Introduction
Kidney tumor is one of the most common causes of
cancer-related cases of mortality, with >270,000 people expected
to be diagnosed with kidney cancer every year (1). According to global cancer statistics in
2013, RCC is the most prevalent subtype of kidney cancer,
accounting for approximately 80–90% of cases, and a continued
increase in the incidence of RCC has been reported (1,2). Although
surgical excision remains the curative treatment when RCC has not
metastasized, a large proportion of patients will relapse or
develop metastatic disease (3).
However, treatment options for renal cancer are limited since it
responds poorly to chemotherapy and radiotherapy (4). Therefore, understanding the molecular
mechanisms of RCC cell invasion and metastasis is imperative for
early diagnosis and treatment of RCC.
NEDD9 was first identified in neuronal precursor
cells as a downregulated gene during the development of the mouse
central nervous system. It belongs to the Crk-associated substrate
family, which is essential for pro-metastasis behavior in several
types of solid tumor (5). Growing
evidence has identified NEDD9 as a tumor-promoting factor in
melanomas (6), glioblastomas
(7), breast carcinoma (8,9),
colorectal cancer and lung cancers (10). It serves a critical function in
regulating cell proliferation, migration, invasion and survival
(11,12). Silencing of NEDD9 in vivo
significantly reduced tumor progression in NEDD9(−/-) mice
(13). Furthermore, a recent study
indicated that miR-145 may exhibit tumor suppressive functions
through regulating NEDD9 oncogenic genes in RCC (12). However, the expression and biological
function of NEDD9 in RCC remains largely unknown.
In a previous study by the current group, it was
reported that NEDD9 is overexpressed in RCC (11). In the present study, NEDD9 expression
was evaluated in a large cohort of patients with RCC, and its
clinical and biological significance was analyzed in a
retrospective manner.
Materials and methods
Clinical RCC specimens and RCC cell
lines
The present study recruited a total of 68
pathologically confirmed patients with RCC who underwent complete
resection of the metastatic lesions and 6 normal renal tissues were
from the tumor bearing kidneys, between January 2013 and December
2015 at the Department of Urology, the Second Hospital and Qilu
Hospital of Shandong University (Jinan, China). Follow-up
information was obtained from a review of the patients' medical
records. All patients had no preoperative radiotherapy,
chemotherapy or immunotherapy. A total of 20 pairs of fresh clear
cell RCC (ccRCC) tissues and matched normal renal tissues were also
stored at −80°C immediately after resection for protein extraction
and reverse transcription-quantitative polymerase chain reaction
(RT-qPCR) analysis. Detailed clinicopathological information of the
patients is listed in Table I. For
all samples, pathologic tumor stage, N stage, TNM stage,
histological grade and renal vein invasion were reevaluated and
determined according to the tumor node metastasis staging system
(14). The study was reviewed and
approved by the Ethical Committee of the Second Hospital of
Shandong University. Written informed consent was provided by all
patients for the use of their samples and data in the study.
 | Table I.Association between NEDD9 expression
and clinicopathological factors in renal cell carcinoma. |
Table I.
Association between NEDD9 expression
and clinicopathological factors in renal cell carcinoma.
| Clinicopathological
factor | n | Overexpressing NEDD9,
n (%) | P-value |
|---|
| Age (years) |
|
| 0.495 |
|
>60 | 35 | 24 (68.57) |
|
|
<60 | 33 | 20 (60.61) |
|
| Sex |
|
| 0.701 |
| Male | 52 | 33 (63.46) |
|
|
Female | 16 | 11 (68.75) |
|
| T stage |
|
| 0.011 |
|
T1 | 23 | 11 (47.82) |
|
|
T2 | 15 | 8 (53.33) |
|
|
T3 | 21 | 18 (85.71) |
|
|
T4 | 9 | 7 (77.78) |
|
| N stage |
|
| 0.414 |
|
N0 | 56 | 35 (62.50) |
|
|
N1,2 | 12 | 9 (75) |
|
| TNM stage |
|
| 0.045 |
| I,
II | 53 | 31 (58.49) |
|
| III,
IV | 15 | 13 (86.67) |
|
| Histological
grade |
|
| 0.406 |
|
G1,2 | 33 | 23 (69.70) |
|
|
G3,4 | 35 | 21 (60) |
|
| Renal vein
invasion |
|
| 0.147 |
| Yes | 11 | 5 (45.45) |
|
| No | 57 | 39 (68.42) |
|
Human RCC cell lines 786-O and Caki1 were purchased
from the American Type Culture Collection (Manassas, VA, USA). The
cells were cultured at 37°C in RPMI 1640 medium containing 10%
fetal bovine serum (FBS; Invitrogen; Thermo Fisher Scientific,
Inc., Waltham, MA, USA), 100 U/ml penicillin (Sigma-Aldrich; Merck
KGaA, Darmstadt, Germany), and 100 µg/ml streptomycin
(Sigma-Aldrich; Merck KGaA).
