Introduction
Ovarian cancer is one of the most lethal
gynecological malignancies (1). In
the US, approximately 22,280 new cases are diagnosed every year,
and approximately 14,240 women were predicted to have died of
ovarian cancer in 2016 (2). Despite
the rapid development of surgical and chemotherapeutic techniques,
the 5-year survival rate of patients at an advanced stage of
ovarian cancer is <30% due to drug resistance, relapse, and the
lack of effective early screening and diagnostic methods (3). Therefore, determining the molecular
mechanism underlying epithelial ovarian cancer is an essential and
important topic for cancer treatment.
cAMP response element-binding 5 (CREB5) is a
transcriptional factor in eukaryotic cells, and it functions as a
protein-coding gene. Its product is a member of the cAMP response
element (CRE)-binding protein family. The encoded protein
specifically binds to CRE as a homodimer or a heterodimer with
c-Jun or CRE-BP1, and functions as a CRE-dependent trans-activator
(4,5).
Members of this family contain zinc-finger and bZIP DNA-binding
domains. Few studies have suggested that the up-regulation of CREB5
is negatively correlated with the prognosis of colorectal cancer
and non-small cell lung cancer (6,7). However,
the relationship between CREB5 and the clinical prognosis of
ovarian cancer still needs to be verified. Therefore, here we
investigated CREB5 expression and its relationship with clinical
prognosis in epithelial ovarian cancer by analyzing its expression
level in cancer cell lines and cancer tissues.
Materials and methods
Cell lines and cell culture
Human ovarian surface epithelial cells (HOSEpiC)
(8) and immortalized normal ovarian
surface epithelial cells (IOSE80) were purchased from ScienCell
Research Laboratories (San Diego, CA, USA) and cultured in ovarian
epithelial cell medium. normal ovarian-adjacent tissues were
purchased from ScienCell Research Laboratories in 2016 and this
cells were grown in 1:1 combination of two media, Medium 199
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and
MCDB 105 (Cell Applications Inc., San Diego, CA, USA) with 10% FBS
in a humidified atmosphere containing 5% CO2 at 37°C.
Ovarian cancer cell lines including COV362.4, A2780, TOV21G, EFO27,
COV644, SKOV3, EFO21, TOV112D, OV90, and OV56 were purchased from
the European Collection of Authenticated Cell Cultures (ECACC) in
2016 and cultured in Dulbecco's modified Eagle's medium
(Invitrogen; Thermo Fisher Scientific, Inc.), supplemented with 10%
fetal bovine serum (HyClone, Logan, UT, USA).
Patient information and tissue
specimens
A total of 125 paraffin-embedded and archived
epithelial ovarian cancer samples, which were histopathologically
and clinically diagnosed at the Sun Yat-sen University Cancer
Center from 2001 to 2006, were analyzed in this retrospective
study. All tumors were staged according to the International
Federation of Gynaecology and Obstetrics standards (FIGO). Ten
freshly collected epithelial ovarian cancer tissues and normal
ovarian-adjacent tissues were frozen and stored in liquid nitrogen
until further use, including FIGO stage I+II (n=31), stage III
(n=70), stage IV (n=24). For the use of the clinical materials for
research purposes, prior patient consent and approval were obtained
from the Institutional Ethical Board (IRB) in the First Affiliated
Hospital of Sun Yat-sen University.
Quantitative polymerase chain reaction
(qPCR)
RNA extraction and qPCR
Total RNA from cultured cells was extracted from ten
fresh ovarian cancer tissue and adjacent normal tissue samples
using TRIzol reagent according to the manufacturer's instructions.
