Introduction
The diagnosis and classification of acute leukemia
rely on an array of multidisciplinary approaches, including
analyses of morphology, immunophenotype, cytogenetics and molecular
genetics. Using these criteria, the majority of acute leukemia
cases can be assigned into a specific lineage (1). However, there is a rare type of acute
leukemia that exhibits concurrent characteristics of myeloid (My)
and T- or B-lymphoid lineages. In the 2008 WHO classification
(2), this type of leukemia was termed
leukemia of ambiguous lineage, which includes acute
undifferentiated leukemia and mixed-phenotype acute leukemia (MPAL)
(3).
MPAL is defined as a type of acute leukemia
expressing antigens of more than one lineage, which is impossible
to assign to any one lineage with certainty (4). The diagnosis of MPAL should exclude
cases either by genetic or by clinical features that can be
classified into another category. For example, acute myeloid
leukemia (AML) with t(8;21), t(15;17) and inv(16) can also express lymphoid-associated
markers (1). In addition, MPALs with
rearrangement of t (9;22)(q34;q11.2) or t (v;11q23) should be
diagnosed as MPALs with BCR, RhoGEF and GTPase activating
protein-ABL proto-oncogene 1, non-receptor tyrosine kinase
(BCR-ABL1) or mixed-lineage leukemia (MLL) rearrangement (1). MAPLs can be classified as bilineage and
biphenotypic acute leukemias (4).
Bilineage acute leukemias usually contain two distinct blast
populations, each of which expresses lymphoid or myeloid lineage
markers. In biphenotypic acute leukemia, the blast cells are
characterized as one population that expresses myeloid and T- or
B-lymphoid markers (3,5). The association and differentiation
between bilineage and biphenotypic leukemia remains unclear.
Bilineage T lymphoid and myeloid (T/My) malignancy
is rare. To the best of our knowledge, between 1988 and 2015, only
9 publications with a total 18 cases of T/My bilineage MPALs were
reported on PubMed (https://www.ncbi.nlm.nih.gov/pubmed) (6–14). In the
majority of cases, leukemic blast cells were detected initially in
the peripheral blood (PB) and bone marrow (BM), meeting the
diagnostic criteria of leukemia. Only 2 cases of T/My bilineage
malignancy presenting initially with extramedullary infiltration
were reported (15,16). The present study reports the case of a
31-year-old man with bilineage T/My malignancy who initially
presented with cervical lymph node enlargement beyond the diagnosis
of leukemia in the PB and in the BM. The present study also reviews
the cellular origin, development and therapeutic strategies of
extramedullary T/My bilineage malignancy.
Case report
A 31-year-old man with painless enlargement of the
cervical lymph node was admitted to a local hospital in November
2014. At 2 months prior to admission, the patient had experienced a
sore throat with dysphagia. The subjective pain disappeared and the
sense of dysphagia was all alleviated following antibiotic
treatment. By the end of October 2014, a dry cough without sputum
plus stretch pain on the upper breast was noted. The patient was
diagnosed with pharyngitis in a local hospital and was treated
again with antibiotics. However, no improvement was obtained. The
patient was transferred to another hospital and lymph node
enlargement was detected in the bilateral cervical and bilateral
supraclavicular regions by ultrasonic examination. Visible blood
flow in these enlarged lymph nodes was detected by color Doppler
flow imaging. The patient then underwent an ultrasound-guided
fine-needle aspiration biopsy. A large number of lymphocytes with
nuclear granular chromatin and nuclear division were found. Chest
computed tomography (CT) revealed multiple enlarged lymph nodes in
the regions of the mediastinum, retroperitoneum and each side of
the neck. Positron emission tomography (PET)-CT scans detected
multiple lymphadenopathy at the bilateral cervical,
supraclavicular, thoracic entrance, mediastinum and post-peritoneal
areas, with increased fluorodeoxyglucose (FDG) uptake. Few
effusions were observed in right pleural cavity. The patient was
suspected as having non-Hodgkin's lymphoma and underwent a right
cervical lymph node biopsy.
Immunohistochemical (IHC) analysis was then used to
examine protein expression. The lymph node sample was fixed in 10%
formalin for >6 h at 25°C, then embedded in paraffin wax for
further hematoxylin and eosin (HE) and IHC staining. Thin sections
(4–5-µm thick) of paraffin-embedded tissue were adhered to slides.
To prevent non-specific binding of the antibody to the tissue, each
section was blocked with 1% rabbit serum (Abcam, Cambridge, UK.,
Cat. No. ab7487) for 10 min at 25°C. Primary antibodies (all
primary antibodies are listed below) were added to each section and
incubated for 30 min at 25°C, then Bond Polymer Refine Detection
reagent (Leica Biosystems, Newcastle, UK, cat. no. DS9800) was
added for 16 mins at 25°C. The sections were then incubated with
3,3′-Diaminobenzidine (DAB) system (Maxim Biotech, Inc., Rockville,
MD, USA; cat. no. DAB-1031) and then stained with 0.1% hematoxylin
for 5 min at 25°C. All slides were observed under a light
microscope.
The primary antibodies applied in diagnosis of this
patient: Anti-cluster of differentiation (CD)2 (cat. no. MAB-0207,
dilution 1:100), anti-CD3 (cat. no. Kit-0003, dilution 1:100),
anti-CD4 (cat. no. RMA-0620, dilution 1:100), anti-CD5 (cat. no.
Kit-0033, dilution 1:50), anti-CD7 (cat. no. RMA-0739, dilution
1:100), anti-CD8 (cat. no. RMA-0514, dilution 1:100), anti-CD10
(cat. no. MAB-0668, dilution 1:200), anti-CD20 (cat. no. Kit-0001,
dilution 1:600), anti-CD34 (cat. no. Kit-0004, dilution 1:400),
anti-CD43 (cat. no. MAB-0032, dilution 1:200), anti-CD56 (cat. no.
Kit-0028, dilution 1:200), anti-CD68 (cat. no. Kit-0026, dilution
1:400), anti-CD79a (cat. no. MAB-0258, dilution 1:200), anti-CD99
(cat. no. MAB-0059, dilution 1:200), anti-CD117 (cat. no. Kit-0029,
dilution 1:200), anti-Bcl-2 (cat. no. MAB-0014, dilution 1:100),
anti-chromogranin A (cat. no. MAB-0202, dilution 1:400),
anti-epithelial membrane antigen (cat. no. Kit-0011, dilution
1:200), anti-Epstein-Barr virus latent membrane protein 1 (cat. no.
MAB-0063, dilution 1:100), anti-Ki-67 (cat. no. MAB-0672, dilution
1:100), anti-myeloperoxidase polyclonal antibody (cat. no.
RAB-0379, dilution 1:400), anti-paired box protein (Pax-5) (cat.
no. MAB-0706, dilution 1:50), anti-thyroid transcription factor-1
(TTF-1) (cat. no. MAB-0599, dilution 1:200) (all Maxim Biotech,
Inc.) and anti-terminal deoxynucleotidyl transferase polyclonal
antibody (Ascend Biotechnology, Guangzhou, China, cat. no. AP0221,
dilution 1:100),
IHC staining results assessment: the cells without
brown color were determined as negative (−), the cells with light
brown color were determined as semi-positive (+/-), the cells shown
brown color were determined as positive (+).
Immunohistochemical analysis of the cervical lymph
node showed myeloperoxidase (MPO)-positive neoplastic cells that
were CD3−, CD4−, CD20−,
CD79a−, Epstein-Barr virus latent membrane protein
1−, paired box protein Pax-5−, epithelial
membrane antigen−, Ki-67+ (90%), terminal
deoxynucleotidyl transferase (TdT)−, CD7+/−,
CD10−, CD34−, CD117−,
CD5−, chromogranin A−, thyroid transcription
factor-1− and B-cell lymphoma-2+. BM aspirate
showed high lymphocytic proliferation with the existence of
prolymphocytes.