RNA preparation and RT-qPCR
Total RNA was extracted from frozen ccRCC tissues
using a commercial guanidinium isothiocyanate-based kit (Buffer RZ,
Tiangen Biotech Co., Ltd., Beijing, China), following the
manufacturer's protocol. The amount of RNA in the sample was
quantified spectrophotometrically at a 260 nm wavelength. First
strand cDNA was synthesized using 2 µg total RNA and M-MLV
retroviridase (Takara Bio, Inc., Otsu, Japan) at 37°C for 15 min,
then 85°C for 5 sec. Oligonucleotide primers were synthesized by
Sangon Biotech Co., Ltd. (Shanghai, China). Primer sequences used
were as follows: NEDD9 forward, 5′-GGGTAAAAAGGTGATAACCCCCGT-3′ and
reverse, 5′-TGCTGATGAGGGAGGGATGTCGT-3′; β-actin forward,
5′-TCCATCATGAAGTGTGACGT-3′ and reverse,
5′-GAGCAATGATCTTGATCTTCAT-3′. qPCR was performed in the ABI PRISM
7500 Sequence Detection system (Applied Biosystems; Thermo Fisher
Scientific, Inc.) using SYBR® Fast qPCR Mix (Takara Bio,
Inc.). Relative expression level was determined using the
2−ΔΔCq method (15). NEDD9
expression in cell lines were evaluated by semi-quantitative PCR,
with β-actin used as the control. The reaction conditions were as
follows: 95°C for 30 sec, 64°C for 25 sec and 72°C for 30 sec, for
a total of 35 cycles. Agarose gel electrophoresis was performed
after the reaction, and the products were observed using an
ultraviolet imaging system. The experiments were performed in
triplicate.
Immunohistochemistry
Immunohistochemistry assay were performed on tissues
collected from 68 pathologically confirmed patients with RCC and 6
normal renal tissues. Immunohistochemistry experiments were
performed as described previously (11). In brief, representative samples were
placed into 4% paraformaldehyde overnight and then 5-mm paraffin
sections were prepared for the experiments. Sections were
deparaffinized in xylene, rehydrated which was hydrate by placing
in 95, 70, 50 and 30% ethanol for 2 min each, and endogenous
peroxidase activity was quenched by 3% hydrogen peroxide in
methanol. The sections were submerged in 10 mM citrate buffer (pH
6.0) and microwaved for 8–15 min for antigen retrieval.
Non-specific binding was blocked by incubation with normal goat
serum (Beijing Zhongshan Golden Bridge Biotechnology Co., Ltd.,
Beijing, China) for 1 h at room temperature, and the slides were
incubated with NEDD9 rabbit monoclonal primary antibodies (cat. no.
ab37161; 1:200; Abcam, Cambridge, MA, USA) at 4°C overnight. After
washing, sections were incubated with horseradish
peroxidase-conjugated goat anti-rabbit IgG at room temperature for
1 h (cat. no. SPN-9001; 1:50; Beijing Zhongshan Golden Bridge
Biotechnology Co., Ltd.). Color was developed with the DAB
Horseradish Peroxidase Color Development kit and imaged using
bright field light microscopy.
IHC evaluation
Slides were evaluated by two independent
pathologists without any prior information of the
clinicopathological variables and survival data. NEDD9 expression
was scored using a reproducible semiquantitative method considering
staining intensity (0, no staining of the tumor cells; 1, light
yellow staining; 2, moderate to deep yellow staining; and 3, brown
staining) and the percentage of positive staining (0, 0–5%; 1,
6–25%; 2, 26–50%; 3, 51–75%; and 4, >76%). Conflicting scores
were resolved by consensus. Cases with combined scores of 0–4 were
defined as the low-expression group, and cases with scores of 5–7
were defined as the high-expression group.
NEDD9 small interfering (si)RNA
transfection
NEDD9 small interfering (si)RNA transfection was
performed on 786-O and Caki1 cells. Knockdown of NEDD9 expression
was performed by RNA interference using specific siRNA
oligonucleotides. The target sequence was:
5′-UCCCAUGCAGGAGACUGCCUCCAGU-3′. The chemically modified siRNAs
targeting NEDD9 and scramble siRNA were purchased from Invitrogen
(Thermo Fisher Scientific, Inc.). Cells were transfected with
either NEDD9 or control siRNA with a concentration of 100 nM using
Lipofectamine RNAiMAX reagent (Invitrogen; Thermo Fisher
Scientific, Inc.), following the manufacturer's protocol. All
experiments were performed 72 h after transfection.
