Introduction
Nasopharyngeal carcinoma (NPC) is commonly observed
in southern China, particularly in the Pearl River delta area and
the Xijiang River basin in the Guangdong and Guangxi provinces,
with an incidence rate as high as 25–50 per 100,000 between 2000
and 2002 (1,2). The etiology of NPC is considered a
result of unique interactions among environmental and genetic
factors, but a detailed mechanism is yet to be elucidated. Studies
have shown that gene polymorphisms not only influence gene
expression or the function of the encoded protein, but may also
affect individual susceptibility to NPC or the severity of the
disease (3).
In a New England Journal of Medicine article
published in 1971, Judah Folkman first proposed the hypothesis that
angiogenesis was essential to cancer cell growth, and that
angiogenesis inhibition could be a target for cancer therapy
(4). A substantial body of
preclinical and clinical evidence shows that vascular endothelial
growth factor (VEGF) and its receptors serve a crucial role
in tumor-related angiogenesis (5,6). Vascular
endothelial growth factor receptor-2 (VEGFR2, also known as
kinase insert domain-containing receptor) is considered to be the
most important type of receptor for angiogenesis during tumor
invasion (7). Numerous studies have
shown that VEGFR2, not VEGF or VEGFR1, is
associated with tumor prognosis and response to therapy (8,9). Specific
single nucleotide polymorphisms (SNPs) of VEGFR2 that
modulate angiogenesis have been associated with an increased risk
of malignancy and higher cancer aggression (10–12). The
rs2071559 polymorphism is located in the promoter region of
VEGFR2 and certain studies have found that this polymorphism
affects mRNA and protein expression (13). However, little is known about the
association between rs2071559 and the etiology, prognosis and
tumor-node-metastasis (TNM) stage of NPC. The aim of the present
study was to determine whether the rs2071559 polymorphism in the
VEGFR2 gene is associated with the risk of developing NPC
and associated cancer invasiveness.
Materials and methods
Study subjects
A total of 171 incident NPC cases were consecutively
recruited from the First Affiliated Hospital of Guangxi Medical
University of Nanning City (Nanning, China) between June 2009 and
June 2010. All subjects were genetically unrelated and were from
Nanning and the surrounding regions in China. The inclusion
criteria were as follows: i) Newly diagnosed NPC; ii) no previous
history of malignant tumors of other organs; iii) pathologically
diagnosed as NPC. The histological type was confirmed by 2
independent pathologists according to the World Health Organization
classification (14). TNM stage
designation was performed according to the definitions of the 2009
edition of Union for International Cancer Control-American Joint
Committee on Cancer clinical staging criteria (15). There was no ambiguity in the
diagnosis. The control group was comprised of 184 unrelated healthy
peripheral blood donors who visited the general health check-up
division at the same hospital. Selection criteria for the control
group included having no evidence of any personal or family history
of cancer or other serious illness. The control group was
comparable to the NPC cases with respect to age, sex and ethnicity.
General information about the healthy controls was extracted from a
standard questionnaire. Written informed consent was obtained from
all subjects. All participants agreed to participate in the study
and provided peripheral blood samples. The present study was
approved by the Ethics Committee of Guangxi Medical University.
Genotype analysis
Genomic DNA was extracted from peripheral blood
using a commercially available kit, according to the manufacturer's
protocol (AxyPrep Blood Genomic DNA Miniprep kit; Axygen; Corning
Incorporated, Corning, NY, USA). The rs2071559 polymorphism was
detected using TaqMan probe-based polymerase chain reaction (PCR)
and the PCR primers were designed by us based on the GenBank
reference sequence and primer 5.0 (16). This method was carried out by PCR
amplification using the primers: rs2071339 forward,
5′-GCTCTTAATCAGAAAACGCACTTG-3′ and reverse,
5′-GGCTAGGCAGGTCACTTCAAA-3′; the allele T TaqMan probe 5′-FAM-CAG
TTC GCC AAC ATT CCC GCT-TAMRA-3′; and the allele C TaqMan probe
5′-HEX-AGT TCG CCA GCA TTC CCG CT-TAMRA-3′ in a 10 µl reaction
mixture containing ~80 ng genomic DNA, 0.45 µl of each primer, and
0.25 µl of each probe. Reactions were cycled with the following
parameters: Preheat at 60°C for 1 min and then at 95°C for 10 min,
followed by 40 cycles of 95°C for 5 sec and 60°C for 31 sec, and
finally, a final post-read for 1 min. Genotype analysis was
automatically generated by Applied Biosystems® 7500
Real-Time PCR system (Thermo Fisher Scientific, Inc., Waltham, MA,
USA). We did not repeat the experiment, but validated it by direct
sequencing.