For flow cytometric analysis of cell membrane
antigens, bone marrow and hydrothorax cells were directly incubated
with antibodies [anti-CD2-Fluorescein isothiocyanate (FITC; cat.
no. 347593), anti-CD3-Phycoerythrin (PE; cat. no. 347347),
anti-CD3- Allophycocyanin (APC; cat. no. 340440), anti-CD5-FITC
(cat. no. 347303), anti-CD7-FITC (cat. no. 347483), anti-CD7-PE
(cat. no. 340581), anti-CD8-FITC (cat. no. 347313), anti-CD10-PE
(cat. no. 340921), anti-CD11b-APC (cat. no. 340937), anti-CD13-PE
(cat. no. 347837), anti-CD15-FITC (cat. no. 332778), anti-CD16-FITC
(cat. no. 335035), anti-CD22-FITC (cat. no. 347573), anti-CD25-FITC
(cat. no. 347643), anti-CD33-PE (cat. no. 347787), anti-CD34-APC
(cat. no. 340441), anti-CD38-APC (cat. no. 345807),
anti-CD45-Peridinin chlorophyll protein (PerCP) (cat. no. 347464),
anti-CD56-PE (cat. no. 347747), anti-CD64-PE (cat. no. 644385),
anti-CD79a-PE (cat. no. 340579), anti-MPO-FITC (cat. no. 340580),
anti-MPO-PE (cat. no. 341642), anti-HLA-DR-PE (cat. no. 347367),
anti-TdT-FITC (cat. no. 347194), anti-T cell receptor (TCR)-αβ-FITC
(cat. no. 347773), anti-TCR-γδ-PE (cat. no. 347907) (all BD
Biosciences)] for 15 min at 22°C in dark, then FACS lysing solution
(BD Biosciences, Franklin Lakes, NJ, USA, cat. no. 349202) was
added to lyse red blood cells. Following two washes with PBS, the
cells were resuspended and detected by FACSCanto II (BD
Biosciences). The corresponding isotype controls [Mouse IgG1-κ
Isotype control-FITC (cat. no. 555748), Mouse IgG1-κ Isotype
control-PE (cat. no. 555749), Mouse IgG1-κ Isotype control- PerCP
(cat. no. 559425), Mouse IgG1-κ Isotype control-APC (cat. no.
550854), Mouse IgG2a-κ Isotype control-FITC (cat. no. 553456),
Mouse IgG2a-κ Isotype control-APC (cat. no. 555576), Mouse IgM-κ
Isotype control-FITC (cat. no. 555583) (all BD Biosciences)] were
used to avoid non-specific binding of the mouse original antibodies
to sample cells in the analysis. FACS data were analyzed by BD
FACSDiva software v.8.0.1 (BD Biosciences).
Flow cytometric analysis of intracytoplasmic (cyCD3
and MPO) or intranuclear (terminal deoxynucleotidyl transferase)
antigens, bone marrow and hydrothorax cells were fixed and
permeabilized with the FIX & PERM kit (Caltag; Thermo Fisher
Scientific, Inc., Waltham, MA, USA; cat. no. GAS-003) according to
the manufacturer's protocol, and incubated with primary antibodies
for 15 min at 22°C in the dark. After washing twice with PBS, the
cells were resuspended and detected using a FACSCanto II flow
cytometer (BD Biosciences). FACS data were analyzed by BD FACSDiva
software v. 8.0.1 (BD Biosciences).
Flow cytometric analysis of BM cells revealed that
3% of the nuclear cells were blast cells, 0.9% of which were
myeloblasts with positive staining of CD34 and CD13. The rest of
the blast cells in the BM were immature T lymphocytes expressing
CD2, CD7, CD10, CD13, CD38 and cyCD3 (cytoplasmic CD3) (Fig. 1A). The patient was diagnosed with
myeloid sarcoma and transferred to Huashan Hospital (Shanghai,
China) for further diagnosis and treatment in December 2014.
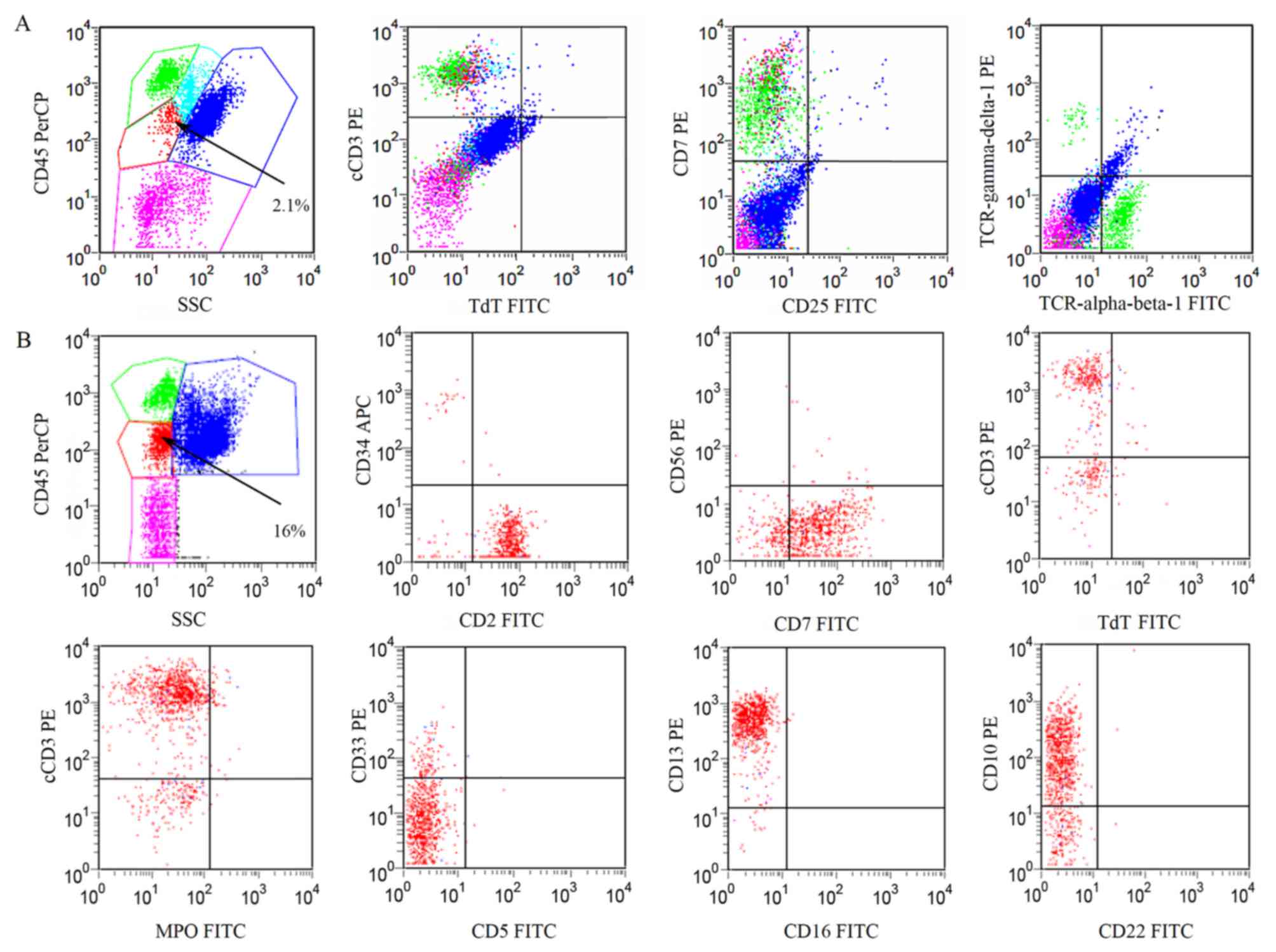 | Figure 1.Immunophenotype expression profile of
abnormal cells in the BM. Fluorescence-activated cell sorting
analysis indicated that phenotypically abnormal nucleated cells
(red dots) in the BM were present at (A) 2.1% in November 2014 and
(B) 16% in December 2014, with expression of CD2, CD7, CD10, CD13
and cyCD3, but without the expression of TCRα, TCRβ, TCRγ and TCRδ.
CD, cluster of differentiation; cyCD3, cytoplasmic CD3; MPO,
myeloperoxidase; TdT, terminal deoxynucleotidyl transferase; TCR, T
cell receptor; PerCP, peridinin chlorophyll protein complex; SSC,
side scatter channel; FITC, fluorescein isothiocyanate; PE,
phycoerythrin; APC, allophycocyanin; BM, bone marrow. |
Upon admission, the physical examination revealed a
body temperature of 36.5°C and a body weight of 78.5 kg. The
patient was found to exhibit superficial lymphadenopathy to a size
of 24×15 mm in the right neck region. The enlarged lymph nodes were
palpated and appeared tough, with no adhesion to the surrounding
tissue, and no broken skin or swelling of the local skin. The
patient exhibited no bleeding tendency and no hepatosplenomegaly.
