Introduction
Glioblastoma multiforme (GBM; World Health
Organization grade IV glioma) is the most common type of primary
brain tumor worldwide (1). In spite
of radical surgery combined with concomitant chemoradiation therapy
based on temozolomide, the median survival rate of patients with
GBM remains ~1 year (2). Furthermore,
clinical trials have demonstrated only limited benefits of targeted
regimens, indicating that the identification of the genetic and
molecular characteristics of GBM is required to develop more
effective treatment strategies (2–4).
Genetic studies including a large number of patients
with GBM have revealed various genetic alterations in GBM,
including isocitrate dehydrogenase gene mutations and mutations in
the promoter region of telomerase reverse transcriptase gene
(TERT) (4,5). Of these, TERT promoter mutations
have been identified in a subpopulation of GBM and were revealed to
be significantly associated with poor clinical prognosis (2,5,6). In addition, these mutations were
associated with increased expression of TERT (7,8). These
results indicated the potential for personalized therapy against
TERT in GBM on the basis of mutation status.
In vitro and in vivo preclinical
models derived from surgical samples of GBMs have revealed the
molecular and functional features of the parental tumors, and may
represent the GBM population experimentally (9). Therefore, patient-derived preclinical
models exhibiting GBMs with or without TERT promoter
mutations may enable experimental examination of personalized
TERT-targeted treatments. In the present study, a patient-derived
GBM preclinical model library, including GBMs with and without
TERT promoter mutations, was established, and preclinical
and clinical implications were determined.
Materials and methods
GBM patients, primary cell culture and
stereotactic transplantation
Surgical specimens and clinical records were
obtained from 25 patients with primary GBM from May 2004 to June
2006 at the Samsung Medical Center (Seoul, Korea). All tissue
samples were collected with written informed consent under a
protocol approved by the Institutional Review Board of the Samsung
Medical Center (Seoul, Korea). GBMs were pathologically diagnosed
by specialized neuropathologists, on the basis of the World Health
Organization criteria (10). For
genomic analysis, parts of the specimens were snap-frozen and
preserved in liquid nitrogen until use. Genomic DNA and mRNA were
extracted using the DNeasy Kit (Qiagen GmbH, Hilden, Germany) and
the RNeasy kit (Qiagen GmbH, Hilden, Germany).
Sections of the surgical samples were enzymatically
dissociated into single cells as previously described (11). Dissociated GBM cells were cultured in
neurobasal media (Gibco; Thermo Fisher Scientific, Inc., Waltham,
MA, USA) containing 1X N2 and 2X B27 supplements (Invitrogen;
Thermo Fisher Scientific, Inc.), human recombinant basic fibroblast
growth factor and epidermal growth factor (25 ng/ml each; R&D
Systems, Inc., Minneapolis, MN, USA) (12). Because clonogenic growth as
neurospheres is an in vitro indicator of the self-renewal of
GBM cells, sphere formation (diameter, ≥50 µm), within 4 weeks, was
used to identify the in vitro sphere formation capacity of
dissociated GBM cells. For neurosphere formation, cells were seeded
at a range of 1–200 cells per well. Following 4 weeks, the number
of wells without spheres were counted and analyzed.
Dissociated GBM cells were stereotactically (2 mm
left and 1 mm anterior to the bregma, 2 mm deep from the dura)
transplanted into the brains of immuno-deficient NOG mice (between
2.5×104 and 1.0×105 cells/10 µl Hank's
Balanced Salt solution for each mice, n=4–9 for each sample)
(13). For the intracranial
injection, 250 6–8 week-old female immune-deficient NOG mice (4–9
mice per group) were obtained from Orient Bio, Inc. (Seongnam,
Korea). Mice were housed under controlled temperature (22±2°C) and
a 12 h light/dark cycle in laminar flow cabinets with filtered air,
and handled using aseptic procedures and food and water were
provided ad libitum. The average weight of the mice prior to the
procedure was 20 g. Mice were sacrificed and their brains were
processed for pathological diagnosis, following the procedures
previous reported (9). Animal
experiments were approved by the Institutional Review Boards of the
Samsung Medical Center and conducted in accordance with the
National Institutes of Health Guide for the Care and Use of
Laboratory Animals.
TERT promoter mutation analysis
Genomic DNA was isolated from tumor samples a using
QIAamp DNA mini kit (Qiagen GmbH). The TERT promoter region
was amplified from isolated genomic DNA using the polymerase chain
reaction (PCR), as described previously (14). PCR was performed using AmpliTaq Gold
DNA polymerase (Applied Biosystems; Thermo Fisher Scientific,
Inc.). PCR products were electrophoresed on 1.5% agarose gel and
stained with ethidium bromide to validate the sizes of the bands.
Subsequently, direct DNA sequencing for the TERT promoter
region was performed using an ABI 3730 DNA sequencer by Bionics
Co., Ltd. (Seoul, Korea). The sequences for TERT were as
follows: Sense, 5′-CACCCGTCCTGCCCCTTCACCTT-3′ and antisense,
5′-GGCTTCCCACGTGCGCAGCAGGA-3′.