Complementary DNA (cDNA) was amplified and quantified using an ABI
Prism 7500 Sequence Detection system (Applied Biosystems; Thermo
Fisher Scientific, Inc.) and SYBR-Green I (Molecular Probes;
Invitrogen; Thermo Fisher Scientific, Inc.). qPCR was used to
quantify the CREB5 mRNA levels. The RT-PCR conditions for genes
were set at 95°C for 10 min, followed by 40 cycles at 95°C for 20
sec, 60°C for 30 sec and 72°C for 1 min. We examined the CREB5
expression by qPCR and the expression of the mRNA was defined based
on Ct, and relative expression levels were calculated as 2-[(Cq of
CREB5)-(Cq of GAPDH)] after normalization with reference to the
expression of housekeeping gene GAPDH, where Cq represents
quantification cycle. The CREB5 sequences were
5′-AGATGGTCCTCTGTTGGGAA-3′ (forward) and
5′-TGGACACGGTTATGAGAATGA-3′ (reverse), and those for GAP DH were
5′-AATGAAGGGGTCATTGATGG-3′ (forward) and 5′-AAGGTGAAGGTCGGAGTCAA-3′
(reverse).
Western blotting
Cell lysates from cell lines and fresh tissue
samples were prepared using cold RIPA buffer. Fresh tissue samples
were milled to powder in liquid nitrogen and lysed by SDS-PAGE
sample buffer. The protein samples (30 µg) were separated on 7.5%
SDS polyacrylamide gels, transferred to polyvinylidene fluoride
(PVDF) membranes (Immobilon P; EMD Millipore, Bedford, MA, USA),
and blocked with 10% nonfat dried milk (NFDM) freshly in
Tris-buffered saline containing 0.1% Tween-20 (TBST) for 1 h at
room temperature. After blocking, the membranes were incubated with
anti-CREB5 monoclonal antibodies (1:1,000; GTX44660; BD
Biosciences, Franklin Lakes, NJ, USA) overnight at 4°C. Then, the
membranes were rinsed thrice with TBST for 10 min each time, and
incubated with horseradish peroxidase-conjugated goat anti-rabbit
IgG (1:2,000; ABIN2474414; Santa Cruz Biotechnology, Inc., Dallas,
TX, USA) for 2 h for 2 h at room temperature. Then, bound
antibodies were detected using an enhanced chemiluminescence
detection system (Amersham Pharmacia Biotech, Piscataway, NJ, USA)
according to the manufacturer's instructions. GADPH (1:1,000;
ab9485; Santa Cruz Biotechnology, Inc.) was chosen as the loading
control. The signals of western blotting bands of figures were
quantified by densitometry, which determined by comparing the ratio
in IOSE80, i.e., the ratio CREB5/GAPDH, in IOSE80 was considered as
1.0.
Immunohistochemical assay
The CREB5 protein expression levels in the human
ovarian cancer tissues were detected by immunohistochemical
analysis. Briefly, 4 µm-thick paraffin-embedded sections were baked
at 60°C for 1 h, deparaffinized with xylene, rehydrated, and
microwaved in EDTA antigen retrieval buffer. Next, high tension was
used for antigen retrieval, and the specimens were treated with 3%
hydrogen peroxide in methanol to quench endogenous peroxidase
activity, followed by incubation with 1% bovine serum albumin
(36102ES10; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) to block
nonspecific binding for 20 min at room temperature, and incubation
with anti-CREB5 monoclonal antibodies (1:100; ABIN599087; Santa
Cruz Biotechnology, Inc.) at 4°C overnight. Normal goat serum
(Abcam, ab7481) was used as the negative control and we incubate
normal goat serum at 4 degrees Celsius overnight. After washing,
the tissue sections were treated with biotinylated anti-mouse
secondary antibody (1:100; PA128568; Sigma-Aldrich; Merck KGaA),
then incubated with streptavidin horseradish peroxidase complex
(Sigma-Aldrich; Merck KGaA) for 30 min at 37°C, immersed in
3-amino-9-ethyl carbazole. the biotinylated anti-mouse secondary
antibody and the streptavidin horseradish peroxidase complex are
part of the streptavidin-biotin kit and the catalogue number of the
streptavidin-biotin kit is SP2002. The sections were then
counterstained with 10% Mayer's hematoxylin, dehydrated, and
mounted in Crystal Mount. biotin-labeled anti-mouse IgG goat
antibody (dilution 1:100; Sigma-Aldrich; Merck KGaA) as the control
IgG. The distribution, positive intensity, and positive ratio of
CREB5 were observed by microscopy by two independent pathologists
who were blinded to the clinical parameters. The staining results
were scored on the basis of the following criteria: i) percentage
of positive tumor cells in tumor tissue: 0 (0%), 1 (1–10%), 2
(11–50%), 3 (51–70%), and 4 (71–100%); and ii) staining intensity:
0 (none), 1 (weak), 2 (moderate), 3 (strong). The staining index
was calculated as the staining intensity score × proportion of
positive tumor cells (range, 0–12). A final score of ≥5 was
considered high expression, the catalogue number of the
streptavidin-biotin kit is SP2002, cells were visualized in an
Olympus BX51 fluorescence microscope (Olympus, Tokyo, Japan).