Routine blood tests showed 15×109/l white blood cells
(WBCs) (normal range, 3.5×109-9.5×109/l),
consisting of 35% neutrophils (normal range, 51–75%), 2.0%
eosinophils (normal range, 0.5–5%), 38% monocytes (normal range,
3–8%) and 18% lymphocytes (normal range, 20–40%). The hemoglobin
level was 148 g/l (normal range, 130–175 g/l) and the platelet
count was 145×109/l (normal range,
125×109-350×109/l). The serum lactate
dehydrogenase (LDH) level was slightly increased at 284 IU/l
(normal range, 125–225 IU/l). A peripheral blood smear detected 39%
mature monocytes with abnormally shaped nuclei. No blast cells, but
metamyelocytes, promonocyte-like cells and prolymphoid cells were
observed in the PB smear at 1% of nucleated cells (Fig. 2A). A BM smear found markedly increased
nucleated cells at the percentage of 2% myeloblasts (normal range,
0–1%) and 2% prolymphoids (normal range, 0–1.5%) cells (Fig. 2B and C). Immunological analysis of the
BM revealed that the percentage of abnormal cells increased to 16%
of nucleated cells, with expression of CD2, CD7, CD10, CD13, CD38
and cyCD3 (Fig. 1B). Moderate right
pleural effusion was detected in the patient. The cellular
hydrothorax smear demonstrated a large number of lymphocytes and
monocytes with morphological variation (Fig. 2D). The cellular immunophenotypic
analysis of the pleural effusion indicated that there were two
populations of CD45 low-expression abnormal cells. One expressed
myeloid-associated antigens, including MPO, CD15, CD33, CD13 and
CD11bdim (Fig. 3A). The
other expressed T lymphoid-associated markers, including cyCD3, CD2
and CD7 (Fig. 3B). The two
populations were each found to have minimal co-expression of
markers specific to their counterpart. CyCD3+ cells
co-expressed myeloid marker CD13, while MPO+ cells
co-expressed lymphoid marker CD7. The two populations were
CD34− and CD117− (Fig. 3).
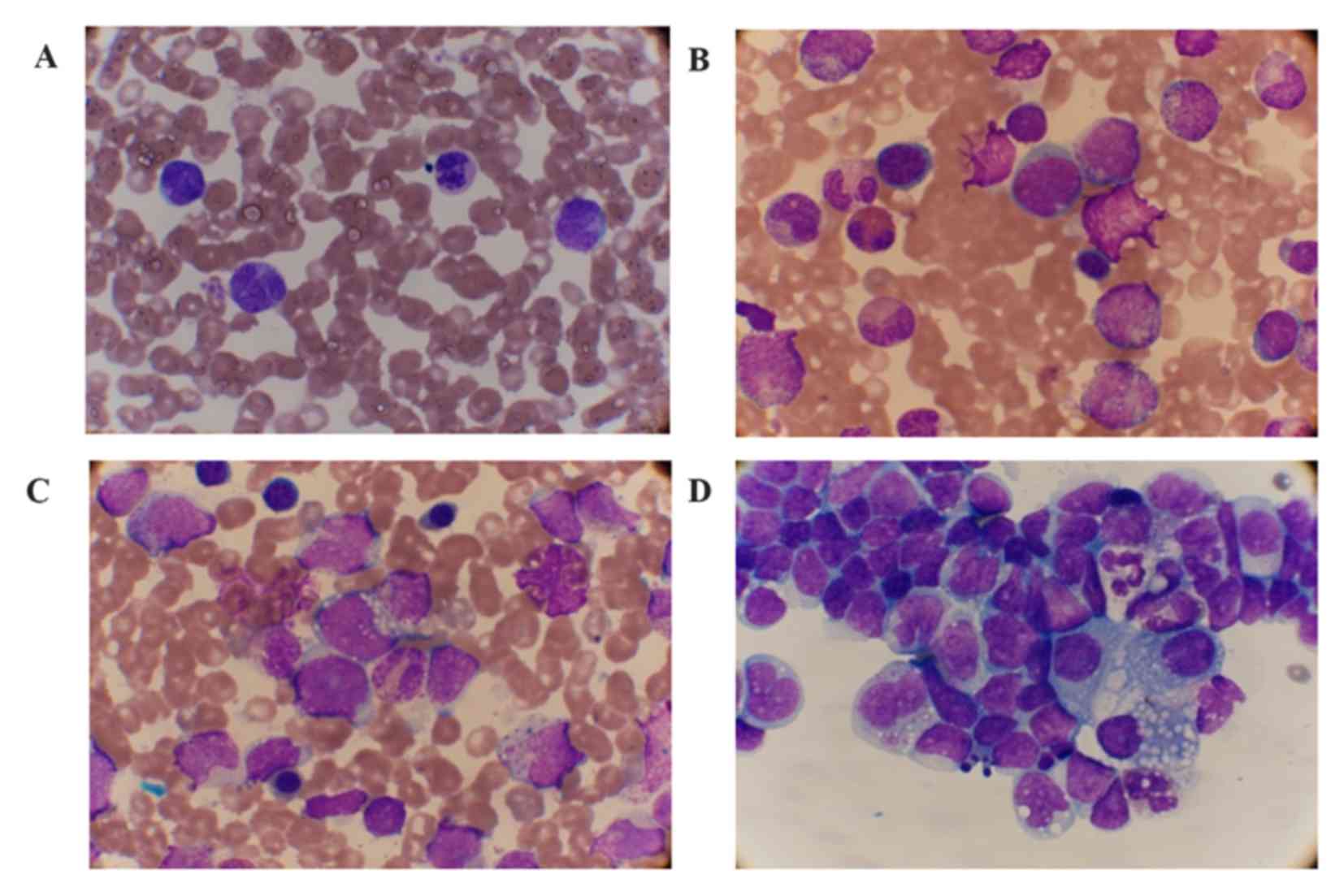 | Figure 2.Cellular morphology of the PB, BM and
hydrothorax smear (Wright stain). (A) Mature monocytes with
abnormal-shaped nuclei and 1% myelocytes and metamyelocytes were
observed in the PB (magnification, ×1,000). (B) 2% myeloblasts, 17%
promyelocytes and (C) 2% prolymphoid cells (arrow) were observed in
the BM (magnification, ×1,000). (D) Lymphocytes and monocytes with
morphological variation were observed in the smear of the
hydrothorax (magnification, ×1,000). PB, peripheral blood; BM, bone
marrow. |
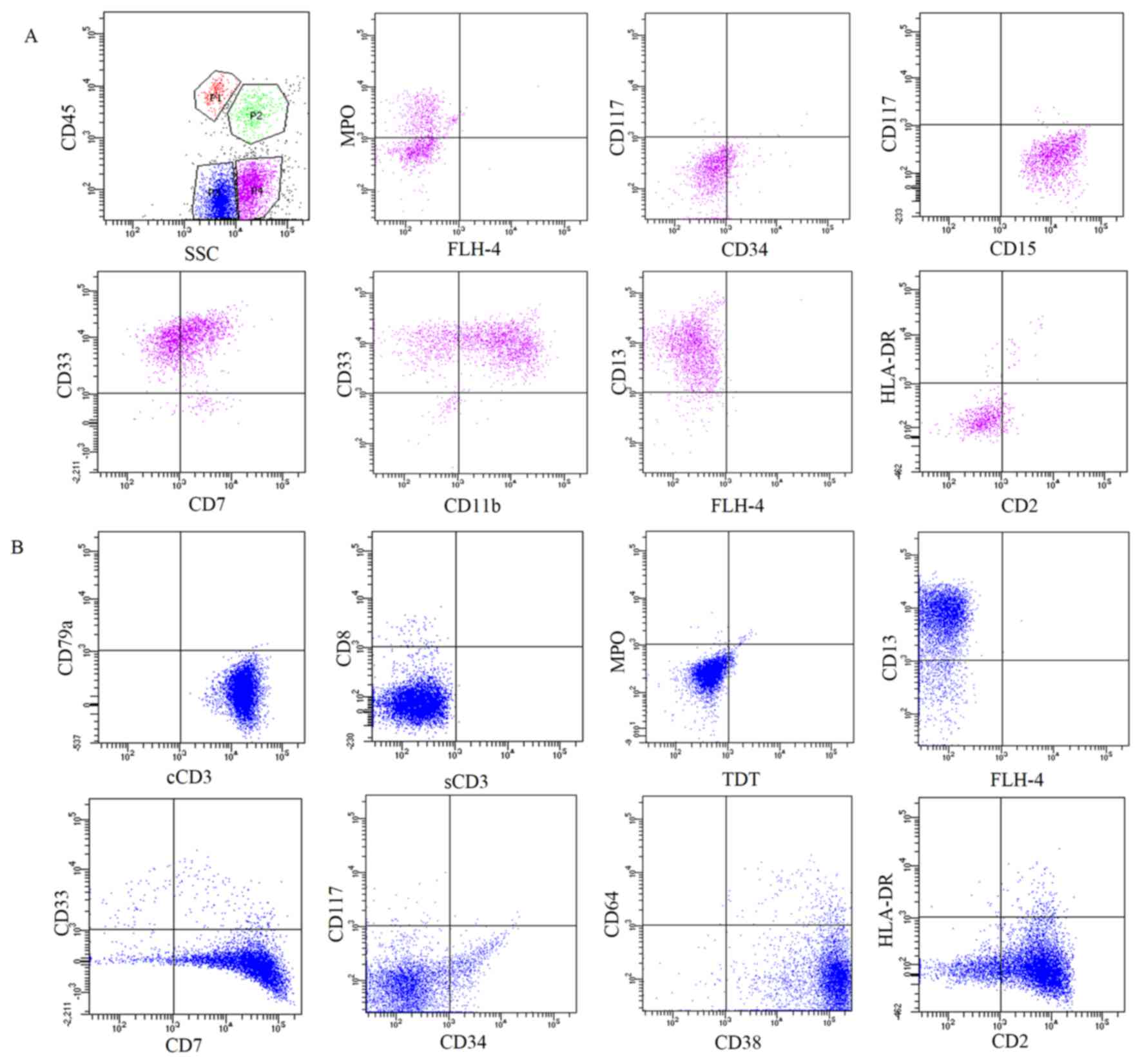 | Figure 3.Immunophenotype expression profile of
the cells in the thoracic fluid. Fluorescence-activated cell
sorting analysis of the cells in the pleural effusion indicated
that there were two populations of CD45 low-expression abnormal
cells. (A) One expressed myeloid-associated antigens, including
MPO, CD15, CD33, CD13 and CD11bdim. (B) The other
expressed T lymphoid-associated markers, including cyCD3, CD2 and
CD7. CD, cluster of differentiation; MPO, myeloperoxidase; cyCD3,
cytoplasmic CD3; HLA-DR, human leukocyte antigen-antigen D-related;
SSC, side scatter complex; FLH-4, fluorescence height-4, (no
antibody added). |
The pathological cervical lymph node sample was
reexamined. Coexistence of
CD3+/CD7+/CD2+ T lymphoid and
MPO+ myeloid neoplastic cells was revealed under the
microscope in the same region (Fig.