Statistical analysis
The χ2 test and Student's t-test were
used to compare differences in sex and age distribution between
nasopharyngeal carcinoma patients and the healthy control group.
The Hardy-Weinberg equilibrium was verified by the calculation of
genotype frequencies. Univariate and multivariate logistic
regression analyses were used to estimate the correlation between
rs2071559 polymorphism and NPC risk by computing the odds ratio
(OR) and 95% confidence interval (CI), while accounting for
confounding factors such as sex and age. In all statistical tests,
P<0.05 was considered to indicate a statistically significant
difference. All statistical analyses were performed with
statistical analysis system software (SPSS 16.0; SPSS, Inc.,
Chicago, IL, USA).
Results
qPCR
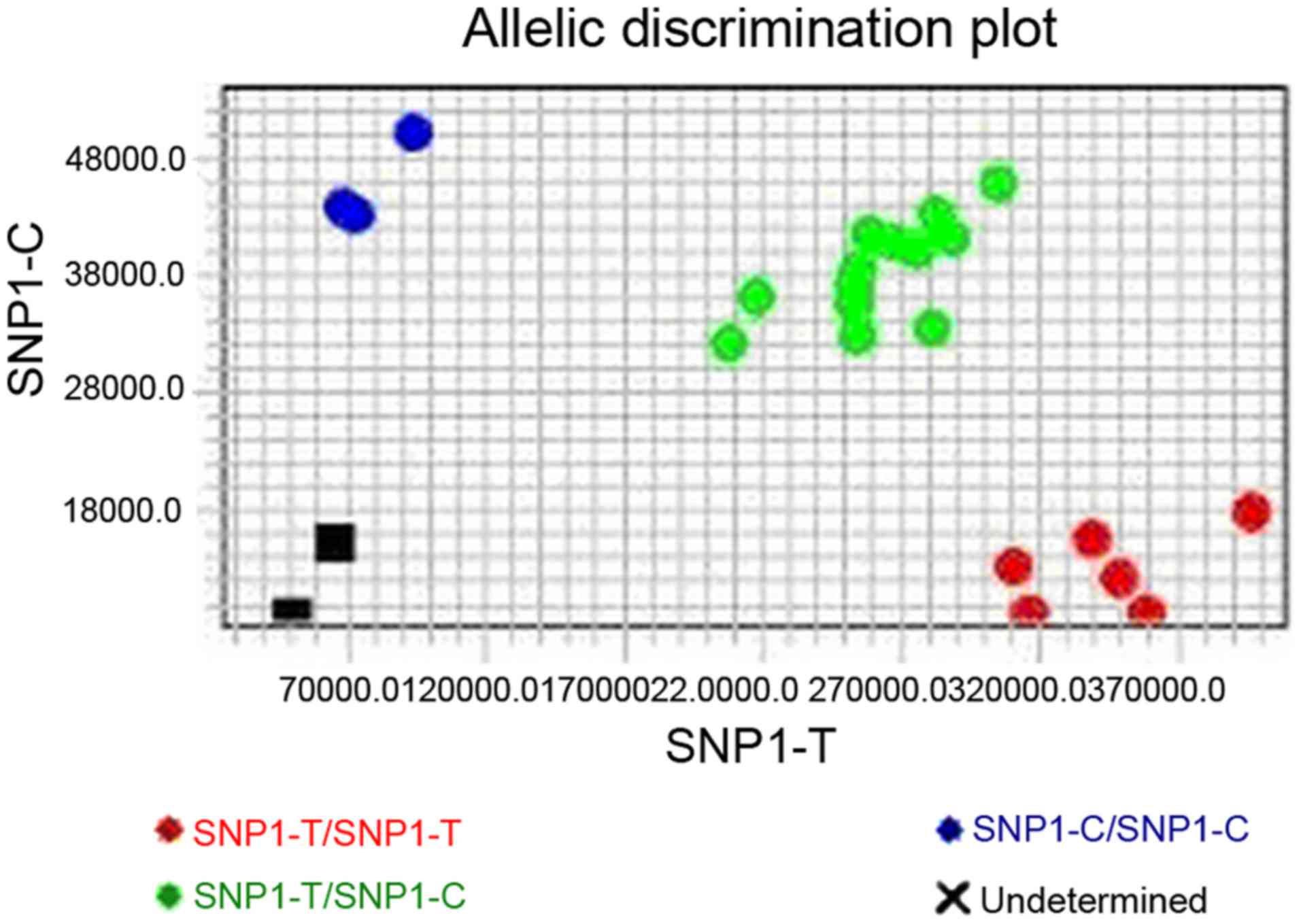
qRT-PCR with TaqMan probes was used to identify the
genotype of the rs2071559 polymorphism in patients with NPC and
healthy controls (Fig. 1). The C and
T alleles were each associated with a different fluorescent label.
The fluorescence signal generated by homozygotes was small, while
heterozygotes produce 2 different fluorescence signals. Although
the amplification efficiency of each sample is not exactly the
same, the genotypes cluster together.
Sequencing of rs2071559 genotypes
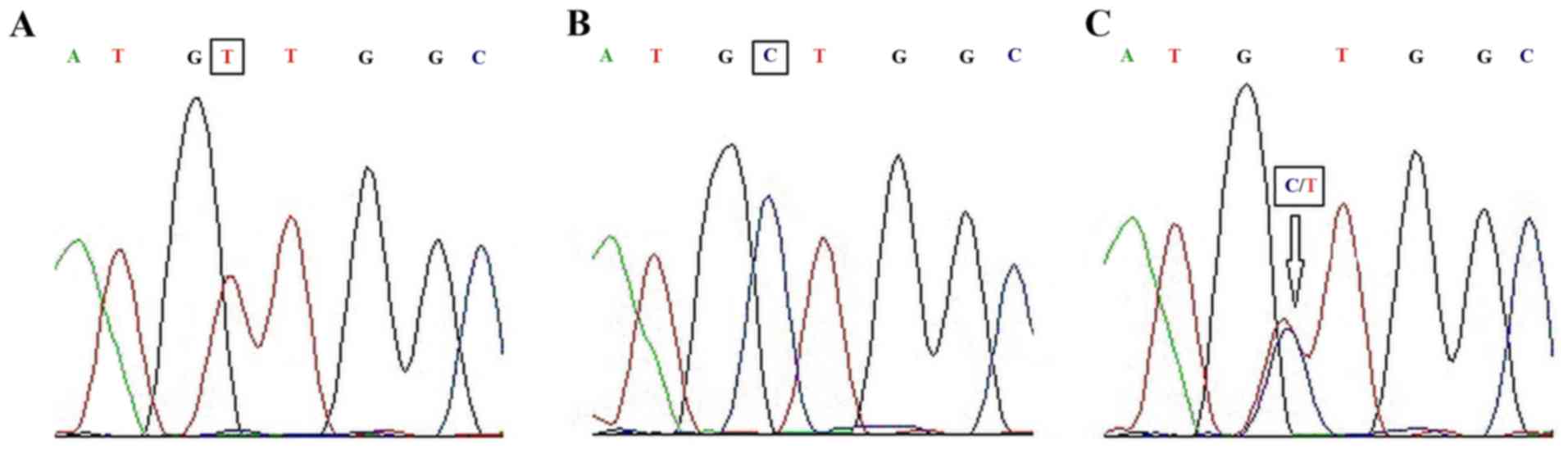
In order to verify the accuracy and reliability of
the qPCR results, the present study sequenced each allele
separately (Fig. 2). The 3 DNA
sequences in Fig. 2 represent the TT,
TC, and CC genotypes. The sequencing results validated the qPCR
results.