Relative telomere length
determination
Telomere length was analyzed using quantitative PCR
(qPCR) (7990HT Fast Real-time, Applied Biosystems; Thermo Fisher
Scientific, Inc.). For telomere length determination, DNA were
extracted from cells. To quantitatively determine telomere length
relative to nuclear DNA, specific primers for telomere (T) and
nuclear DNA-encoded β-globin (S) were selected according to a
previous study (14). The β-globin
gene was used as the reference gene. qPCR was performed using a
LightCycler 480 II system (Roche Diagnostics, Basel, Switzerland)
with SYBR PCR master mix (Toyobo Life Science, Osaka, Japan). For
telomere PCR, there were 18 cycles at 95°C for 15 sec and at 54°C
for 2 min. Relative telomere length was determined by calculating
T/S values using the formula T/S=2−∆Cq, where
∆Cq=average Cqtelomere-average Cqβ-globin.
Each measurement was repeated in triplicate and 5 serially diluted
control samples were included in each experiment. The primer
sequences used in this experiment were follows: β-globin sense,
5′-TGTGCTGGCCCATCACTTTG-3; β-globin antisense,
5′-ACCAGCCACCACTTTCTGATAGG-3′; telomere sense,
5′-CGGTTTGTTTGGGTTTGGGTTTGGGTTTGGGTTTGGGTT-3′; and telomere
antisense, 5′-GGCTTGCCTTACCCTTACCCTTACCCTTACCCTTACCCT-3′.
GBM subtype prediction
According to The Cancer Genome Atlas data of the
mRNA expression of 840 genes in 173 patients with GBM, the subtypes
(proneural, neural, classical and mesenchymal) of the 25 GBMs used
in the present study were determined (15). The Cancer Genome Atlas data were
downloaded from tcga-data.nci.nih.gov/docs/publications/gbm_exp. The
Nearest Template Prediction algorithm was used to predict the class
of a sample with statistical significance (false discovery rate,
<0.2) using a predefined set of markers specific to multiple
classes (GenePattern Modules; version 2; Broad Institute) (15). For the dataset in the present study,
the 788 overlapping genes, out of the 840 genes, were used to
predict the subtype.
Genetic alteration analysis
Microarray-based comparative genomic hybridization
(aCGH) was performed using the Agilent SurePrint G3 Human CGH
4×180K arrays (Agilent Technologies, Inc., Santa Clara, CA, USA).
aCGH feature extraction files were processed and normalized fold
changed of matched normal samples using Agilent Genomic WorkBench
7.0.4.0 (Agilent Technologies, Inc.). The DNAcopy R package
(version 1.48.0; performed in Bioconductor) was used to estimate
DNA copy numbers (16). From the copy
numbers at the segment level, the copy number for each gene was
measured using the mean value of the copy numbers of all exonic
segments of the gene (9).
Somatic mutations were detected using the Agilent
SureSelect kit (Agilent Technologies, Inc.) to capture exonic DNA
fragments. The Illumina HiSeq 2000 was used to generate
2×101 bp paired-end reads (Illumina, Inc., San Diego,
CA, USA). The sequenced reads in FASTQ files were mapped to the
human reference genome assembly (hg19), using the Burrows-Wheeler
Aligner (version 0.6.2) (17). The
initial alignment BAM files were subjected to regular preprocessing
prior to mutation calling: Sorting reads by genomic coordinates,
removing duplicated reads, locally realigning reads around
potential small indels and recalibrating base quality scores using
SAMtools (18), Picard (version 1.73;
Broad Institute), and Genome Analysis ToolKit (GATK version 2.5.2;
Broad Institute). For mutation calling, MuTect (GATK version 1.1.4;
Broad Institute) and SomaticIndelDetector (GATK version 2.2; Broad
Institute) were used to make high-confidence predictions regarding
somatic mutations from the tumor and paired blood. Copy number data
were obtained using the ngCGH python package (version 0.4.4) to
generate aCGH-like data from whole exome sequencing (WES) data. The
matched blood WES data were used as a reference to calculate
fold-changes in copy numbers in tumors. In cases without matched
blood WES, created ‘pseudo-normal’ profile blood WES data were
generated using the same sequencing platform and analysis pipeline
as were used for the tumor data. Downstream analysis (segmentation
and calculation of copy number) were conducted as described for
aCGH data.
Using the Illumina TruSeq RNA Sample Prep kit
(Illumina, Inc.), RNA-seq libraries were prepared for all cases.
The trimmed reads in FASTQ files were aligned with hg19 using GSNAP
(version 2012-12-20) with two output formats: GSNAP native format
(exon-skipping analysis) and SAM format (point mutation analysis)
(19). The resulting GSNAP native
format files were analyzed to isolate the ‘split’ reads spanning
non-canonical splicing junctions, with a minimal anchor of five
nucleotides on each exon. In cases demonstrating plural reading
splits between two exons, the event was termed a skipped exon event
between the two exons. The SAM format files were sorted using the
same preprocessing procedures as those applied for the WES data,
with the exception of local realignments were restricted to exonic
regions to prevent the mislabeling of normal splicing events as
misaligned indels. Potential point mutations were identified using
UnifiedGenotyper (GATK version 1.2.0; Broad Institute).