4A-D). Myeloid neoplastic cells showed pleomorphic nuclei and
distinct nucleoli with positive immunostaining for MPO. Lymphoid
neoplastic cells showed ovoid nuclei, finely dispersed chromatin
and inconspicuous nucleoli, with positive immunostaining of
lymphoid-associated marker, containing CD3, CD4, CD99, CD43 and TdT
(Fig. 4E-M). The boundary between two
populations was distinguishable by staining for MPO (Fig. 4A and N) and CD3 (Fig. 4D and G). The two populations exhibited
high expression of Ki-67 (Fig. 4O).
Cytogenetic analysis found that 1/20 of the examined BM cells
possessed the translocation t (12;13)(q10;p10), while the remainder
were of a normal karyotype. Investigation into genetic mutations
associated with acute leukemia, including AML12/ETO, PML/RARα,
PLZF/RARα, NPM/RARα, CBFβ/MYH11, TLS/ERG, DEK/CAN, NPM/MLF1,
dupMLL, MLL/AF6, MLL/AF9, MLL/AF10, MLL/AF17, MLL/ELL, EVI1, HOX11,
FIP1L1/PDGFRα, ETV6/PDGFRβ and BCR/ABL (forms P190, P210 and P230),
was negative in the BM. Detection of T-cell receptor (TCR)
rearrangement by polymerase chain reaction analysis and FGFR1
mutation by fluorescence in situ hybridization, as described
previously (17,18), detection in lymph node samples were
negative.
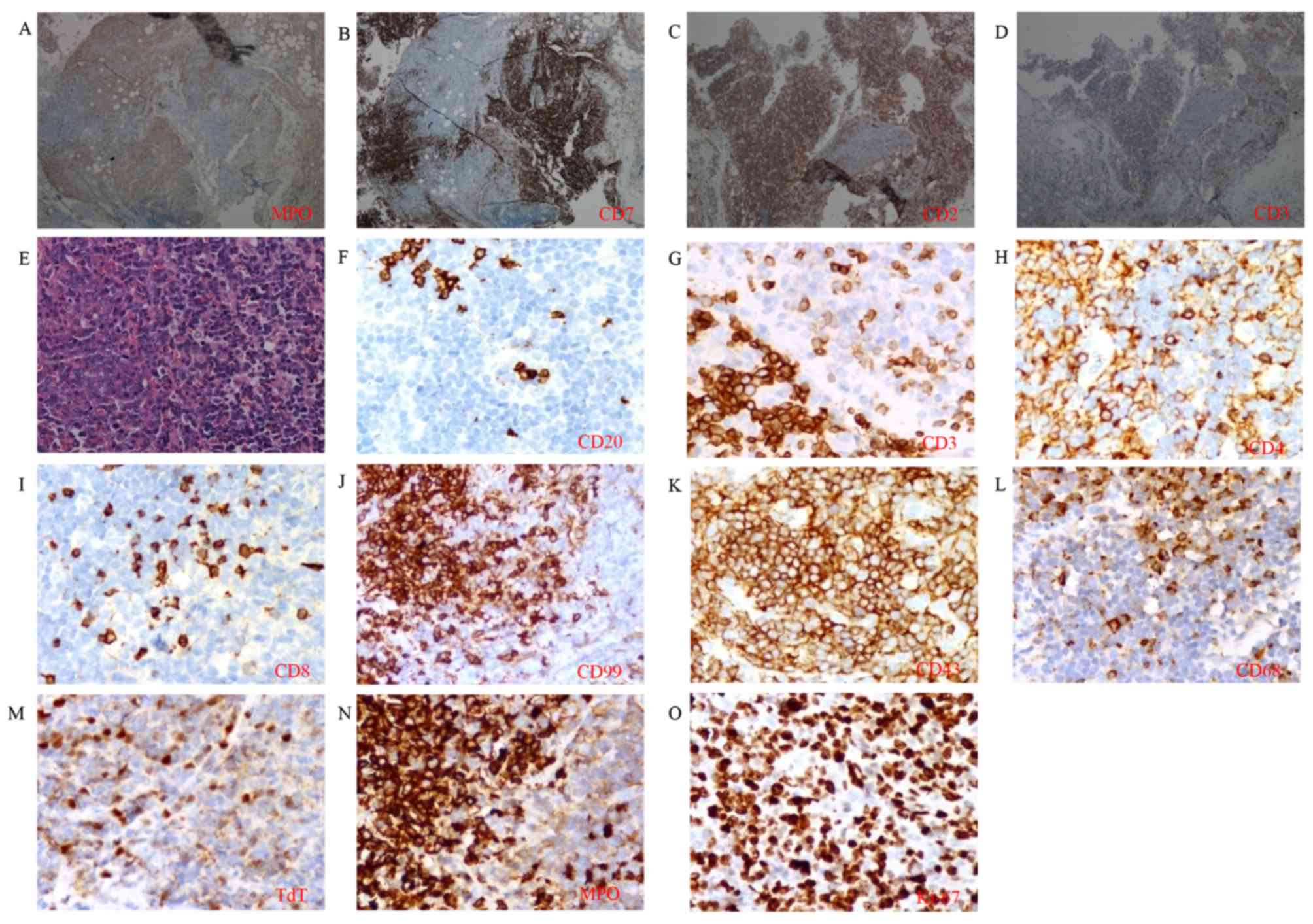 | Figure 4.Immunohistological analysis of the
cervical lymph node. Coexistence of T lymphoid and myeloid
neoplastic cells was found in the same region by staining for (A)
MPO, (B) CD7, (C) CD2 and (D) CD3 (magnification, ×100). (E)
Cellular morphology was distinguishable by hematoxylin and eosin
staining (magnification, ×400). Expression of (F) CD20, (G) CD3,
(H) CD4, (I) CD8, (J) CD99, (K) CD43, (L) CD68, (M) TdT, (N) MPO
and (O) Ki-67 at high magnification was detected (magnification,
×400). CD, cluster of differentiation; MPO, myeloperoxidase; TdT,
terminal deoxynucleotidyl transferase. |
For PCR, total RNA of the bone marrow sample was
extracted using RNAprep Pure Blood kit (Tiangen Biotech, Beijing,
China, cat. no. DP433), and reverse-transcribed to cDNA using the
Reverse Transcription System kit (Promega Corporation, Madison, WI,
USA), according to the manufacturer's protocol. For 20 µl PCR
system, 10 µl GoTaq GreenMaster mix (Promega Corporation), 1.5 µl
cDNA, 1.5 µl primer (primer sequences are listed in Table I) and 7 µl ddH2O was added.