rs2071559 polymorphism and NPC
risk
The differences in age and sex between the 171
patients with NPC and 184 healthy controls were not statistically
significant (P=0.061 and P=0.845, respectively; Table I). The genotype frequencies for the
NPC cases and controls were in agreement with the Hardy-Weinberg
equilibrium (0.055 for NPC and 0.388 for controls). The genotype or
allele distribution of the rs2071559 polymorphism had no
significant association with the risk of NPC following adjustment
for age, sex and ethnicity by multivariate logistic regression
analyses. The P-values were as follows: Co-dominant model, 0.994
and 0.200; dominant model, 0.736; recessive model, 0.253 and allele
model, 0.394 (Table II).
 | Table I.Clinical characteristics of patients
with NPC and healthy controls. |
Table I.
Clinical characteristics of patients
with NPC and healthy controls.
| Characteristics | Patients | Controls | P-value |
|---|
| Sex |
|
| 0.061 |
| Male | 122 | 114 |
|
|
Female | 49 | 70 |
|
| Age, mean ± SD | 44.17±11.34 | 43.99±12.74 | 0.845 |
| Histological
type |
|
|
|
| UCNT | 112 | – | – |
| DNKC | 59 | – |
|
| Tumor size |
|
|
|
| T1 | 2 | – | – |
| T2 | 40 | – |
|
| T3 | 65 | – |
|
| T4 | 64 | – |
|
| Lymph node
status |
|
|
|
| N0 | 16 | – | – |
| N1 | 41 | – |
|
| N2 | 89 | – |
|
| N3 | 25 | – |
|
| Distant
metastasis |
|
|
|
|
Yes | 18 | – | – |
| No | 153 | – |
|
 | Table II.Comparison of NPC patients and
controls by genotype frequencies of rs2071559 polymorphism. |
Table II.
Comparison of NPC patients and
controls by genotype frequencies of rs2071559 polymorphism.
| Variable | Genotype | Control | Patient | Adjusted OR (95%
CI)a | P-value |
|---|
| Co-dominant
model | TT | 66 | 65 | 1 |
|
|
| CT | 93 | 90 | 0.998
(0.633–1.573 | 0.994 |
|
| CC | 25 | 16 | 0.623
(0.302–1.286) | 0.200 |
| Dominant model | TT | 66 | 65 | 1 |
|
|
| CC+CT | 118 | 106 | 0.919
(0.582–1.417) | 0.736 |
| Recessive
model | TT+CT | 159 | 155 | 1 |
|
|
| CC | 25 | 16 | 0.664
(0.325–1.283) | 0.253 |
| Allele model | T | 225 | 220 | 1 |
|
|
| C | 143 | 122 | 0.873
(0.643–1.183) | 0.394 |
rs2071559 polymorphism and the
aggressiveness of NPC
Clinical characteristics of patients with NPC
including tumor size, lymph node metastasis, distant metastasis,
clinical stage and histological type of cancer were analyzed. The
distribution of genotypes and alleles of the rs2071559 polymorphism
for NPC patients with tumor characteristics is shown in Table III. Earlier lymph node status
according to the TNM staging system was significantly associated
with rs2071559 C allele and the related genotypes (OR 0.402, 95% CI
0.193–0.835, P=0.016; and OR 0.347, 95% CI 0.145–0.829, P=0.024,
respectively; Table III). However,
no significant correlations were identified between the genotype or
allele distribution and the primary tumor size, distant metastasis,
clinical stage or histological type (Table III).
 | Table III.Stratification analyses of the
rs2071559 polymorphism with clinical characteristics of NPC
patients. |
Table III.