Statistical analysis
The SPSS statistical package, version 19.0, was used
for statistical analyses (IBM Corp., Armonk, NY, USA).
χ2, Fischer's exact tests, Mann Whitney U tests and
Spearman's correlation analysis were used to analyze the
associations between variables. Survival curves, estimated using
the Kaplan-Meier method (univariate analysis), were compared using
the log-rank test. Overall survival was defined as the time between
diagnosis and mortality (as a result of any cause).
Progression-free survival was defined as the time between diagnosis
and disease recurrence. P<0.05 was considered to indicate a
statistically significant difference. Data are presented as the
mean ± standard deviation.
Results
TERT promoter mutation status of GBMs
is associated with sphere formation capacity
First, TERT promoter mutations were
investigated in the 13 GBMs with in vitro sphere formation
capacity to establish patient-derived GBM preclinical model
libraries, including GBMs with and without TERT promoter
mutations. In parallel, in vivo tumor formation, gene
mutation status and global gene expression were analyzed. Notably,
TERT promoter mutations were identified in 92.3% (12/13)
GBMs (Table IA). All TERT mutations
were revealed to be C228T, but 1 GBM exhibited C228T and C250T
mutations. It was revealed that 1 GBM without TERT promoter
mutation exhibited α-thalassemia/mental retardation syndrome
X-linked (ATRX) amplification, although it was previously
demonstrated that ATRX amplification and TERT
promoter mutation were mutually exclusive (6). On the basis of this, it was hypothesized
that the genetic alteration in ATRX is equivalent to
TERT promoter mutation (6).
 | Table I.Association between TERT
promoter mutation and other gene mutation status in GBMs. |
Table I.
Association between TERT
promoter mutation and other gene mutation status in GBMs.
| A, Sphere
formation-positive GBMs |
|---|
|
|---|
|
|
Sample |
|---|
|
|
|
|---|
| Gene mutation
status | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 |
|---|
| TERT
mutation | + | + | + | + | + | + | + | + |
| + | + | + | + |
|
C228T | + | + | + | + | + | + | + | + |
| + | + | + | + |
|
C250T |
|
|
|
|
| + |
|
|
|
|
|
|
|
| Sphere
formation | + | + | + | + | + | + | + | + | + | + | + | + | + |
| ATRX
amplification |
|
|
|
|
| ND |
|
| + | ND |
|
|
|
| ATRX
mutation | + | + |
|
| + |
| + | + | + | ND |
|
| + |
| EGFR
mutation |
| + |
| + |
|
|
| + |
| ND | + | + | + |
| EGFR
amplification | + | + |
| + | + | ND |
|
| + | ND | + | + | + |
| IDH1
mutation |
|
|
|
|
|
|
|
|
| ND |
|
|
|
| TP53
mutation | + |
|
|
| + |
| + | + |
| ND |
|
|
|
| PTEN
deletion | + |
| + | + | + | ND | + |
| + | ND | + | + | + |
| PTEN
mutation |
|
|
|
| + |
| + |
|
| ND |
|
|
|
| CDKN2A
deletion | + | ND | + | + | + | ND | + | + | + | ND | + | + | + |
|
| B, Sphere
formation-negative GBMs |
|
| Sample |
|
|
|
| Gene mutation
status | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
|
| TERT
mutation |
| + |
|
|
|
|
|
|
| + | + | + |
|
|
C228T |
| + |
|
|
|
|
|
|
| + | + | + |
|
|
C250T |
|
|
|
|
|
|
|
|
|
|
|
|
|
| Sphere
formation |
|
|
|
|
|
|
|
|
|
|
|
|
|
| ATRX
amplification |
|
|
|
|
|
|
|
|
|
|
|
|
|
| ATRX
mutation | ND | ND | ND | ND | ND | ND |
|
| ND | ND |
| + |
|
| EGFR
mutation | ND | ND | ND | ND | ND | ND |
|
| ND | ND | + |
|
|
| EGFR
amplification | + | + |
|
|
|
| + |
|
|
| + | + |
|
| IDH1
mutation | ND | ND | ND | ND | ND | ND |
|
| ND | ND |
|
|
|
| TP53
mutation | ND | ND | ND | ND | ND | ND |
|
| ND | ND |
| + |
|
| PTEN
deletion |
|
|
|
|
| + | + |
|
|
| + | + |
|
| PTEN
mutation | ND | ND | ND | ND | ND | ND |
| + |
|
|
| + |
|
| CDKN2A
deletion | + | + |
|
|
| + | + |
| + |
| + | + |
|
The TERT promoter mutation rate (92.3%) in
GBMs with in vitro sphere formation capacity was not
expected because TERT promoter mutations were observed in
between 28–84% of GBMs (20).