Following PCR amplification (thermocycling conditions are listed in
Table I) of the target gene, gene
expression was analyzed by DNA gel electrophoresis. Total DNA of
paraffin-embedded lymph node tissue sample was extracted using a
QIAamp DNA Mini kit (Qiagen GmbH, Hilden, Germany). For 20 µl PCR
system, 10 µl GoTaq GreenMaster mix (Promega Corporation), 2 µl
DNA, 2 µl primer mix and 6 µl dH2O were added. Following
PCR amplification of the target gene, the gene expression was
analyzed by DNA gel electrophoresis.
 | Table I.Primer sequences and thermocycling
conditions of fusion gene detection by PCR. |
Table I.
Primer sequences and thermocycling
conditions of fusion gene detection by PCR.
|
|
| Primer
sequence |
|
|---|
|
|
|
|
|
|---|
| Fusion gene | Round | Forward | Reverse | Thermocycling
condition |
|---|
| AML12/ETO | 1 | 5′-
CTACCGCAGCCATGAAGAACC −3 |
5′-AGAGGAAGGCCCATTGCTGAA −3′ | 94°C 60 sec→55°C 50
sec→72°C 60 sec, ×30 cycles |
|
| 2 | 5′-
ATGACCTCAGGTTTGTCGGTCG −3′ | 5′-
TGAACTGGTTCTTGGAGCTCCT-3′ | 94°C 60 sec→55°C 50
sec→72°C 60 sec, ×30 cycles |
| PML/RARα | 1 | 5′-
CAGTGTACGCCTTCTCCATCA −3′ |
5′-GCTTGTAGATGCGGGGTAGA −3′ | 94°C 60 sec→55°C 50
sec→72°C 60 sec, ×30 cycles |
|
| 2 | 5′-
TCAAGATGGAGTCTGAGGAGG −3′ | 5′-
CTGCTGCTCTGGGTCTCAAT −3′ | 94°C 60 sec→55°C 50
sec→72°C 60 sec, ×30 cycles |
| TLS/ERG | 1 |
5′-CTATGGACAGCAGGACCGTG-3′ | 5′
-CATAGTAGTAACGGAGGGCG-3′ | 94°C 60 sec→60°C 60
sec→72°C 60 sec, ×40 cycles |
|
| 2 |
5′-GGTGGCTATGAACCCAGAGG-3′ |
5′-CCTCGTCGGGATCCGTCATC-3′ | 94°C 60 sec→60°C 60
sec→72°C 60 sec, ×25 cycles |
| DEK/CAN | 1 |
5′-CCTACAGATGAAGAGTTAA-3′ | 5′
-TCTTCCTCTGTTGGTTGATG-3′ | 94°C 60 sec→56°C 60
sec→72°C 90 sec, ×30 cycles |
|
| 2 |
5′-GGCCAGTGCTAACTTGG-3′ |
5′-GTGTCTCTCGCTCTGG-3′ | 94°C 60 sec→50°C 45
sec→72°C 2 min, ×25 cycles |
| DupMLL | 1 |
5′-GTCGAAGTGGAAGAGGGAAAAG-3′ | 5′
-GGATAGTCTTCGCTCTTCATGAACA-3′ | 95°C 30 sec→59°C 30
sec→72°C 30 sec, ×20 cycles |
|
| 2 |
5′-AAGCTTCACTCTGACCATCACTGT −3′ |
5′-GTGTCTCTCGCTCTGG-3′ | 95°C 30 sec→59°C 30
sec→72°C 20 sec, ×29 cycles |
| MLL/AF6 | 1 |
5′-CCTACAGATGAAGAGTTAA-3′ | 5′
-TCTTCCTCTGTTGGTTGATG-3′ | 95°C 30 sec→59°C 30
sec→72°C 30 sec, ×20 cycles |
|
| 2 |
5′-GGCCAGTGCTAACTTGG-3′ |
5′-GTGTCTCTCGCTCTGG-3′ | 95°C 30 sec→59°C 30
sec→72°C 20 sec, ×29 cycles |
| MLL/AF9 | 1 |
5′-GTTCCCGATATGGATGATGAAGAAG-3′ | 5′
-TCGTGATCCCACTCCAAGTCT-3′ | 95°C 30 sec→59°C 30
sec→72°C 30 sec, ×20 cycles |
|
| 2 |
5′-AGGAAGGATTAGAAGATATTGACGAAGA-3′ |
5′-CATCTTGGACAGCAAATTTCACA −3′ | 95°C 30 sec→59°C 30
sec→72°C 20 sec, ×29 cycles |
| MLL/AF10 | 1 |
5′-CCATCTTCCAGGAGCGAGATC-3′ | 5′
-TGTCATCATATTTGGCAGGTTTTT-3′ | 95°C 30 sec→59°C 30
sec→72°C 30 sec, ×20 cycles |
|
| 2 |
5′-GTCGAAGTGGAAGAGGGAAAAG-3′ |
5′-GTGAGGAAGCGCTTGAGCTT-3′ | 95°C 30 sec→59°C 30
sec→72°C 20 sec, ×29 cycles |
| MLL/AF17 | 1 |
5′-CACCCCAGGAACCTCCAGTA-3′ | 5′
-TTGTCGCGGCCGATGT-3′ | 95°C 30 sec→59°C 30
sec→72°C 30 sec, ×20 cycles |
|
| 2 |
5′-CCAAGTTTGGTGGTCGCAAT-3′ |
5′-GTTGGAGAGGTAGAAGGAGAACGT-3′ | 95°C 30 sec→59°C 30
sec→72°C 20 sec, ×29 cycles |
| MLL/ELL | 1 |
5′-CACCCCAGGAACCTCCAGTA-3′ | 5′
-GCTCTTGCTCTCGGAGTCTTTG-3′ | 95°C 30 sec→59°C 30
sec→72°C 30 sec, ×20 cycles |
|
| 2 |
5′-AGTAAAGAAAGGACGTCGATCGA-3′ |
5′-GTCACCTTGTGTGGCTTGGA-3′ | 95°C 30 sec→59°C 30
sec→72°C 20 sec, ×29 cycles |
| BCR/ABL | 1 |
5′-GCAGCAGAAGAAGTGTTTCAG-3′ |
5′-GTGATTATAGCCTAAGACCCG-3′ | 94°C 60 sec→55°C 50
sec→72°C 60 sec, ×30 cycles |
|
| 2 |
5′-GGAGCTGCAGATGCTGACCAAC-3′ |
5′-TCAGACCCTGAGGCTCAAAGTC-3′ | 94°C 60 sec→55°C 50
sec→72°C 60 sec, ×30 cycles |
| PLZF/RARα | 1 |
5′-GTGGGCATGAAGTCAGAGAGC-3′ | 5′
-TGGATGCTGCGGCGGAAGAAG-3′ | 95°C 30 sec→58°C 30
sec→72°C 60 sec, ×25 cycles |
|
| 2 |
5′-GTGGGCATGAAGTCAGAGAGC-3′ |
5′-GGTCACCTTGTTGATGATGCAG-3′ | 95°C 30 sec→58°C 30
sec→72°C 60 sec, ×25 cycles |
| NPM/RARα | 1 |
5′-GTTGCACATTGTTGAAGCAGAGG-3′ | 5′
-TGGATGCTGCGGCGGAAGAAG-3′ | 95°C 30 sec→58°C 30
sec→72°C 60 sec, ×25 cycles |
|
| 2 |
5′-ACGAAGGCAGTCCAATTAAAGTAAC-3′ |
5′-GGTCACCTTGTTGATGATGCAG-3′ | 95°C 30 sec→58°C 30
sec→72°C 60 sec, ×25 cycles |
| CBFβ/MYH11 | 1 |
5′-TTTGAAGGCTCCCATGATTCTG-3′ | 5′
-GAGCTGGATGTTGAGAGTGGAGAT-3′ | 95°C 30 sec→58°C 30
sec→72°C 60 sec, ×25 cycles |
|
| 2 |
5′-TGGGCTGTCTGGAGTTTGATG-3′ |
5′-AGGTCCCCTTCCAGCTTCTTCT-3′ | 95°C 30 sec→58°C 30
sec→72°C 60 sec, ×25 cycles |
| EVI1 | 1 |
5′-CCACTAAGCGAAAGGATGAGAAG-3′ | 5′
-CGTCGAATCAAGACCTGCTTC-3′ | 95°C 30 sec→58°C 30
sec→72°C 60 sec, ×25 cycles |
|
| 2 |
5′-TGCCGTGTTAGGTTTGCAGAC-3′ |
5′-GAACATAGAGGGCACTGACTGTAAG-3′ | 95°C 30 sec→58°C 30
sec→72°C 60 sec, ×25 cycles |
| HOX11 | 1 |
5′-GGGCGTCAACAACCTCACTG-3′ | 5′
-CTTCCCCTGGATGGAGAGTAAC-3′ | 95°C 30 sec→58°C 30
sec→72°C 60 sec, ×25 cycles |
|
| 2 |
5′-GTCTGCCGTCTCCACTTTGTC-3′ |
5′-GCGCATCGGTCATTTTGAG-3′ | 95°C 30 sec→58°C 30
sec→72°C 60 sec, ×25 cycles |
| FIP1L1/PDGFRα | 1 |
5′-GATACCTGGAAAGCTTACTGTG-3′ | 5′
-TTGGTGCAGGCTCCCAGCAAG-3′ | 95°C 30 sec→58°C 30
sec→72°C 60 sec, ×25 cycles |
|
| 2 |
5′-CCTTCTTTGTTCAAGACTGGGC-3′ |
5′-TGATGATGTAAATGGGGCCTGAC-3′ | 95°C 30 sec→58°C 30
sec→72°C 60 sec, ×25 cycles |
| ETV6/PDGFRβ | 1 |
5′-GTGCTCTATGAACTCCTTCAGC-3′ | 5′
-CATAAGGGCTTGCTTCTCACTG-3′ | 95°C 30 sec→58°C 30
sec→72°C 60 sec, ×25 cycles |
|
| 2 |
5′-ACACGCGTGATCCAGCTGATG-3′ |
5′-CATGGGGTCCACGTAGATGTAC-3′ | 95°C 30 sec→58°C 30
sec→72°C 60 sec, ×25 cycles |
| NPM/MLF1 | 1 |
5′-GTTGCACATTGTTGAAGCAGAGG-3′ | 5′
-CATAAGGGCTTGCTTCTCACTG-3′ | 95°C 30 sec→58°C 30
sec→72°C 60 sec, ×25 cycles |
|
| 2 |
5′-ACACGCGTGATCCAGCTGATG-3′ |
5′-CATGGGGTCCACGTAGATGTAC-3′ | 95°C 30 sec→58°C 30
sec→72°C 60 sec, ×25 cycl |
The patient was diagnosed with a hemato-lymphoid
neoplasm showing separate differentiation toward T lymphoblastic
and myeloid lineage, and was administered a combined induction
chemotherapy regimen (chemotherapy cycle 1: 4 mg/day vindesine
intravenous on day 1, 10 mg/day idarubicin intravenous on days 1–3,
180 mg/day cytarabine intravenous on days 1–7 and 80 mg/day
methylprednisolone intravenous on days 1–7) in December 2014.
Subsequently, 1 day after the final dose of chemotherapy, the chest
