Up-regulation of ALG-2
cancer tissue in hepatomas and lung. Am J Pathol. 163:81–89. 2003.
View Article : Google Scholar : PubMed/NCBI
|