Accordingly, the present study hypothesized that TERT
promoter mutation is associated with the in vitro sphere
formation capacity of GBMs. To test the hypothesis, TERT
promoter mutations in GBMs without in vitro sphere formation
capacity were subsequently analyzed.
TERT promoter mutations are decreased
in GMBs without sphere formation capacity
TERT promoter mutations were identified in
33.3% (4/12) GBMs without in vitro sphere formation capacity
(Table IB). All mutations were C228T
(Table IB). This frequency was
significantly decreased compared with that in GBMs with in
vitro sphere formation capacity (P=0.004). Other preclinical
characteristics and genetic changes were not associated with
TERT promoter mutation in GBMs without in vitro
sphere formation capacity (data not shown).
TERT promoter mutation is associated
with age and sex
Out of 25 GBMs (Table
II), TERT promoter mutation was demonstrated in 64.0%
(16/25). TERT promoter mutation was significantly associated
with increased age (P=0.050) and sex; being more prevalent in male
patients (P<0.001). Notably, GBMs with in vivo
tumorigenic potential demonstrated a significantly increased
TERT promoter mutation rate (P=0.004) compared with those
without. Although the values were not statistically significant,
TERT promoter mutations were at an increased frequency in
GBMs with epidermal growth factor receptor (EGFR) gene
mutation (P=0.117), EGFR amplification (P=0.102), cyclin
dependent kinase inhibitor 2A (CDKN2A) deletion (P=0.116)
and phosphatase and tensin homolog deletion (P=0.102).
 | Table II.Clinicopathological and experimental
characteristics of GBMs with or without TERT promoter
mutation. |
Table II.
Clinicopathological and experimental
characteristics of GBMs with or without TERT promoter
mutation.
|
| TERT
promoter mutation, n (%) |
|
|---|
|
|
|
|
|---|
| Variable | + | − | P-value |
|---|
| Age, years | 56.5±8.3 | 47.2±14.6 | 0.050 |
| Sex |
|
| <0.001 |
|
Male | 12 (100) | 0 (0) |
|
|
Female | 4 (30.8) | 9 (69.2) |
|
| Tumor size, cm | 4.70±1.63 | 4.79±0.75 | 0.943 |
| ATRX
mutation |
|
| 1.00 |
| + | 7 (87.5) | 1 (12.5) |
|
| − | 6 (75.0) | 2 (25.0) |
|
| ATRX
amplification |
|
| 0.391 |
| + | 0 (0) | 1 (100) |
|
| − | 14 (63.6) | 8 (36.4) |
|
| EGFR
mutation |
|
| 0.117 |
| + | 11 (91.7) | 1 (8.3) |
|
| − | 4 (57.1) | 3 (42.9) |
|
| EGFR
amplification |
|
| 0.102 |
| + | 10 (76.9) | 3 (23.1) |
|
| − | 4 (40.0) | 6 (60.0) |
|
| CDKN2A
deletion |
|
| 0.116 |
| + | 12 (70.6) | 5 (29.4) |
|
| − | 1 (20.0) | 4 (80.0) |
|
| PTEN mutation |
|
| 1.00 |
| + | 3 (75.0) | 1 (25.0) |
|
| − | 10 (83.3) | 2 (16.7) |
|
| PTEN
deletion |
|
| 0.102 |
| + | 10 (76.9) | 3 (23.1) |
|
| − | 4 (40.0) | 6 (60.0) |
|
| Subtype |
|
| 0.176 |
|
Classical | 6 (100) | 0 (0) |
|
|
Mesenchymal | 2 (40.0) | 3 (60.0) |
|
|
Proneural | 3 (60.0) | 2 (40.0) |
|
| Not
determined | 5 (55.6) | 4 (44.4) |
|
| In vivo
tumor formation |
|
| 0.004 |
| + | 12 (80.0) | 3 (20.0) |
|
| − | 0 (0) | 5 (100) |
|
Telomere length is not associated with
TERT promoter mutation status
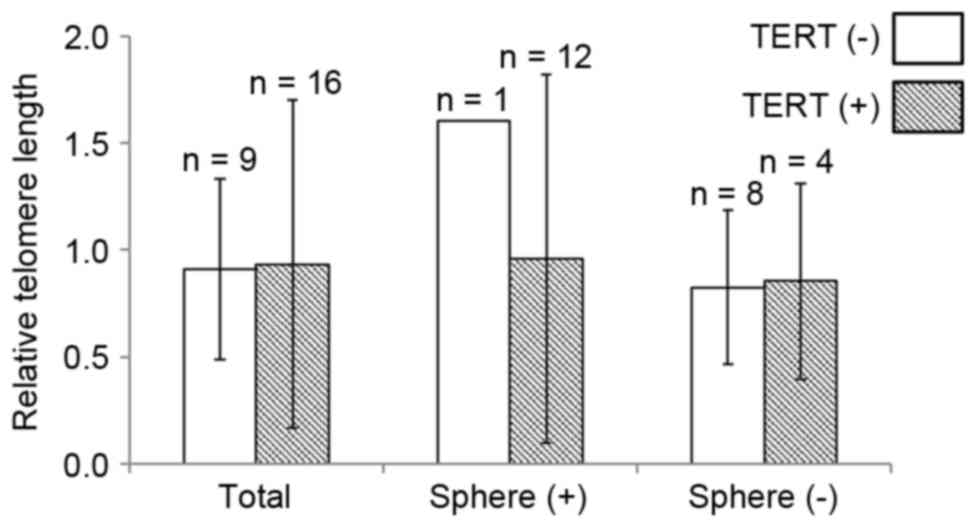
Relative telomere length (0.92±0.49) was analyzed in
the GBMs. However, the length did not differ according to
TERT promoter mutation status (0.91±0.42 vs. 0.93±0.77;
P=0.598; Fig. 1). When the GBMs were
divided into two groups according to in vitro sphere
formation capacity, TERT promoter mutation revealed limited
association with the relative telomere length (Fig. 1).