CT scan indicated that the pleural effusion was significantly
reduced. The superficial lymph nodes shrank and the cough symptom
improved. The number of WBCs and the proportion of monocytes
decreased to 6.99×109/l and 4%, respectively. The
patient then received chemotherapy of hyper-CVAD A regimen
(chemotherapy cycle 2: 1 g/day cyclophosphamide intravenous on days
1–3, 4 mg/day vindesine intravenous on days 4 and 11, 15 mg/day
idarubicin intravenous on days 4, 40 mg/day dexamethasone
intravenous on days 1–4 and 11–14), hyper-CVAD B regimen
(chemotherapy cycle 3: 1.8 g/day methotrexate intravenous on day 1,
6 g/day cytarabine intravenous on days 2–3) and the induction
regimen (chemotherapy cycle 4: identical to cycle 1) repeatedly as
consolidation treatment. Immediately prior to the 4th cycle of
chemotherapy, PET-CT was reviewed and all the enlarged lymph nodes
had normalized. A hypermetabolic lesion in the mediastinum remained
detectable, but at a markedly decreased level compared with that
previously. The proportion of abnormal T cells in the BM was
reduced to 0.3%. The matched unrelated allogeneic hematopoietic
stem cell transplantation (allo-HSCT) was postponed on account of
invasive Cryptococcus infection in the right lung. However,
in late June 2015, the neoplasm relapsed with leukemic symptoms;
24% of abnormal T cells and 44% of CD45− myeloid blasts
were detected in the BM, with infiltrations in the cervical lymph
nodes and the posterior pharyngeal wall. The repeat use of the
initial induction regimen and more intensive regimen (4 mg/day
vindesine intravenous on days 1 and 8, 12 mg/day mitoxantrone
intravenous on days 1–3, 1 g/day cytarabine intravenous on days 1,
3 and 5, and methylprednisolone intravenous 60 mg/day on days 1–9,
40 mg/day on days 10–15 and 3,750 U peg-asparaginase intramuscular
on day 15), all failed. The patient succumbed in mid-September
2015.
The requirement for patient consent for publication
was waived by the Institutional Review Board of Huashan
Hospital.
Discussion
The patient in the current study presented with
extra-medullary infiltration as the initial symptom. Notably, two
distinct neoplastic populations, cyCD3+ T lymphoid cells
and MPO+ myeloid cells, were detected in the pleural
effusion and lymph nodes. None of the cells expressed the
pluripotent markers CD34 or CD117. A certain minimal degree of
cross-expression of markers specific to their lineage counterpart
was detected. CyCD3+ T lymphoid cells coexpressed the
myeloid antigen CD13, while MPO+ myeloid lineage cells
coexpressed lymphoid marker CD7. However, the fact that none of the
populations at the initial stage fitted the diagnostic criteria of
acute leukemia within either the BM or the PB raised a difficulty
in providing an accurate diagnosis (1,4). To
clarify the origin of the neoplastic cells and the differentiation
stage of mutational occurrence should assist in forming an accurate
diagnosis and choosing the therapeutic strategy.
The symmetric model of lineage commitment
differentiation from HSCs, in which clear division of lymphoid and
myeloid commitment is the first step of lineage restriction, used
to be widely accepted. In this model, common myeloid progenitors
(CMPs) and common lymphoid progenitors (CLPs) are symmetrically
derived from the same multipotent progenitors (MPPs). CMPs then
gradually differentiate into all types of myeloid offspring,
including granulocytes, monocytes, erythrocytes and megakaryocytes.
Similarly, CLPs differentiate into mature lymphoid cells, including
T cells, B cells and natural killer cells (19).
However, previous studies indicated that HSCs
differentiate into lymphoid lineage and myeloid lineages
asymmetrically. All MPPs subsets contribute to lymphoid lineage
differentiation. The most primitive fms-like tyrosine kinase
3lo/vascular cell adhesion protein 1+
(Flt3loVCAM-1+) MPPs were indicated to be
able to give rise to CLPs and CMPs. However, the most advanced
Flt3hiVCAM-1− MPPs only gave rise to CLPs
(20,21), suggesting that lymphoid
differentiation occupies the backbone of the differentiation
process, whereas myeloid differentiation is a lateral branch. Two
other stages specified as granulocyte/macrophage-lymphoid bipotent
progenitors and lymphoid-specified progenitors sit between MPPs and
CLPs (22). This model provides
certain clues to explain the occurrence of T/My
bilineage/biphenotype malignancies.
Thymic T lymphocytes mature from
CD4−CD8− double-negative (DN) cells to
CD4+CD8+ double-positive (DP) cells, then to
the fully mature CD4 or CD8 single positive (SP) cells. The DN
stage can be subdivided into 4 substages depending on the lack of
CD4 and CD8 surface expression and differential expression of CD25
and CD44: CD44+CD25− (DN1),
CD44+CD25+ (DN2),
CD44−CD25+ (DN3), and
CD44−CD25− (DN4). In addition, the
differential expression of CD117 in DN2 cells enabled the
establishment of two further subsets, DN2a
(CD4−CD8−CD44+CD25+CD117hi)
and DN2b
(CD4−CD8−CD44+CD25+CD117int)
(23,24). In the thymic T cell maturation
process, the myeloid differentiation potential is retained until
the middle of the DN2 stage, while the rearrangement of TCR starts
from the DN2b stage (25). Meanwhile,
the cellular phenotypes were orchestrated with different maturation
stages.
In the 1980s and the 1990s, a class of T-ALL with
myeloid differentiation features, known as T stem cell
leukemia/lymphoma (T-SCL/lymphoma) was reported in the literature
(26–30). T-SCL/lymphoma cells are
CD7+/CD4−/CD8−, representing a
portion of T lymphocytes that are going to migrate from the BM to
the thymus (29). This indicated that
the neoplastic cells of T-SCL/lymphoma originated from the HSCs,
which maintained the ability to differentiate into lymphoid and
myeloid lineages (26,30). In almost all the reported
T-SCL/lymphoma cases, the patients presented with a giant mass in
the mediastinum or lymph nodes, with coexpression of T lymphoid and
myeloid phenotypes on blast cells (27,29).