Western blot analysis
Western blot analysis was performed as described
previously (11). Total proteins
extracted from frozen tissues and cells lines transfected for 72 h
and then incubated with RIPA lysis buffer (DBI Bioscience,
Shanghai, China), following the manufacturer's protocol. After
protein quantitation using a BCA Protein Quantitative kit (DBI
Bioscience), 100 µg protein per lane were separated by SDS-PAGE
(10% gel) and blotted onto polyvinylidene difluoride membranes. The
membranes were blocked with 5% skim milk and then incubated
overnight at 4°C with anti-NEDD9 (1:1,000; cat. no. ab37161; Abcam)
or anti-tubulin (cat. no. sc-8035; 1:1,000; Santa Cruz
Biotechnologies, Inc., Dallas, TX, USA), followed by incubation
with horseradish peroxidase-conjugated IgG goat anti-rabbit
H&L; (cat. no. ab97051; 1:2,000; Abcam) and goat anti-mouse IgG
H&L (cat. no. ab6708; 1:2,000; Abcam) at room temperature for 1
h. An enhanced chemiluminescence kit (Merck KGaA) was used for
detection using FluorChem™ Q software (version 3.4.0.0; Protein
Simple, San Jose, CA, USA). The experiments were performed in
triplicate.
Cell migration and invasion assay
The cell migration assay was performed using a
24-well transwell chamber with a pore size of 8 µm. In brief,
1×104 cells were seeded in the upper chambers in 200 µl
serum-free Dulbecco's modified Eagle's medium (DMEM; Corning Inc.,
Corning, NY, USA), and 750 µl of 10% FBS-DMEM (Invitrogen; Thermo
Fisher Scientific, Inc.) was added into the lower wells. After 24 h
at 37°C, cells that had migrated to the bottom of the membrane were
fixed and stained with 0.1% crystal violet in methanol in 15 min at
room temperature. To quantify the cells, three independent fields
per well were photographed under phase contrast microscopy. The
number of cells per field were counted and averaged. For the cell
invasion assay, the insert of the pore was coated with 50 µl
Matrigel (dilution at 1:2; BD Biosciences, Franklin Lakes, NJ,
USA). All experiments were performed in triplicate.
Wound healing assay
A scratch wound-healing assay was performed to
determine cell migration. Cells were seeded (5×105) into
6-well plates the day before siRNA transfection. At 24 h after
transfection, wounds were prepared using a 200 µl pipette tip