Statistical analysis
All data were statistically analyzed using SPSS 18.0
statistical software. The relationship between CREB5 expression and
clinicopathological characteristics was analyzed using the
chi-square test. The Kaplan-Meier method was used to establish
survival curves, and the survival differences were compared using
the log-rank test. Survival data were evaluated using univariate
and multivariate Cox regression analyses. P<0.05 was considered
to indicate a statistically significant difference.
Results
CREB5 expression profile
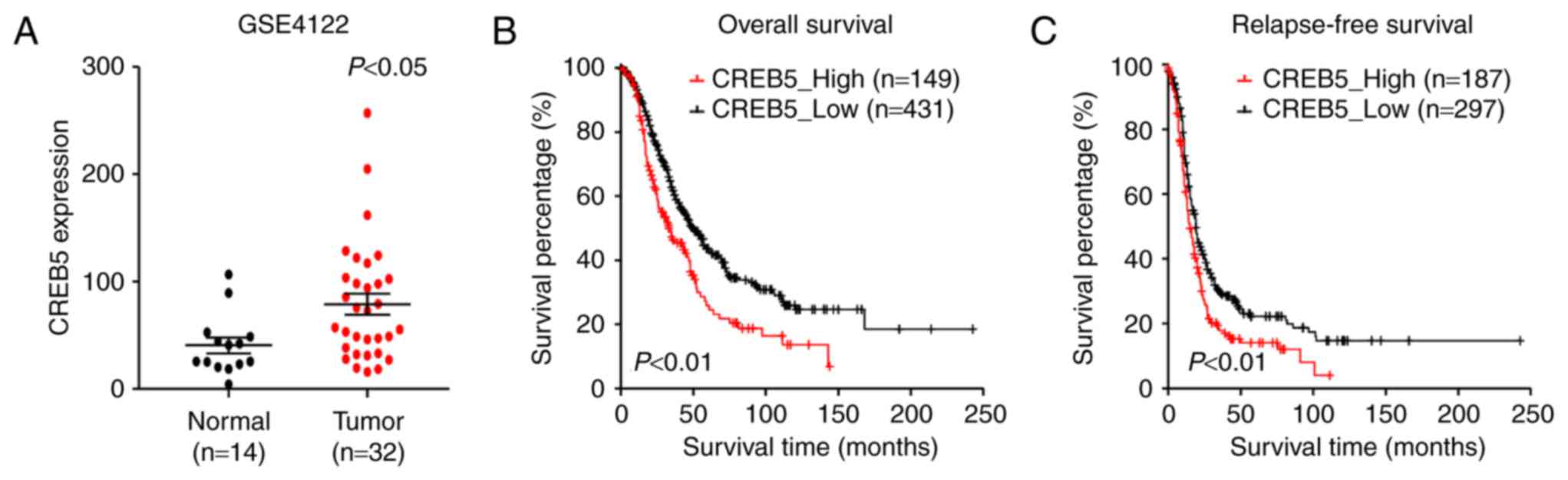
We analyzed previously published CREB5 expression
profiles obtained from 32 patients with epithelial ovarian cancer
(GSE4122). The results of our analysis revealed that CREB5 was
significantly upregulated in the 32 epithelial ovarian cancer
tissues compared to the 14 normal HOSEpiC cell lines analyzed
(Fig. 1A). Importantly, in patients
of the GSE4122 dataset, higher CREB5 expression was associated with
shorter relapse-free survival time and overall survival time,
whereas lower CREB5 expression was associated with longer
relapse-free survival and overall survival (P<0.05; Fig. 1B and C). In conclusion, these results
indicate a possible link between CREB5 overexpression and the
progression of human epithelial ovarian cancer.