However, with molecular and immunological progress in the studies
of hematopoietic diseases, the diagnostic criteria depending only
on the expression of CD7, CD4 and CD8 were far from accurate.
In the 2008 WHO classification (2), T-precursor neoplasms were classified
into pro-T, pre-T, cortical T and medullary T subtypes according to
the neoplastic phenotypes. Expression of cyCD3, CD34, TdT, surface
CD3 (sCD3), CD4 and CD8 can assist in distinguishing between T
subtype differentiation stages (31).
However, the origin of T/My MPALs remains unclarified.
Since 2009, a novel subtype of T-ALL, early T-cell
precursor-acute lymphoblastic leukemia (ETP-ALL), has been
reported. ETP-ALL is a high-risk subset and comprises ~15% of all
T-ALL cases (32). ETPs represent
thymocytes recently immigrating from the BM to the thymus and thus
retaining the multilineage differentiation potential (33–35). ETPs,
immunophenotypically characterized as CD5−/weak,
CD1a− and CD8−, with positive expression of
myeloid markers (human leukocyte antigen-antigen D-related, CD13,
CD33, CD11b or CD65) and stem cell markers (CD34 and CD117)
(36), were indicated to be at the
DN1 differentiation stage (25).
With the latest understanding of the T lymphoid
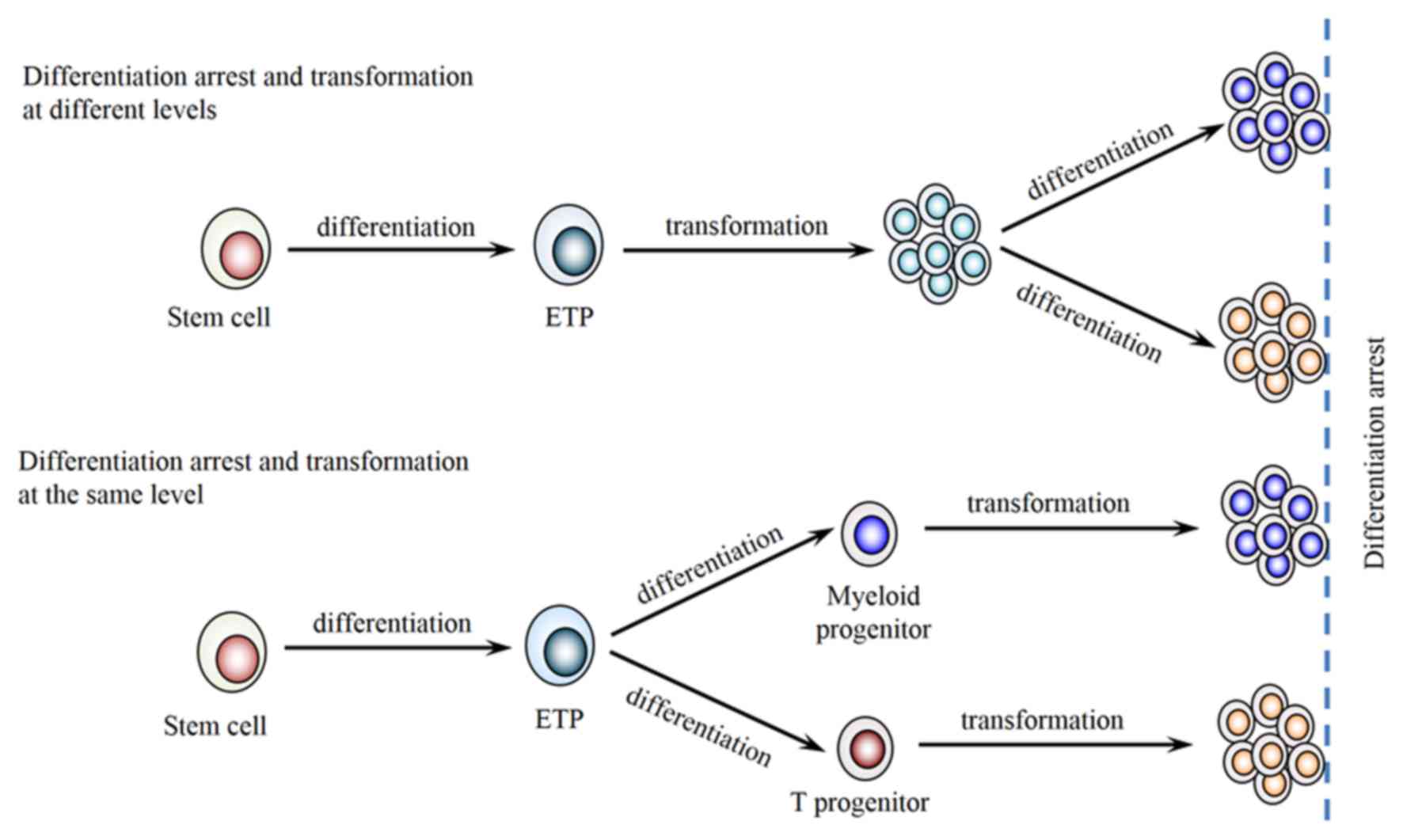
differentiation process, we hypothesized that the neoplastic cells
in the present case were transformed from early DN2a stage, which
retained further differentiation potential into T lymphoid and
myeloid lineages, resulting in two distinct populations in the same
extramedullary site (Fig. 5). The
patient was thus diagnosed with a hemato-lymphoid neoplasm showing
separate differentiation toward T lymphoblastic and myeloid
lineages. The disease progressed to develop leukemic symptoms at
relapse, where blasts were detected in the BM and extramedullary
site. However, there remains a lack of accurate nomenclature to
name this type of T/My bilineage malignancy with extramedullary
infiltration at the initial stage. The pathological development
mechanism of this hematological malignant entity requires further
investigation.
In treating bilineal or biphenotypic MPALs, there
are no generally accepted regimens of induction chemotherapy that
can cover the lymphoid and/or the myeloid lineage. In a previous
study, 20 cases of MPAL, including 1 case of T/My bilineage
leukemia, were reported to be treated with prednisone, vincristine,
L-asparaginase and daunorubicin, and successfully achieved complete
remission after initial induction therapy (7). The fludarabine, cytarabine and
idarubicine protocol (9) and
ALL-based induction chemotherapy (10) were previously reported as used in
treating MPALs in different medical centers. However, it has been
indicated that a large proportion of MPALs are resistant and
refractory to conventional chemotherapy. Allo-HSCT, particularly
early in the disease course, is thus far the best treatment
strategy (37). Allo-HSCT was delayed
in the current patient due to invasive lung Cryptococcus
infection. The disease relapsed and progressed rapidly, and the
patient did not obtain the opportunity to achieve repeat
remission.
In conclusion, the present case initially showed
extramedullary infiltration of distinct T lymphoid and myeloid
populations beyond the diagnosis of leukemia in the PB or in the
BM. The disease progressed to leukemia at relapse stage, indicating
the very early stage of T/My bilineage MPALs. Clarification of the
origin of the neoplastic cells and the differentiation stage of
mutational occurrence is crucial to choose the appropriate
treatment strategy.
Acknowledgements
The authors would like to thank Kindstar Global
Corporation (Shanghai, China) for performing PCR and FISH
experiments. The present study was partially supported by the
National Natural Science Foundation of China (grant nos 31371480
and 91542109) and the Foundation of the Science and Technology
Commission of Shanghai Municipality (grant no. 16XD1400600).
References
|
1
|
Weinberg OK and Arber DA: Mixed-phenotype
acute leukemia: Historical overview and a new definition. Leukemia.