scratched through the wells. The cell migration speed was
calculated by measuring the distance migrated in 24 h. Images were
obtained using a phase-contrast microscope at different time points
(0 and 24 h) after scratch. All experiments were performed in
triplicate.
Statistical analysis
Data are presented as the mean ± standard error of
the mean. All statistical calculations were performed using SPSS 18
statistical software (SPSS, Inc., Chicago, IL, USA). The relative
NEDD9 expression level in the matched tumor and normal renal
tissues was calculated using a paired t-test. The Chi-square test
was used to evaluate the association between NEDD9 expression
profiles and clinicopathological factors. Survival analysis was
performed using the Kaplan-Meier method and compared using the
log-rank test. P<0.05 was considered to indicate a statistically
significant difference.
Results
Expression of NEDD9 in clinical RCC
samples
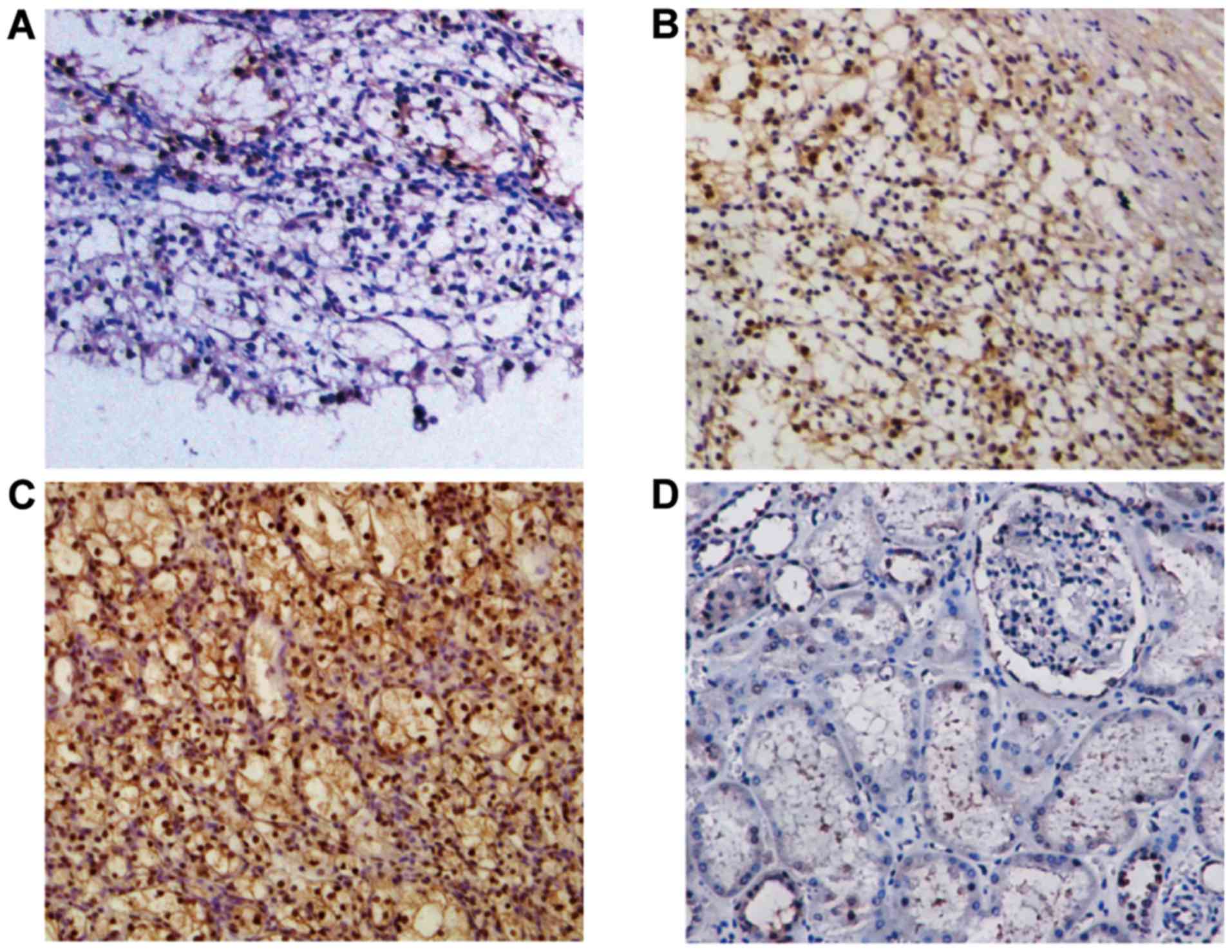
To investigate the abnormalities of NEDD9 expression
in RCC, NEDD9 expression was analyzed in 68 RCC tissues and 6
normal tissues using immunohistochemistry (Fig. 1). It was identified that NEDD9 protein
was expressed in all RCC tissues RCC tissues and NEDD9 protein was
highly expressed in 44 out of the 68 (64.71%) NEDD9-positive
patients with RCC, whereas normal renal tissues were 83.33% (5 of
6) negative, with a small number of samples exhibiting very low
NEDD9 expression in the cytoplasmic region. These observations
suggested that NEDD9 expression may be associated with RCC.
Association between NEDD9 expression
levels and patient survival
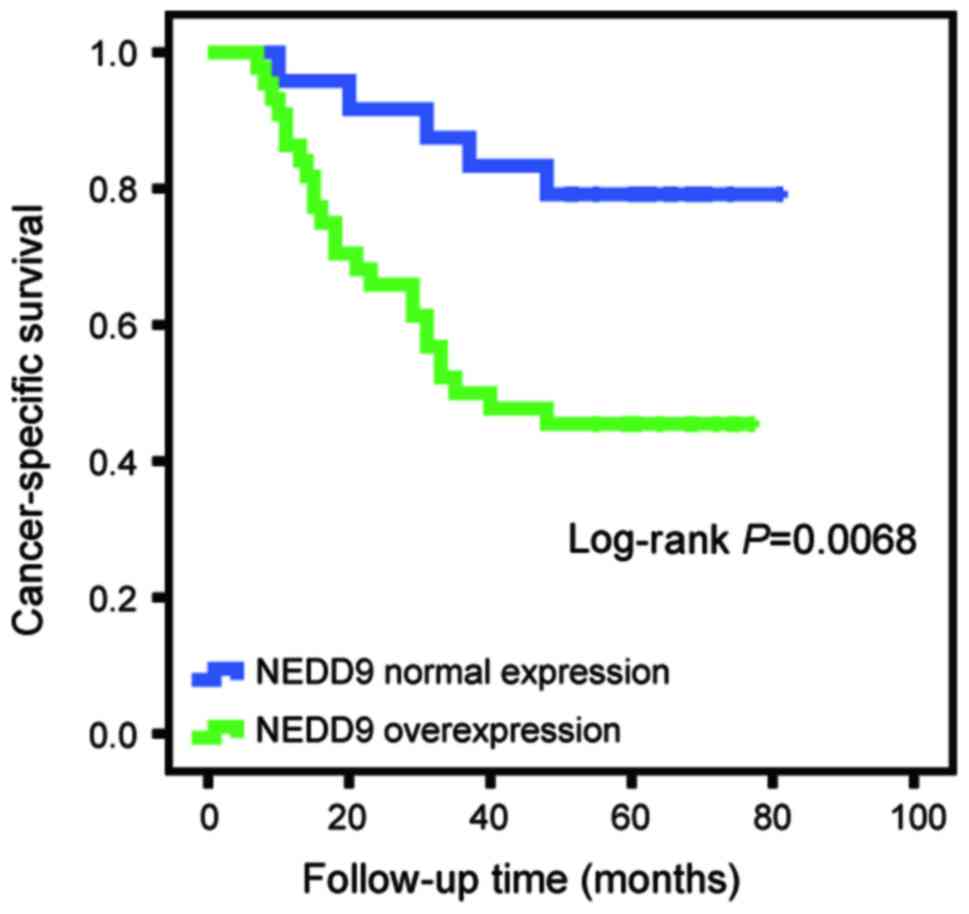
The association between aberrant NEDD9 expression
and clinicopathological factors was investigated. It was identified
that the NEDD9 staining level was significantly associated with
pathological tumor stage (P=0.011; Table
I) and advanced clinical TNM stage (P=0.045). No significant
associations were observed between NEDD9 expression and patient
sex, age or histological type (Table
I). The log-rank test revealed that the survival time of
patients with RCC overexpressing NEDD9 (37.5±6.49 months) was
significantly shorter compared with patients with normal NEDD9
expression (57.67±7.24 months; P=0.0068; Fig. 2).