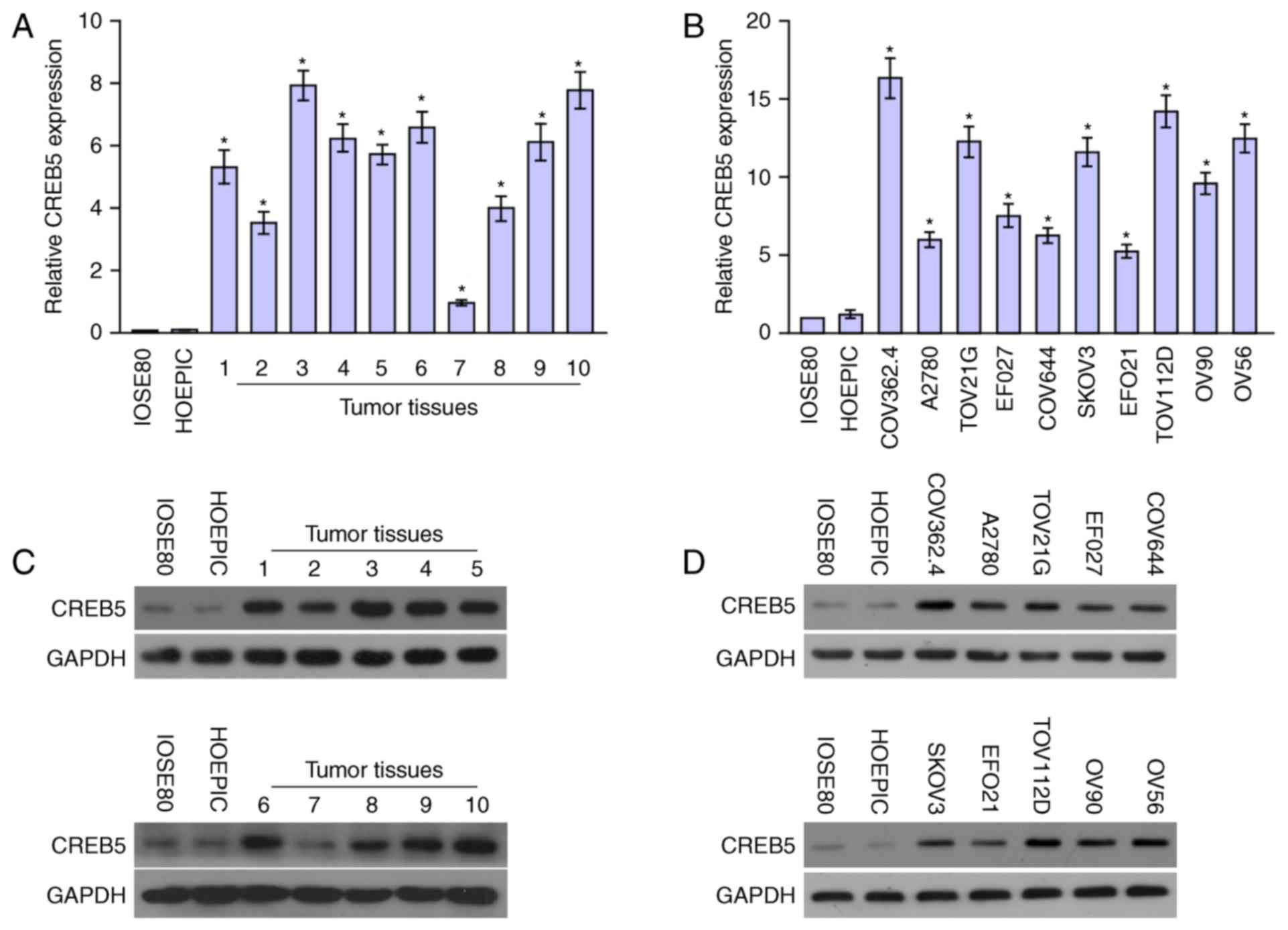
CREB5 expression in epithelial ovarian
cancer cell lines and tissues
CREB5 mRNA and protein expression levels were
determined in ten epithelial ovarian cancer cell lines and ten
epithelial ovarian cancer samples relative to HOSEpiC and IOSE80
cells were determined using qPCR and western blot analysis.