24:1844–1851. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Swerdlow SH, Campo E, Harris NL, Jaffe ES,
Pileri SA, Stein H, Thiele J and Vardiman JW: WHO Classification of
Tumours of Haematopoietic and Lymphoid Tissues. 4th edition. IARC;
Lyon: pp. 150–155. 2008
|
|
3
|
Béné MC and Porwit A: Acute leukemias of
ambiguous lineage. Semin Diagn Pathol. 29:12–18. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Béné MC: Biphenotypic, bilineal, ambiguous
or mixed lineage: Strange leukemias! Haematologica. 94:891–893.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Nishiuchi T, Ohnishi H, Kamada R, Kikuchi
F, Shintani T, Waki F, Kitanaka A, Kubota Y, Tanaka T and Ishida T:
Acute leukemia of ambiguous lineage, biphenotype, without CD34, TdT
or TCR-rearrangement. Intern Med. 48:1437–1441. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tota G, Coccaro N, Zagaria A, Anelli L,
Casieri P, Cellamare A, Minervini A, Minervini CF, Brunetti C,
Impera L, et al: ADAMTS2 gene dysregulation in T/myeloid mixed
phenotype acute leukemia. BMC Cancer. 14:9632014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bachir F, Zerrouk J, Howard SC, Graoui O,
Lahjouji A, Hessissen L, Bennani S, Quessar A and El Aouad R:
Outcomes in patients with mixed phenotype acute leukemia in
Morocco. J Pediatr Hematol Oncol. 36:e392–e397. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sharma P, Lall M, Jain P, Saraf A,
Sachdeva A and Bhargava M: A Bi-Lineal acute leukemia (T/Myeloid,
NOS) with complex cytogenetic abnormalities. Indian J Hematol Blood
Transfus. 29:119–122. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Colovic M, Colovic N, Jankovic G, Kurtovic
Kraguljac N, Vidovic A, Djordjevic V and Bogdanovic A: Mixed
phenotype acute leukemia of T/myeloid type with a prominent
cellular heterogeneity and unique karyotypic aberration 45,XY,
dic(11;17). Int J Lab Hematol. 34:290–294. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Oliveira JL, Kumar R, Khan SP, Law ME,
Erickson-Johnson M, Oliveira AM, Ketterling RP and Dogan A:
Successful treatment of a child with T/myeloid acute bilineal
leukemia associated with TLX3/BCL11B fusion and 9q deletion.
Pediatr Blood Cancer. 56:467–469. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Weir EG, Ali Ansari-Lari M, Batista DA,
Griffin CA, Fuller S, Smith BD and Borowitz MJ: Acute bilineal
leukemia: A rare disease with poor outcome. Leukemia. 21:2264–2270.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kobayashi N, Matsuda K, Sakashita K,
Matsuzaki S, Iwasaki R and Koike K: Bilineage acute leukemia of
T-lymphoid and myeloid lineages. Haematologica. 89:1139–1141.
2004.PubMed/NCBI
|
|
13
|
Akashi K, Shibuya T, Harada M, Morioka E,
Oshima K, Kimura N, Takeshita M, Kurokawa M, Kikuchi M and Niho Y:
Acute ‘bilineal-biphenotypic’ leukaemia. Br J Haematol. 74:402–407.
1990. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Akashi K, Harada M, Shibuya T, Morioka E,
Okamura T, Asano Y, Taniguchi S, Teshima T, Kikuchi M and Niho Y:
Clinical characteristics of hybrid leukemia: Report of five cases.
Leuk Res. 14:145–153. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Licci S, Canal F, Tos Dei AP, Fedrigo M,
Gherlinzoni F, Brenna A, Zanatta L and Rossi S: Aleukemic
granulocytic sarcoma with associated T-cell lymphoblastic lymphoma
in the same lymph node: Morphologic features and molecular
signatures. Leuk Lymphoma. 49:1411–1415. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Schwarz J, Trnková Z, Bedrlíková R,
Jirásek A, Záková D, Trnený M, Sedlácková E, Kodetová D, Valenta J,
Stöckbauer P, et al: Aleukemic granulocytic sarcoma with AML1/ETO
fusion gene expression and clonal T cell populations. Leuk Res.
25:1137–1142. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
van Dongen JJ, Langerak AW, Brüggemann M,
Evans PA, Hummel M, Lavender FL, Delabesse E, Davi F, Schuuring E,
García-Sanz R, et al: Design and standardization of PCR primers and
protocols for detection of clonal immunoglobulin and T-cell
receptor gene recombinations in suspect lymphoproliferations:
Report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia.
17:2257–2317. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Patnaik MM, Gangat N, Knudson RA, Keefe
JG, Hanson CA, Pardanani A, Ketterling RP and Tefferi A: Chromosome
8p11.2 translocations: Prevalence, FISH analysis for FGFR1 and
MYST3, and clinicopathologic correlates in a consecutive cohort of
13 cases from a single institution. Am J Hematol. 85:238–242. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lai AY and Kondo M: T and B lymphocyte
differentiation from hematopoietic stem cell. Semin Immunol.
20:207–212. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lai AY, Lin SM and Kondo M: Heterogeneity
of Flt3-expressing multipotent progenitors in mouse bone marrow. J
Immunol. 175:5016–5023. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lai AY and Kondo M: Asymmetrical lymphoid
and myeloid lineage commitment in multipotent hematopoietic
progenitors. J Exp Med. 203:1867–1873. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Adolfsson J, Månsson R, Buza-Vidas N,
Hultquist A, Liuba K, Jensen CT, Bryder D, Yang L, Borge OJ, Thoren
LA, et al: Identification of Flt3+ lympho-myeloid stem cells
lacking erythro-megakaryocytic potential a revised road map for
adult blood lineage commitment. Cell. 121:295–306. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Koch U and Radtke F: Mechanisms of T cell
development and transformation. Annu Rev Cell Dev Biol. 27:539–562.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ceredig R and Rolink T: A positive look at
double-negative thymocytes. Nat Rev Immunol. 2:888–897. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kawamoto H, Ikawa T, Masuda K, Wada H and
Katsura Y: A map for lineage restriction of progenitors during
hematopoiesis: The essence of the myeloid-based model. Immunol Rev.
238:23–36. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nagano M, Kimura N, Akiyoshi T, Nishimura
J, Kozuru M, Okamura J, Katsuno M, Yoshida T, Takeshita M,
Tachibana K, et al: T-stem cell leukemia/lymphoma with both myeloid
lineage conversion and T-specific delta recombination. Leuk Res.
21:763–773. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Katsuno M, Abe Y, Taguchi F, Yufu Y,
Sadamura S, Goto T, Takatsuki H, Nishimura J, Hirata J, Akiyoshi T,
et al: CD7+ stem cell leukemia/lymphoma. Features of a subgroup
without circulating blast cells. Cancer. 72:99–104. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Silva ML, de Oliveira MS, Valente AN,
Abdelhay E, Bouzas LF, Laun L and Ribeiro RC: CD7+, CD4-/CD8- acute
leukemia with t(11;14)(p15;q11) in a child. Cancer Genet Cytogenet.
56:171–176. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kurtzberg J, Waldmann TA, Davey MP, Bigner
SH, Moore JO, Hershfield MS and Haynes BF: CD7+, CD4-, CD8- acute
leukemia: A syndrome of malignant pluripotent lymphohematopoietic
cells. Blood. 73:381–390. 1989.PubMed/NCBI
|
|
30
|
Hershfield MS, Kurtzberg J, Harden E,
Moore JO, Whang-Peng J and Haynes BF: Conversion of a stem cell
leukemia from a T-lymphoid to a myeloid phenotype induced by the
adenosine deaminase inhibitor 2′-deoxycoformycin. Proc Natl Acad
Sci USA. 81:pp. 253–257. 1984, View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Onciu M: Acute lymphoblastic leukemia.
Hematol Oncol Clin North Am. 23:655–674. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang J, Ding L, Holmfeldt L, Wu G,
Heatley SL, Payne-Turner D, Easton J, Chen X, Wang J, Rusch M, et
al: The genetic basis of early T-cell precursor acute lymphoblastic
leukaemia. Nature. 481:157–163. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bell JJ and Bhandoola A: The earliest
thymic progenitors for T cells possess myeloid lineage potential.
Nature. 452:764–767. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wada H, Masuda K, Satoh R, Kakugawa K,
Ikawa T, Katsura Y and Kawamoto H: Adult T-cell progenitors retain
myeloid potential. Nature. 452:768–772. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Rothenberg EV, Moore JE and Yui MA:
Launching the T-cell-lineage developmental programme. Nat Rev
Immunol. 8:9–21. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Coustan-Smith E, Mullighan CG, Onciu M,
Behm FG, Raimondi SC, Pei D, Cheng C, Su X, Rubnitz JE, Basso G, et
al: Early T-cell precursor leukaemia: A subtype of very high-risk
acute lymphoblastic leukaemia. Lancet Oncol. 10:147–156. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Shimizu H, Saitoh T, Machida S, Kako S,
Doki N, Mori T, Sakura T, Kanda Y, Kanamori H, Miyawaki S, et al:
Allogeneic hematopoietic stem cell transplantation for adult
patients with mixed phenotype acute leukemia: Results of a
matched-pair analysis. Eur J Haematol. 95:455–460. 2015. View Article : Google Scholar : PubMed/NCBI
|