To explore the association between telomere length
and clinicopathological parameters, GBMs were divided into two
groups according to the median value (0.92) of relative telomere
length. In the analysis, telomere length was not associated with
any clinicopathological characteristics or molecular changes (data
not shown).
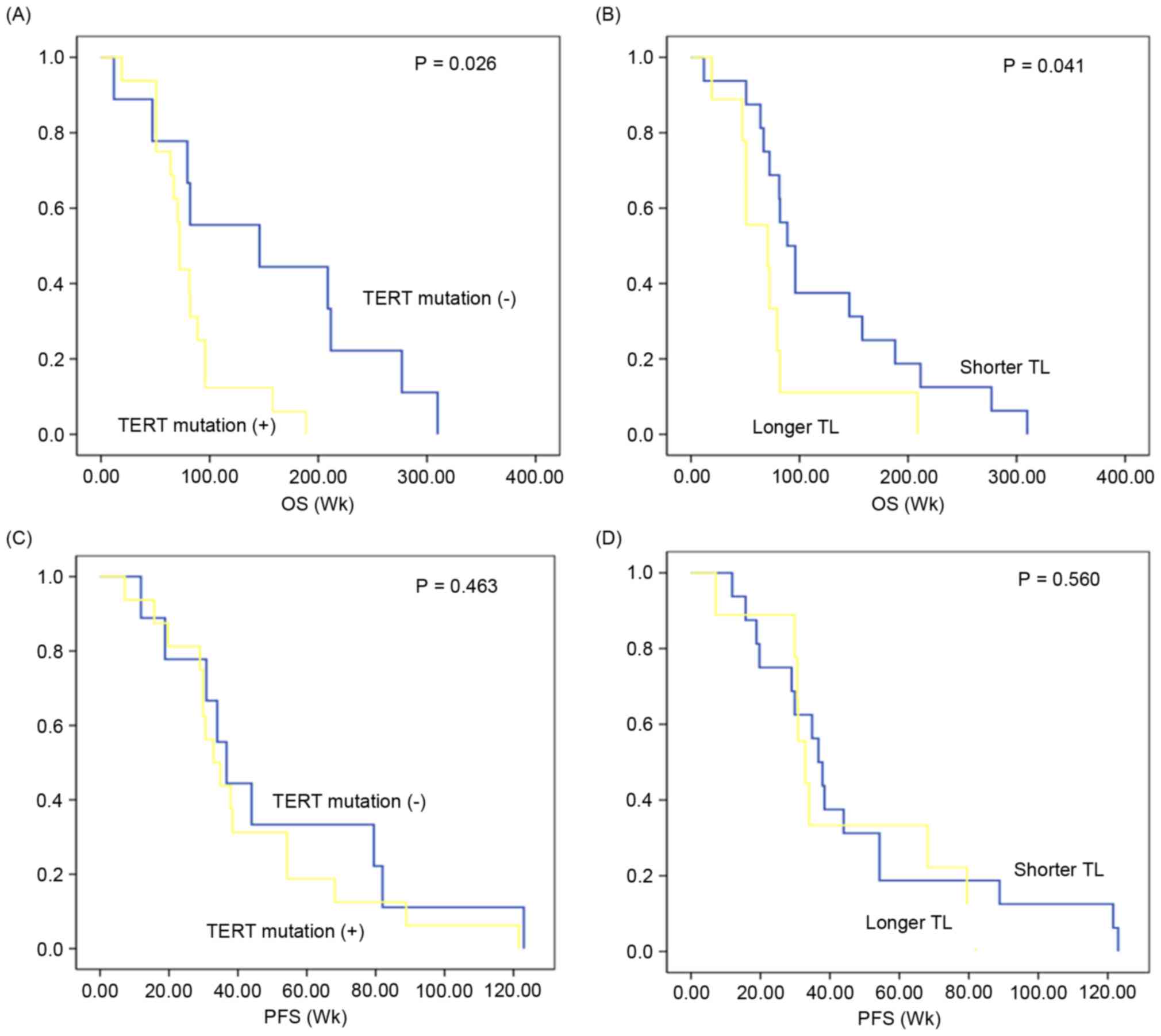
Positive TERT promoter status is
associated with poor survival
The clinical prognosis of TERT promoter
mutation-positive GBMs (n=16) was compared with that of TERT
promoter mutation-negative GBMs (n=9). The median overall survival
in GBMs exhibiting TERT promoter mutation was 81.7 [95%
confidence interval (CI), 61.71–101.85] weeks, which was
significantly decreased compared with that in GBMs without
TERT promoter mutation (median, 152.6 weeks; 95% CI,
84.05–221.16; P=0.026; Fig. 2A).