Comparison of the results showed that CREB5 mRNA and protein
levels were differentially upregulated in all ten epithelial
ovarian cancer samples compared with the normal tissues and cells
(Fig. 2). Collectively, the result
demonstrated that CREB5 was upregulated in epithelial ovarian
cancer.
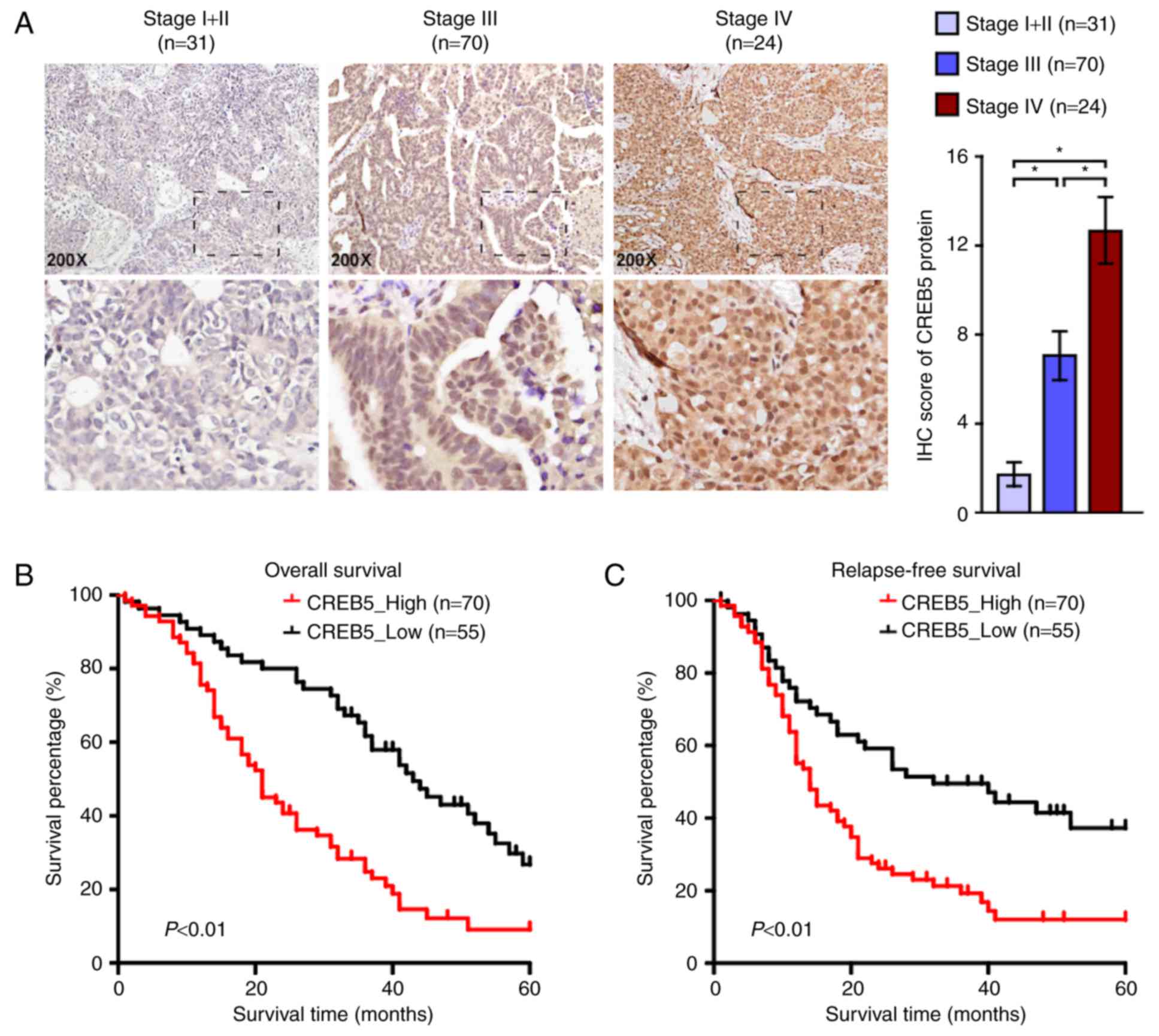
CREB5 protein expression in epithelial
ovarian cancer tissues and association with clinical features
Based on the immunohistochemistry score, we found
that CREB5 expression in epithelial ovarian cancer tissues was
positively correlated with FIGO stage. Immunohistochemical staining
revealed that CREB5 distribution was localized to the nuclei
(Fig. 3A). We also explored the
relationship between CREB5 and clinical and pathological
characteristics and found that CREB5 expression was positively
correlated with FIGO stage and pelvic lymph node metastasis
(P<0.001). There was no significant correlation between CREB5
expression and histological type or patient age at surgery
(P>0.05, Table I), suggesting that
epithelial ovarian cancer progression was associated with increase
in CREB5 expression. We examined the relationship between CREB5
expression and clinical outcomes (P<0.05; Table I). As shown in Fig. 3B and C, patients with high/low CREB5
expression presented different recurrence and survival states.