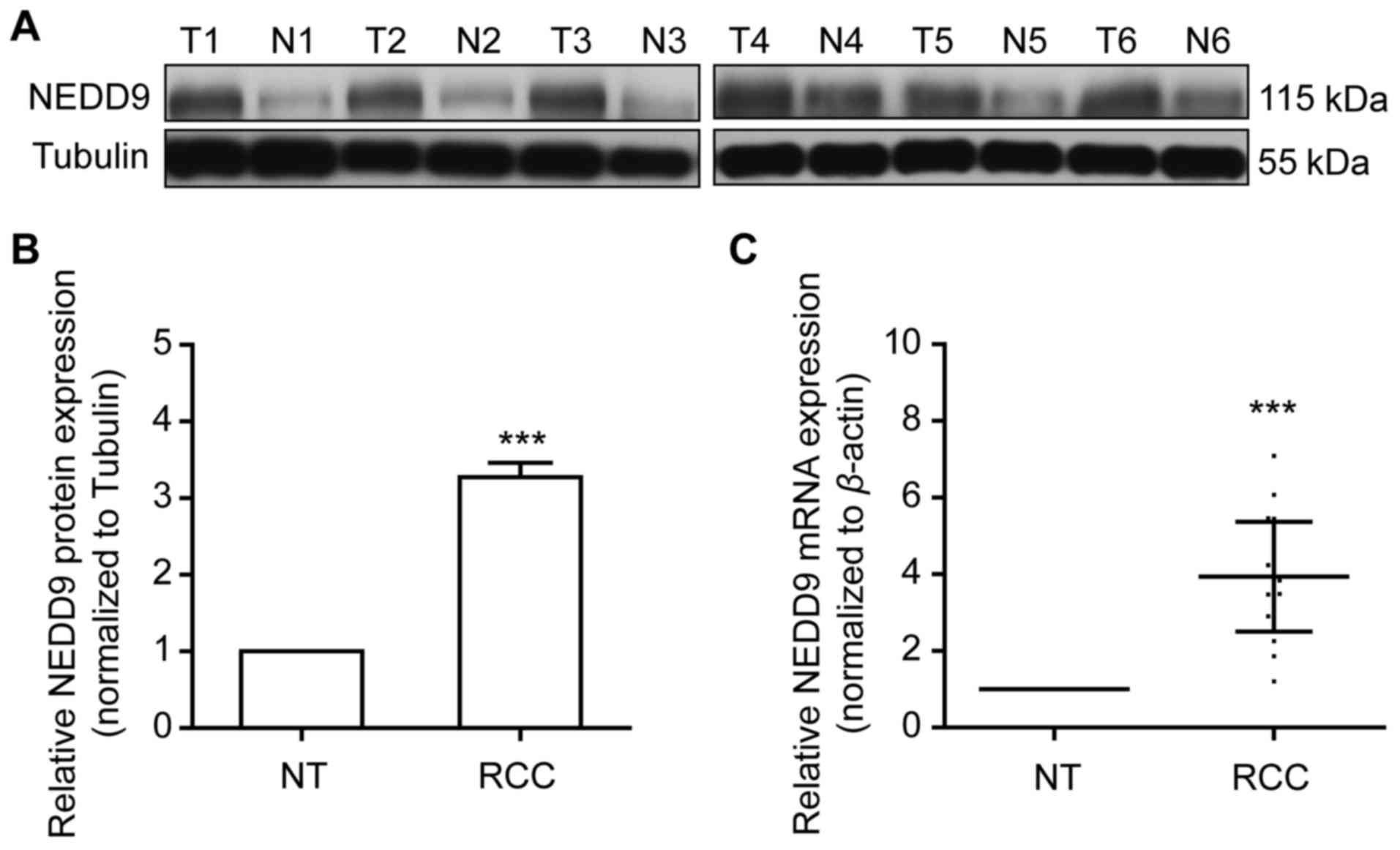
Increased NEDD9 expression in RCC
tissues
NEDD9 protein expression level was examined using
western blotting in 20 pairs of frozen clinical ccRCC samples and
matched normal renal tissues. NEDD9 expression was significantly
higher in ccRCC tissues compared with matched normal renal tissues
(P<0.001; Fig. 3A and B).