Stratification analyses of the
rs2071559 polymorphism with clinical characteristics of NPC
patients.
|
| Dominant model | Allele model |
|---|
|
|
|
|
|---|
|
Characteristics | OR (95% CI) | P-value | OR (95% CI) | P-value |
|---|
| Age (years) |
|
|
| 0.233 |
|
≤40 | 1 |
| 1 |
|
|
>40 | 1.240
(0.648–2.370) | 0.618 | 1.365
(0.848–2.198) |
|
| Sex |
|
|
| 0.456 |
|
Male | 1 |
| 1 |
|
|
Female | 0.752
(0.383–1.478) | 0.486 | 0.801
(0.488–1.314) |
|
| Nation |
|
|
| 0.708 |
|
Han | 1 |
| 1 |
|
|
Minority | 0.668
(0.340–1.311) | 0.296 | 0.884
(0.540–1.448) |
|
| Histological
type |
|
|
| 0.553 |
|
DNKC | 1 |
| 1 |
|
|
UCNT | 0.764
(0.395–1.475) | 0.508 | 0.850
(0.535–1.350) |
|
| Tumor size |
|
|
| 0.484 |
|
T1-3 | 1 |
| 1 |
|
| T4 | 1.283
(0.673–2.444) | 0.516 | 1.198
(0.760–1.888) |
|
| Lymph node
status |
|
|
| 0.016 |
|
N0-2 | 1 |
| 1 |
|
| N3 | 0.347
(0.145–0.829) | 0.024 | 0.402
(0.193–0.835) |
|
| Distant
metastasis |
|
|
| 0.997 |
| M0 | 1 |
| 1 |
|
| M1 | 0.832
(0.180–3.840) | 0.873 | 1.002
(0.328–3.059) |
|
| Stage grouping |
|
|
| 0.543 |
|
I–III | 1 |
| 1 |
|
| IV | 0.665
(0.289–1.529) | 0.385 | 0.809
(0.438–1.495) |
|
Discussion
To the best of our knowledge, this is the first
study targeting the association between the rs2071559 polymorphism
and patients with NPC in a cohort from south China. The present
study found that patients with NPC with the rs2071559 C allele and
the related genotypes appeared to have lower risk of N3 lymph node
metastases according to the TNM staging system, implying that the
rs2071559 C allele may decrease the migratory ability and suppress
the aggressiveness of NPC cells.
Bioinformatic analysis suggested that the rs2071559
polymorphism is located in the binding site for transcriptional
factor E2F (involved in cell cycle regulation and interacts with Rb
p107). The transition from the T allele to the C allele leads to
the disappearance of the binding site for transcriptional factor
E2F, which suppresses transcriptional activity and reduces the
expression of VEGFR2 (13).
Consequently, An et al (17)
found that the VEGFR2 polymorphism was only associated with
mRNA expression and had no association with protein expression or
tumorigenesis. Zhang et al (18) demonstrated that the promoter variant
rs2071559 T was associated with reduced susceptibility to
atherothrombotic stroke; however, it was inversely associated with
the development of coronary artery lesion in Kawasaki disease
patients (19), implying that the T
allele could increase neovascularization. In addition, as suggested
by the data of Kariž and Petrovič (20), the rs2071559 CC genotype is a risk
factor for myocardial infarction in Caucasians with type 2 diabetes
mellitus.
Tumor growth and metastasis is dependent on the
neovascularization of its surroundings. Depletion of the tumor
blood supply inhibits growth and metastasis (21). Studies have demonstrated that the
VEGFR2 signaling pathway performs an important role in tumor
neovascularization and could serve as a promising target for
anti-tumor therapy. Duff et al (22) showed that VEGFC and
VEGFR2 were co-expressed and correlated with a number of
metastatic lymph nodes. Another study has also shown that
VEGFA could induce tumor neovascularization and lymph node
lymphangiogenesis, and even promote lymphatic metastasis, through
the VEGFR2 pathway (23).