Patients with higher CREB5 expression had significantly longer
disease-free survival and overall survival than those with low
CREB5 expression. Results of the univariate logistic regression and
stepwise multivariate analyses showed that the FIGO stage and high
CREB5 expression levels were significant risk factors for
epithelial ovarian cancer (P<0.001; Table II). Taken together, our findings
suggest that CREB5 may be an independent prognostic biomarker in
patients with epithelial ovarian cancer.
 | Table I.The relationship between CREB5 and
clinical pathological characteristics in 125 patients with ovarian
cancer. |
Table I.
The relationship between CREB5 and
clinical pathological characteristics in 125 patients with ovarian
cancer.
|
|
| CREB5 expression |
|
|---|
|
|
|
|
|
|---|
| Parameters | Number of cases | High (n=70) | Low (n=55) | P-values |
|---|
| Age (years) |
|
|
|
|
|
<55 | 62 | 40 | 32 | 0.391 |
| ≥55 | 63 | 30 | 33 |
|
| Histological
type |
|
|
|
|
|
Serous | 96 | 58 | 38 | 0.193 |
|
Mucinous | 10 | 4 | 6 |
|
| Clear
cell | 8 | 6 | 2 |
|
|
Endometrioid | 9 | 4 | 5 |
|
|
Others | 2 | 1 | 1 |
|
| FIGO stage |
|
|
|
|
| I or
II | 31 | 10 | 21 | 0.001 |
| III | 70 | 40 | 30 |
|
| IV | 24 | 20 | 4 |
|
| Pelvic lymph node
metastasis |
|
|
|
|
| Yes | 86 | 56 | 30 | 0.003 |
| No | 39 | 14 | 25 |
|
| Recurrence |
|
|
|
|
| Yes | 88 | 57 | 31 | 0.003 |
| No | 37 | 13 | 24 |
|
| Survival status |
|
|
|
|
| Dead | 94 | 58 | 36 |
|
| NO | 31 | 12 | 19 | 0.036 |
 | Table II.Univariate and multivariate analysis
of factors associated with overall survival in 125 ovarian cancer
patients. |
Table II.
Univariate and multivariate analysis
of factors associated with overall survival in 125 ovarian cancer
patients.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Characteristics | HR (95% CI) | P-values | HR (95% CI) | P-values |
|---|
| Age (years) | 0.993
(0.978–1.008) | 0.370 | 0.993
(0.979–1.008) | 0.373 |
| Histologic grade | 0.860
(0.528–1.398) | 0.542 | 0.993
(0.609–1.620) | 0.978 |
| FIGO stage | 3.961
(2.218–7.070) | <0.001 | 3.940
(2.164–7.175) | <0.001 |
| CREB5 | 2.629
(1.697–4.072) | <0.001 | 2.722
(1.723–4.299) | <0.001 |
Discussion
Despite several advances in treatment, including
chemotherapy and cytoreductive surgery, ovarian cancer remains to
be the most lethal malignant gynecological cancer. However, most
patients with ovarian cancer that respond to initial therapy
subsequently relapse within 5 years (9), and the 5-year survival rate is
approximately 2.4–23% (10,11). In general, >50% of treated patients
experience tumor recurrence and ultimately succumb to this
malignancy (12,13). Therefore, it is important to identify
novel markers in epithelial ovarian cancer, which can advance and
initiate more individualized treatment.