Consistent with the NEDD9 protein expression profile, in ccRCC
tissues, NEDD9 mRNA level was also significantly elevated, as
detected by RT-qPCR analysis (P<0.001; Fig. 3C). The results indicated that NEDD9 is
overexpressed in RCC specimens.
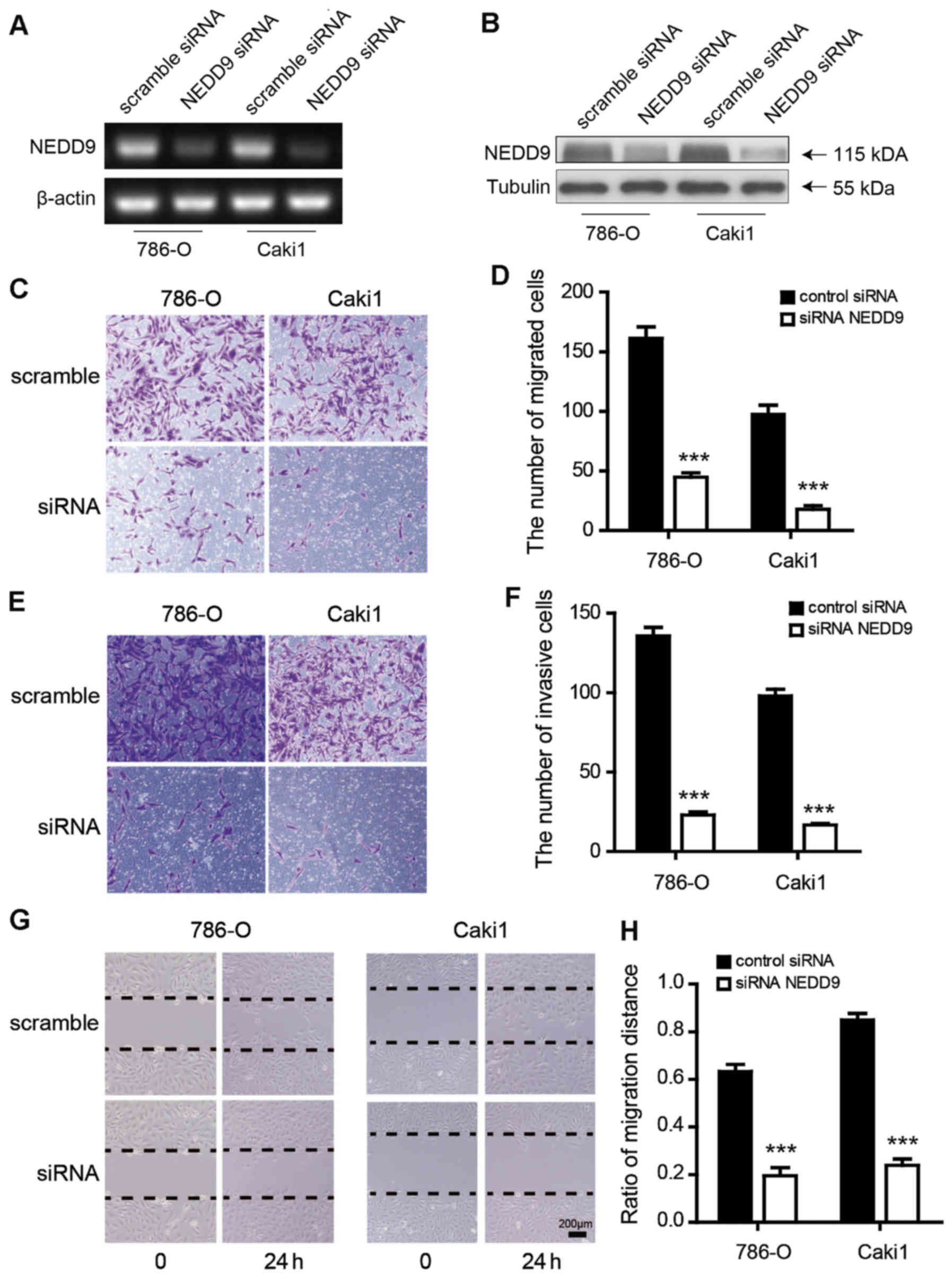
NEDD9 knockdown suppresses cell
migration and invasion
First, it was demonstrated that NEDD9 siRNA could
reduce NEDD9 mRNA and protein expression in different RCC cell
lines (Fig. 4A and B). Next,
transwell assays were performed to investigate whether migration
and invasion of RCC cells would be attenuated by reduction of NEDD9
expression after transfection with NEDD9 siRNA. The results
indicated that NEDD9 siRNA significantly reduced the migratory and
invasive capability of 786-O and Caki1 RCC cells (P<0.0001;
Fig. 4C-F). In addition, a wound
healing assay indicated that the migratory capacity of NEDD9
siRNA-treated cells was significantly decreased at 24 h after
scratch (ratios of wound closure: Scramble vs. NEDD9 siRNA, 786-O:
0.633±0.043 vs. 0.200±0.032, P<0.001; Caki1: 0.850±0.022 vs.