According to the median value of telomere length (0.92), GBMs were
stratified into longer and shorter groups to analyze the prognostic
value of telomere length. Overall survival in patients with GBM
with a longer telomere length (median, 75.70 weeks; 95% CI,
40.65–110.75) was reduced compared with those with shorter telomere
length (median, 125.04 weeks; 95% CI, 84.02–166.05; P=0.041;
Fig. 2B). In contrast,
progression-free survival did not differ according to TERT
promoter mutation (median, 51.19 vs. 43.32; 95% CI, 27.56–74.82 vs.
29.09–57.54; P=0.463; Fig. 2C) and
telomere length (median, 47.43 vs. 43.88; 30.47–64.40 vs.
26.88–60.88; P=0.560; Fig. 2D]. When
survival analysis was performed separately according to other
variables [in vitro sphere formation capacity, in
vivo tumor formation, age (<60 vs. ≥60), sex and subtype],
there was no significant prognostic difference in overall survival
or progression-free survival (data not shown).
Discussion
The present study revealed that TERT promoter
mutations in GBMs are significantly associated with the in
vitro sphere formation capacity and in vivo tumorigenic
potential of dissociated GBM cells. In vitro and in
vivo preclinical models using GBM cells primarily cultured from
surgical samples have provided an improved understanding of the
biology of the disease (9). Several
preclinical characteristics of primary cultured GBM cells,
including in vitro sphere formation capacity and in
vivo tumorigenic potential, were revealed to be associated with
clinical aggressiveness in corresponding patients (9). The associations may be utilized to
determine molecular and/or functional mechanisms of clinical
aggressiveness of GBMs.
In the present study, in vitro and in
vivo preclinical GBM models exhibiting TERT promoter
mutation-positive and -negative GBMs were established. As
preclinical models may summarize the clinicopathological features
of patient with GBMs (9,21–23), the
preclinical models may be utilized to predict the treatment effects
of TERT-targeting therapies for TERT promoter
mutation-positive GBMs, compared with those for TERT
promoter mutation-negative GBMs. In the present study, the majority
of GBMs with in vitro sphere formation capacity exhibited
TERT promoter mutations (92.3%). By contrast, the
TERT promoter mutation rate in GBMs without in vitro
sphere formation capacity (33.3%) was significantly decreased. This
significant difference was observed between GBMs with and without
in vivo tumorigenic potential.
The in vitro sphere-forming assay has been
widely used in stem cell biology as an experimental method for
determining the self-renewal and differentiation potential of stem
cells (9,11). Therefore, a significant association
between TERT promoter mutation and the in vitro
sphere-forming capacity of GBM cells, identified in the present
study, suggests that mutations in the TERT promoter region
are associated with the biology of GBM cells to enhance
self-renewal capacity. Self-renewal capacity is a key feature of
GBM cancer stem cells that exert recurrence following anti-cancer
treatments (24,25). Therefore, TERT promoter
mutations resulting in overexpression of TERT may be
associated with treatment resistance of GBMs. In addition, the
possible association between TERT promoter mutations and
treatment resistance of GBMs is supported by the survival analysis
results in the present study, which revealed that GBMs with
TERT promoter mutation have significantly decreased overall
survival.
Previous studies have demonstrated the negative
clinical impacts of TERT promoter mutation or longer
telomere length in a number of types of cancer, including GBMs
(15,20,21,25,26).
TERT promoter mutations may generate novel binding motifs
for E26 transformation-specific/T-cell factor transcription
factors, and cause two- to four-fold increases in transcriptional
activity and telomere length (14,26,27). These
data suggested that overexpression of TERT by its promoter
mutation may increase the self-renewal capacity of GBM cancer stem
cells and induce poor clinical outcomes. The results of the present
study did not reveal a significant association between TERT
promoter mutation and relative telomere length in the GBM samples.
Previous studies demonstrated that the association between telomere
length and TERT expression is complex and may be regulated
by a number of other factors, including the activities of signaling
pathways and alterations of genes (4,28,29).