Together, these findings shed light on the findings of the present
study. VEGFR2 in patients with NPC with the rs2071559 T
allele may exhibit higher transcriptional activity, subsequently
increasing the migratory ability of NPC cells, in concert with
certain other factors. The present study also found that patients
with NPC with the rs2071559 C allele had decreased lymphatic
metastasis, but no difference in NPC tumorigenesis. Additional
studies are required to develop a detailed mechanism explaining
these observations.
There are also numerous studies that have found that
VEGFR2 affects the tumor response to radio-chemotherapy and
other treatments. Nagy et al (24) found by multivariate analysis that only
the expression of VEGFR2, and not patient age or tumor size,
predicts the response of cervical carcinoma to radio-chemotherapy.
Rydén et al (25) also found
that tumor-specific expression of VEGFR2 was associated with
an impaired response to the adjuvant tamoxifen in breast cancer.
However, certain studies have implied that a decrease in the
expression of VEGFR2 could increase the response of breast
cancer to chemotherapy. Mele et al (26) found that VEGFR2 expression in
patients with breast cancer increased with treatment with either
tamoxifen or epirubicin alone, but decreased in patients receiving
tamoxifen plus epirubicin. Sorafenib, the first FDA-approved
targeted agent for the treatment of advanced renal cell carcinoma
(RCC) and hepatocellular carcinoma (HCC), was initially identified
as a Raf kinase inhibitor (27);
however, it also inhibits VEGFR2. Therefore, Escudier et
al (28) found that rs2071559 can
predict sorafenib efficacy in patients with metastatic RCC, and
Zheng et al (29) demonstrated
that rs2071559 was an independent factor in overall survival rate
of patients with advanced HCC. These observations imply the
potential synergism of such drugs. The risk of serious hematologic
complications, including neutropenia, should be considered if
anti-VEGF drugs are to be administered with chemotherapy
(30). Assessing the maturation state
of the microvessels, however, may allow improved characterization
of the vasculature and could prove useful in guiding
anti-angiogenic treatment (31,32).
There are potential limitations of the present
study. The VEGFR2 gene is a polymorphic gene, and only
rs2071559 was investigated. Other SNPs within VEGFR2 gene
may contribute to gene regulation (1,7,33). Thus, additional studies on other
functional SNPs and haplotypes are required to delineate the
genetic contribution of VEGFR2 polymorphisms to NPC. Second,
the sample size was relatively small. Additional studies with
larger populations are required to define the role of rs2071559
polymorphisms in the onset and course of NPC.
To conclude, the present results suggest that the
rs2071559 polymorphism is associated with lymphatic metastasis of
NPC, and may be a good prognostic indicator in patients with NPC.
However, the precise molecular mechanism should been elucidated in
further studies.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant no. 30960103).
References
|
1
|
Wee JT, Ha TC, Loong SL and Qian CN: Is
nasopharyngeal cancer really a ‘Cantonese cancer’? Chin J Cancer.
29:517–526. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Guo XC, Scott K, Liu Y, Dean M, David V,
Nelson GW, Johnson RC, Dilks HH, Lautenberger J, Kessing B, et al:
Genetic factors leading to chronic Epstein-Barr virus infection and
nasopharyngeal carcinoma in South East China: Study design, methods
and feasibility. Hum Genomics. 2:365–375. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Försti A, Jin Q, Altieri A, Johansson R,
Wagner K, Enquist K, Grzybowska E, Pamula J, Pekala W, Hallmans G,
et al: Polymorphisms in the KDR and POSTN genes: Association with
breast cancer susceptibility and prognosis. Breast Cancer Res
Treat. 101:83–93. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Folkman J: Tumor angiogenesis: Therapeutic
implications. N Engl J Med. 285:1182–1186. 1971. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Carmeliet P and Jain RK: Angiogenesis in
cancer and other diseases. Nature. 407:249–257. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Guo P, Xu L, Pan S, Brekken RA, Yang ST,
Whitaker GB, Nagane M, Thorpe PE, Rosenbaum JS, Huang Su HJ, et al:
Vascular endothelial growth factor isoforms display distinct
activities in promoting tumor angiogenesis at different anatomic
sites. Cancer Res. 61:8569–8577. 2001.PubMed/NCBI
|
|
7
|
Brenner RM, Nayak NR, Slayden OD,
Critchley HO and Kelly RW: Premenstrual and menstrual changes in
the macaque and human endometrium: Relevance to endometriosis. Ann
N Y Acad Sci. 955(60–74): 86–88, 396–406. 2002.