0.24±0.013, P<0.001; Fig. 4G and
H).
Discussion
RCC is the most common carcinoma of the adult kidney
and the incidence of RCC has gradually increased in recent decades
(1). Therefore, understanding the
pathological mechanisms of RCC and identifying treatment targets is
crucial. Previous studies have implicated NEDD9 as a central
component of integrin-dependent signaling cascades, which were
identified to be activate the focal adhesion and Src kinases. NEDD9
thereby regulates activation of Src kinases and exhibits diverse
functions, including apoptosis and cell cycle regulation,
migration, adhesion, invasion and chemotaxis (16–18).
Several studies have revealed aberrant expression of NEDD9 in
various types of tumor, including colorectal cancer, gastric
carcinoma, lung carcinoma and pancreatic carcinoma (12,19,20). To
the best of our knowledge, there are few studies focused on the
role of NEDD9 in RCC, and the correlation of NEDD9 expression with
the clinicopathological factors of RCC have not been
determined.
The current study demonstrated that NEDD9
overexpression widely occurs in kidney cancer. The subcellular
localization of endogenous NEDD9 in RCC tissues was evaluated by
immunohistochemical staining. This indicated an intense staining of
NEDD9 in 64.71% of RCC tissues. Statistical analysis of the
clinicopathological features of patients with RCC in our study
revealed that there was a significant association between NEDD9
overexpression and certain clinicopathological factors, such as
primary tumor stage and tumor, node and metastasis stage.
Furthermore, a high level of NEDD9 expression was significantly
associated with the aggressive characteristics of RCC, such as
pathological tumor stage and TNM stage. These observations are
consistent with Shi et al (21), who reported that high NEDD9 expression
exhibited a significant association with poor prognosis for gastric
cancer patients. In addition, in the present study, three- to
four-fold higher levels of expression of NEDD9 protein and mRNA
were observed in tumor tissues compared with matched normal renal
tissues in 20 paired RCC samples. These findings indicated that the
upregulation of NEDD9 protein is involved in the progression and
metastasis of RCC. To the best of our knowledge, this demonstrates
the clinical and biological significance of NEDD9 in RCC for the
first time.
Tumor metastasis caused by tumor cell invasion or
metastasis is critical for cancer patient survival and it may
reduce patient survival (22). The
migration ability of tumor cells is believed to be the
rate-limiting step in tumor metastasis; the migration of the tumor
cell from the primary site to the surrounding tissue through the
basement membrane is an important invasion function (23,24).
Recently, Feng et al (25)
reported that NEDD9 depletion reduced the Matrigel invasion of
gastric SGC-7901 and GES-1 cells. In the present study, a
significant reduction in cell migration and invasion was observed
in RCC cell lines. These results may explain the current findings
that NEDD9 overexpression was the major defining characteristic of
RCC tumors and was associated with numerous clinicopathological
factors.
In conclusion, the present study has demonstrated
that NEDD9 is upregulated in RCC, and high NEDD9 expression is
associated with poor survival of patients with RCC. Inhibition of
NEDD9 expression leads to attenuated migration and invasion
ability. Therefore, NEDD9 may be a novel target for prevention and
treatment of RCC.
Acknowledgements
The authors are grateful to the Central Research
Laboratory, the Second Hospital of Shandong University (Jinan,
China) for experimental techniques and generous support. This
project was supported by the Seed Foundation of the Second Hospital
of Shandong University (grant no. Y2015010039).
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Baldewijns MM, van Vlodrop IJ, Schouten
LJ, Soetekouw PM, de Bruïne AP and van Engeland M: Genetics and
epigenetics of renal cell cancer. Biochim Biophys Acta.
1785:133–155. 2008.PubMed/NCBI
|
|
3
|
Drucker BJ: Renal cell carcinoma: Current
status and future prospects. Cancer Treat Rev. 31:536–545. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Huang Y, Dai Y, Yang J, Chen T, Yin Y,
Tang M, Hu C and Zhang L: Microarray analysis of microRNA
expression in renal clear cell carcinoma. Eur J Surg Oncol.
35:1119–1123. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shagisultanova E, Gaponova AV, Gabbasov R,
Nicolas E and Golemis EA: Preclinical and clinical studies of the
NEDD9 scaffold protein in cancer and other diseases. Gene.