Previous studies have demonstrated that TERT
promoter is the most common type of mutation in GBMs, suggesting
that it may be an early event in GBM carcinogenesis (2,4,5,30–32). In the present study, TERT
promoter mutations were identified in 64% of GBMs, and were
associated with the age and sex of patients with GBM. These results
are consistent with those of previous studies (4,5). The
association between TERT promoter mutations and other
genetic alterations has been observed in previous studies (4,5). These
studies suggested that TERT promoter mutations revealed a
significant inverse association with isocitrate dehydrogenase 1
mutation and P53 mutation, but a positive association with
EGFR amplification and CDKN2 deletion. Though the
results of the present study were not statistically significant due
to the limited sample size of primary GBMs, TERT promoter
mutations tend to be associated with EGFR amplification and
CDKN2A deletion. This result supports the reliability of our
results.
The present study revealed the association between
TERT promoter mutation and preclinical characteristics of
GBM, including in vitro sphere-forming capacity and in
vivo tumorigenic potential. As TERT promoter mutation is
a prognostic marker of GBM, the identification of preclinical
characteristics of TERT promoter mutations may reveal the
functions of TERT and telomere length in the self-renewal of
GBM cells, and treatment resistance of GBM. Furthermore, the
results may provide a foundation for the development of innovative
telomerase-based therapeutic strategies for treatment-resistant
GBMs.
Acknowledgements
The present study was supported by the Basic Science
Research Program through the National Research Foundation of Korea
funded by the Ministry of Education (grant no.
NRF-2014R1A6A3A04058057). Additional support was supported by the
National Research Foundation of Korea (NRF) Grant funded by the
Korean Government (MSIP) (grant no. NRF-2016R1A5A2945889).
References
|
1
|
Mosrati MA, Malmström A, Lysiak M,
Krysztofiak A, Hallbeck M, Milos P, Hallbeck AL, Bratthäll C,
Strandéus M, Stenmark-Askmalm M and Söderkvist P: TERT promoter
mutations and polymorphisms as prognostic factors in primary
glioblastoma. Oncotarget. 6:16663–16673. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Reitman ZJ, Pirozzi CJ and Yan H:
Promoting a new brain tumor mutation: TERT promoter mutations in
CNS tumors. Acta Neuropathol. 126:789–792. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Verhaak RG, Hoadley KA, Purdom E, Wang V,
Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al:
Integrated genomic analysis identifies clinically relevant subtypes
of glioblastoma characterized by abnormalities in PDGFRA, IDH1,
EGFR, and NF1. Cancer Cell. 17:98–110. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Labussière M, Boisselier B, Mokhtari K, Di
Stefano AL, Rahimian A, Rossetto M, Ciccarino P, Saulnier O,
Paterra R, Marie Y, et al: Combined analysis of TERT, EGFR, and IDH
status defines distinct prognostic glioblastoma classes. Neurology.
83:1200–1206. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Nonoguchi N, Ohta T, Oh JE, Kim YH,
Kleihues P and Ohgaki H: TERT promoter mutations in primary and
secondary glioblastomas. Acta Neuropathol. 126:931–937. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Killela PJ, Reitman ZJ, Jiao Y, Bettegowda
C, Agrawal N, Diaz LA Jr, Friedman AH, Friedman H, Gallia GL,
Giovanella BC, et al: TERT promoter mutations occur frequently in
gliomas and a subset of tumors derived from cells with low rates of
self-renewal. Proc Natl Acad Sci USA. 110:pp. 6021–6026. 2013,
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
George J, Banik NL and Ray SK: Knockdown
of hTERT and concurrent treatment with interferon-gamma inhibited
proliferation and invasion of human glioblastoma cell lines. Int J
Biochem Cell Biol. 42:1164–1173. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Beck S, Jin X, Sohn YW, Kim JK, Kim SH,
Yin J, Pian X, Kim SC, Nam DH, Choi YJ and Kim H: Telomerase
activity-independent function of TERT allows glioma cells to attain
cancer stem cell characteristics by inducing EGFR expression. Mol
Cells. 31:9–15. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Joo KM, Kim J, Jin J, Kim M, Seol HJ,
Muradov J, Yang H, Choi YL, Park WY, Kong DS, et al:
Patient-specific orthotopic glioblastoma xenograft models
recapitulate the histopathology and biology of human glioblastomas
in situ. Cell Rep. 3:260–273. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Louis DN, Ohgaki H, Wiestler OD, Cavenee
WK, Burger PC, Jouvet A, Scheithauer BW and Kleihues P: The 2007
WHO classification of tumours of the central nervous system. Acta
Neuropathol. 114:97–109. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Joo KM, Kim SY, Jin X, Song SY, Kong DS,
Lee JI, Jeon JW, Kim MH, Kang BG, Jung Y, et al: Clinical and
biological implications of CD133-positive and CD133-negative cells
in glioblastomas. Lab Invest. 88:808–815. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pollard SM, Yoshikawa K, Clarke ID, Danovi
D, Stricker S, Russell R, Bayani J, Head R, Lee M, Bernstein M, et
al: Glioma stem cell lines expanded in adherent culture have
tumor-specific phenotypes and are suitable for chemical and genetic
screens. Cell Stem Cell. 4:568–580. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ito M, Hiramatsu H, Kobayashi K, Suzue K,
Kawahata M, Hioki K, Ueyama Y, Koyanagi Y, Sugamura K, Tsuji K, et
al: NOD/SCID/gamma(c)(null) mouse: An excellent recipient mouse
model for engraftment of human cells. Blood. 100:3175–3182. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu T, Wang N, Cao J, Sofiadis A, Dinets
A, Zedenius J, Larsson C and Xu D: The age- and shorter
telomere-dependent TERT promoter mutation in follicular thyroid
cell-derived carcinomas. Oncogene. 33:4978–4984. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Oh YT, Cho HJ, Kim J, Lee JH, Rho K, Seo
YJ, Choi YS, Jung HJ, Song HS, Kong DS, et al: Translational
validation of personalized treatment strategy based on genetic
characteristics of glioblastoma. PLoS One. 9:e1033272014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Olshen AB, Venkatraman ES, Lucito R and
Wigler M: Circular binary segmentation for the analysis of
array-based DNA copy number data. Biostatistics. 5:557–572. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li H and Durbin R: Fast and accurate short
read alignment with Burrow-Wheeler transform. Bioinformatics.