|
|
8
|
Qi L, Robinson WA, Brady BM and Glode LM:
Migration and invasion of human prostate cancer cells is related to
expression of VEGF and its receptors. Anticancer Res. 23:3917–3922.
2003.PubMed/NCBI
|
|
9
|
Waltenberger J, Claesson-Welsh L, Siegbahn
A, Shibuya M and Heldin CH: Different signal transduction
properties of KDR and Flt1, two receptors for vascular endothelial
growth factor. J Biol Chem. 269:26988–26995. 1994.PubMed/NCBI
|
|
10
|
Iughetti P, Suzuki O, Godoi PH, Alves VA,
Sertié AL, Zorick T, Soares F, Camargo A, Moreira ES, di Loreto C,
et al: A polymorphism in endostatin, an angiogenesis inhibitor,
predisposes for the development of prostatic adenocarcinoma. Cancer
Res. 61:7375–7378. 2001.PubMed/NCBI
|
|
11
|
Howell WM, Bateman AC, Turner SJ, Collins
A and Theaker JM: Influence of vascular endothelial growth factor
single nucleotide polymorphisms on tumour development in cutaneous
malignant melanoma. Genes Immun. 3:229–232. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Balasubramanian SP, Brown NJ and Reed MW:
Role of genetic polymorphisms in tumour angiogenesis. Br J Cancer.
87:1057–1065. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang Y, Zheng Y, Zhang W, Yu H, Lou K,
Zhang Y, Qin Q, Zhao B, Yang Y and Hui R: Polymorphisms of KDR Gene
are associated with coronary heart disease. J Am Coll Cardiol.
50:760–767. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Thompson L: World Health Organization
classification of tumours: Pathology and genetics of head and neck
tumours. Ear Nose Throat J. 85:742006.PubMed/NCBI
|
|
15
|
Sobin LH, Gospodarowicz MK and Wittekind
C: TNM Classification of Malignant Tumors. 7th edition.
Wiley-Blackwell; Oxford: 2009
|
|
16
|
Patterson C, Perrella MA, Hsieh CM,
Yoshizumi M, Lee ME and Haber E: Cloning and functional analysis of
the promoter for KDR/flk-1, a receptor for vascular endothelial
growth factor. J Biol Chem. 270:23111–23118. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
An SJ, Chen ZH, Lin QX, Su J, Chen HJ, Lin
JY and Wu YL: The −271G>A polymorphism of kinase insert
domain-containing receptor gene regulates its transcription level
in patients with non-small cell lung cancer. BMC Cancer. 9:1442009.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang W, Sun K, Zhen Y, Wang D, Wang Y,
Chen J, Xu J, Hu FB and Hui R: VEGF receptor-2 variants are
associated with susceptibility to stroke and recurrence. Stroke.
40:2720–2726. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kariyazono H, Ohno T, Khajoee V, Ihara K,
Kusuhara K, Kinukawa N, Mizuno Y and Hara T: Association of
vascular endothelial growth factor (VEGF) and VEGF receptor gene
polymorphisms with coronary artery lesions of Kawasaki Disease.
Pediatr Res. 56:953–959. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kariž S and Petrovič D: Minor association
of kinase insert domain-containing receptor gene polymorphism
(rs2071559) with myocardial infarction in Caucasians with type 2
diabetes mellitus: Case-control cross-sectional study. Clin
Biochem. 47:192–196. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shalaby F, Rossant J, Yamaguchi TP,
Gertsenstein M, Wu XF, Breitman ML and Schuh AC: Failure of
blood-island formation and vasculogenesis in Flk-1 deficient mice.