567:1–11. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kim M, Gans JD, Nogueira C, Wang A, Paik
JH, Feng B, Brennan C, Hahn WC, Cordon-Cardo C, Wagner SN, et al:
Comparative oncogenomics identifies NEDD9 as a melanoma metastasis
gene. Cell. 125:1269–1281. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Natarajan M, Stewart JE, Golemis EA,
Pugacheva EN, Alexandropoulos K, Cox BD, Wang W, Grammer JR and
Gladson CL: HEF1 is a necessary and specific downstream effector of
FAK that promotes the migration of glioblastoma cells. Oncogene.
25:1721–1732. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tornillo G, Defilippi P and Cabodi S: Cas
proteins: Dodgy scaffolding in breast cancer. Breast Cancer Res.
16:4432014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Loskutov YV, Kozyulina PY, Kozyreva VK,
Ice RJ, Jones BC, Roston TJ, Smolkin MB, Ivanov AV, Wysolmerski RB
and Pugacheva EN: NEDD9/Arf6-dependent endocytic trafficking of
matrix metalloproteinase 14: A novel mechanism for blocking
mesenchymal cell invasion and metastasis of breast cancer.
Oncogene. 34:3662–3675. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kondo S, Iwata S, Yamada T, Inoue Y,
Ichihara H, Kichikawa Y, Katayose T, Souta-Kuribara A, Yamazaki H,
Hosono O, et al: Impact of the integrin signaling adaptor protein
NEDD9 on prognosis and metastatic behavior of human lung cancer.
Clin Cancer Res. 18:6326–6338. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang J, Yang WJ, Sun C, Luan Y, Cheng GH,
Li KL and Kong F: siRNA suppression of NEDD9 inhibits proliferation
and enhances apoptosis in renal cell carcinoma. Oncol Res.
22:219–224. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Han T, Yi XP, Liu B, Ke MJ and Li YX:
MicroRNA-145 suppresses cell proliferation, invasion and migration
in pancreatic cancer cells by targeting NEDD9. Mol Med Rep.
11:4115–4120. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Izumchenko E, Singh MK, Plotnikova OV,
Tikhmyanova N, Little JL, Serebriiskii IG, Seo S, Kurokawa M,
Egleston BL, Klein-Szanto A, et al: NEDD9 promotes oncogenic
signaling in mammary tumor development. Cancer Res. 69:7198–7206.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Eble J, Sauter G, Epstein J and Sesterhenn
IE: World health organization classification of tumors pathology
and genetics of the urinary systemandmale genital organs. LARC
Press; Lyon: pp. 12–14. 2004
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-timequantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tikhmyanova N, Tulin AV, Roegiers F and
Golemis EA: Dcas supports cell polarization and cell-cell adhesion
complexes in development. PLoS One. 5:e123692010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nikonova AS, Gaponova AV, Kudinov AE and
Golemis EA: CAS proteins in health and disease: An update. IUBMB
Life. 66:387–395. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
O'Neill GM, Seo S, Serebriiskii IG, Lessin
SR and Golemis EA: A new central scaffold for metastasis: Parsing
HEF1/Cas-L/NEDD9. Cancer Res. 67:8975–8979. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xue YZ, Sheng YY, Liu ZL, Wei ZQ, Cao HY,
Wu YM, Lu YF, Yu LH, Li JP and Li ZS: Expression of NEDD9 in
pancreatic ductal adenocarcinoma and its clinical significance.
Tumour Biol. 34:895–899. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li P, Zhou H, Zhu X, Ma G, Liu C, Lin B
and Mao W: High expression of NEDD9 predicts adverse outcomes of
colorectal cancer patients. Int J Clin Exp Pathol. 7:2565–2570.
2014.PubMed/NCBI
|
|
21
|
Shi R, Wang L, Wang T, Xu J, Wang F and Xu
M: NEDD9 overexpression correlates with the progression and
prognosis in gastric carcinoma. Med Oncol. 31:8522014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wan L, Pantel K and Kang Y: Tumor
metastasis: Moving new biological insights into the clinic. Nat
Med. 19:1450–1464. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zeng G, Gao L and Yu RK: Reduced cell
migration, tumor growth and experimental metastasis of rat F-11
cells whose expression of GD3-synthase is suppressed. Int J Cancer.
88:53–57. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shin KD, Lee MY, Shin DS, Lee S, Son KH,
Koh S, Paik YK, Kwon BM and Han DC: Blocking tumor cell migration
and invasion with biphenyl isoxazole derivative KRIBB3, a synthetic
molecule that inhibits Hsp27 phosphorylation. J Biol Chem.
280:41439–41448. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Feng J, Zhao J, Xie H, Yin Y, Luo G, Zhang
J, Feng Y and Li Z: Involvement of NEDD9 in the invasion and
migration of gastric cancer. Tumour Biol. 36:3621–3628. 2015.
View Article : Google Scholar : PubMed/NCBI
|