25:1754–1760. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li H, Handsaker B, Wysoker A, Fennell T,
Ruan J, Homer N, Marth G, Abecasis G and Durbin R: 1000 Genome
Project Data Processing Subgroup: The sequence Alignment/Map format
and SAMtools. Bioinformatics. 25:2078–2079. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wu TD and Nacu S: Fast and SNP-tolerant
detection of complex variants and splicing in short reads.
Bioinformatics. 26:873–881. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Vinagre J, Pinto V, Celestino R, Reis M,
Pópulo H, Boaventura P, Melo M, Catarino T, Lima J, Lopes JM, et
al: Telomerase promoter mutations in cancer: An emerging molecular
biomarker? Virchows Arch. 465:119–133. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fichtner I, Rolff J, Soong R, Hoffmann J,
Hammer S, Sommer A, Becker M and Merk J: Establishment of
patient-derived non-small cell lung cancer xenografts as models for
the identification of predictive biomarkers. Clin Cancer Res.
14:6456–6468. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
John T, Kohler D, Pintilie M, Yanagawa N,
Pham NA, Li M, Panchal D, Hui F, Meng F, Shepherd FA and Tsao MS:
The ability to form primary tumor xenografts is predictive of
increased risk of disease recurrence in early-stage non-small cell
lung cancer. Clin Cancer Res. 17:134–141. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lee HW, Lee JI, Lee SJ, Cho HJ, Song HJ,
Jeong DE, Seo YJ, Shin S, Joung JG, Kwon YJ, et al: Patient-derived
xenografts from non-small cell lung cancer brain metastases are
valuable translational platforms for the development of
personalized targeted therapy. Clin Cancer Res. 21:1172–1182. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hale JS, Otvos B, Sinyuk M, Alvarado AG,
Hitomi M, Stoltz K, Wu Q, Flavahan W, Levison B, Johansen ML, et
al: Cancer stem cell-specific scavenger receptor CD36 drives
glioblastoma progression. Stem Cells. 32:1746–1758. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Borah A, Raveendran S, Rochani A, Maekawa
T and Kumar DS: Targeting self-renewal pathways in cancer stem
cells: Clinical implications for cancer therapy. Oncogenesis.
4:e1772015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Horn S, Figl A, Rachakonda PS, Fischer C,
Sucker A, Gast A, Kadel S, Moll I, Nagore E, Hemminki K, et al:
TERT promoter mutations in familial and sporadic melanoma. Science.
339:959–961. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Huang FW, Hodis E, Xu MJ, Kryukov GV, Chin
L and Garraway LA: Highly recurrent TERT promoter mutations in
human melanoma. Science. 339:957–959. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chiba K, Johnson JZ, Vogan JM, Wagner T,
Boyle JM and Hockemeyer D: Cancer-associated TERT promoter
mutations abrogate telomerase silencing. Elife. 4:2015. View Article : Google Scholar
|
|
29
|
Ko E, Seo HW, Jung ES, Kim BH and Jung G:
The TERT promoter SNP rs2853669 decreases E2F1 transcription factor
binding and increases mortality and recurrence risks in liver
cancer. Oncotarget. 7:684–699. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Huse JT: TERT promoter mutation designates
biologically aggressive primary glioblastoma. Neuro Oncol. 17:5–6.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Simon M, Hosen I, Gousias K, Rachakonda S,
Heidenreich B, Gessi M, Schramm J, Hemminki K, Waha A and Kumar R:
TERT promoter mutations: A novel independent prognostic factor in
primary glioblastomas. Neuro Oncol. 17:45–52. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Spiegl-Kreinecker S, Lötsch D, Ghanim B,
Pirker C, Mohr T, Laaber M, Weis S, Olschowski A, Webersinke G,
Pichler J and Berger W: Prognostic quality of activating TERT
promoter mutations in glioblastoma: Interaction with the rs2853669
polymorphism and patient age at diagnosis. Neuro Oncol.
17:1231–1240. 2015. View Article : Google Scholar : PubMed/NCBI
|