Nature. 376:62–66. 1995. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Duff SE, Jeziorska M, Rosa DD, Kumar S,
Haboubi N, Sherlock D, O'Dwyer ST and Jayson GC: Vascular
endothelial growth factors and receptors in colorectal cancer:
Implications for anti-angiogenic therapy. Eur J Cancer. 42:112–117.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hirakawa S, Kodama S, Kunstfeld R, Kajiya
K, Brown LF and Detmar M: VEGF-A induces tumor and sentinel lymph
node lymphangiogenesis and promotes lymphatic metastasis. J Exp
Med. 201:1089–1099. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Nagy VM, Buiga R, Brie I, Todor N, Tudoran
O, Ordeanu C, Virág P, Tarta O, Rus M and Bălăcescu O: Expression
of VEGF, VEGFR, EGFR, COX-2 and MVD in cervical carcinoma, in
relation with the response to radio-chemotherapy. Rom J Morphol
Embryol. 52:53–59. 2011.PubMed/NCBI
|
|
25
|
Rydén L, Jirström K, Haglund M, Stål O and
Fernö M: Epidermal growth factor receptor and vascular endothelial
growth factor receptor 2 are specific biomarkers in triple-negative
breast cancer. Results from a controlled randomized trial with
long-term follow-up. Breast Cancer Res Treat. 120:491–498. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mele T, Generali D, Fox S, Brizzi MP,
Bersiga A, Milani M, Allevi G, Bonardi S, Aguggini S, Volante M, et
al: Anti-angiogenic effect of tamoxifen combined with epirubicin in
breast cancer patients. Breast Cancer Res Treat. 123:795–804. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wilhelm SM, Carter C, Tang L, Wilkie D,
McNabola A, Rong H, Chen C, Zhang X, Vincent P, McHugh M, et al:
BAY 43–9006 exhibits broad spectrum oral antitumor activity and
targets the RAF/MEK/ERK pathway and receptor tyrosine kinases
involved in tumor progression and angiogenesis. Cancer Res.
64:7099–7109. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Escudier B, Rini BI, Motzer RJ, Tarazi J,
Kim S, Huang X, Rosbrook B, English PA, Loomis AK and Williams JA:
Genotype correlations with blood pressure and efficacy from a
randomized phase iii trial of second-line axitinib versus sorafenib
in metastatic renal cell carcinoma. Clin Genitourin Cancer.
13:328–337. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zheng YB, Zhan MX, Zhao W, Liu B, Huang
JW, He X, Fu SR, Zhao Y, Li Y, Hu BS and Lu LG: The relationship of
kinase insert domain receptor gene polymorphisms and clinical
outcome in advanced hepatocellular carcinoma patients treated with
sorafenib. Med Oncol. 31:2092014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Novitskiy SV, Csiki I, Huang Y, Johnson
DH, Harth EM, Carbone DP and Dikov MM: Anti-vascular endothelial
growth factor treatment in combination with chemotherapy delays
hematopoietic recovery due to decreased proliferation of bone
marrow hematopoietic progenitor cells. J Thorac Oncol. 5:1410–1415.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hlatky L, Hahnfeldt P and Folkman J:
Clinical application of antiangiogenic therapy: Microvessel
density, what it does and doesn't tell us. J Natl Cancer Inst.
94:883–893. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Vermeulen PB, Gasparini G, Fox SB,
Colpaert C, Marson LP, Gion M, Beliën JA, de Waal RM, Van Marck E,
Magnani E, et al: Second international consensus on the methodology
and criteria of evaluation of angiogenesis quantification in solid
human tumours. Eur J Cancer. 38:1564–1579. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Park HW, Shin ES, Lee JE, Kwon HS, Chun E,
Kim SS, Chang YS, Kim YK, Min KU, Kim YY and Cho SH: Multilocus
analysis of atopy in Korean children using
multifactor-dimensionality reduction. Thorax. 62:265–269. 2007.
View Article : Google Scholar : PubMed/NCBI
|