In the present study, we first identified the
overexpression of CREB5 in cell lines and fresh tissues of
epithelial ovarian cancer at both the mRNA and protein levels.
Immunostaining analysis revealed a positive correlation between
CREB5 expression and FIGO stage. Furthermore, CREB5 overexpression
was found to correlates with epithelial ovarian cancer progression,
patients with higher CREB5 expression had shorter overall survival
time and relapse-free survival time.
Our findings suggest that CREB5 is an independent
prognostic indicator of poor outcomes in epithelial ovarian cancer
patients. CREB5 is a potential therapeutic target for epithelial
ovarian cancer. Furthermore, results of the univariate logistic
regression and stepwise multivariate analyses revealed that FIGO
stage and high CREB5 expression level were significant risk factors
for epithelial ovarian cancer (P<0.001; Table II). Additionally, high CREB5
expression was significantly positively associated with ascending
FIGO stage (Table I; Fig. 3). Thus, according to the results of
the logistic regression analysis, high CREB5 expression is an
independent risk factor for overall survival in patients with
epithelial ovarian cancer. CREB is an important transcription
factor that regulates diverse cellular processes, including cell
differentiation, proliferation, survival, glucose metabolism,
immune regulation, and synaptic plasticity associated with memory
(14–20).
Similar findings have been reported for other
tumors, Qi and Ding analyzed gene expression profiles in colorectal
cancer and studied how CREB5 gene expression levels affect
the molecular events in colorectal cancer, and found that these
molecular events were correlated with tumor metastasis (6). Seo et al reported that CREB and
phosphorylated CREB mRNA and protein levels were significantly
higher in most non-small cell lung cancer cell lines and tumor
specimens than in the normal human tracheobronchial epithelial
cells and adjacent normal lung tissue, respectively (7). Analysis of CREB mRNA expression and gene
copy number showed that CREB overexpression occurred mainly at the
transcriptional level.
CREB could activate the transcription of downstream
genes by binding to CRE. Alternatively spliced transcript variants
encoding different isoforms have been identified. Gene ontology
annotations related to this gene include nucleic acid binding and
sequence-specific DNA binding. The study of Seo et al found
that CREB expression levels in non-small cell lung cancer cells
were highly dependent on the elevated CREB activity for their
survival and cell growth, and inhibition of CREB effectively
suppressed the growth of non-small cell lung cancer cells (7). Kinjo et al reported that a
majority of patients with acute lymphoid and myeloid leukemia
overexpressed CREB in the bone marrow. CREB overexpression was
associated with poor initial outcome of clinical disease in
patients with acute myelocytic leukemia (21). Our study proved that CREB5 was
significantly overexpressed in epithelial ovarian cancer. The CREB5
expression level was significantly correlated with aggressive
features and an unfavorable prognosis in 125 patients with ovarian
cancer. Furthermore, Kaplan-Meier curves and multivariate Cox
proportional hazard regression analysis indicated that CREB5
overexpression was a strong and independent predictor for poor
overall survival in patients with ovarian carcinoma. These findings
revealed that CREB5 might play an important role as an indicator of
poor outcomes in individual patients with ovarian cancer.
In conclusion, our study revealed that CREB5 played
an important role in the development and progression of epithelial
ovarian cancer, and CREB5 overexpression was positively associated
with the ascending FIGO stage and poor survival of epithelial
ovarian cancer. Therefore, we believe that CREB5 may be a potential
target for epithelial ovarian cancer therapy. The results showed in
this manuscript were preliminary data. In future studies, samples
should be enriched, because the present study analyzed only 125
epithelial ovarian cancer cases. We should further verify the role
and function of CREB5 in ovarian cell lines. The gain- and loss-of
function of CREB5 in ovarian cell lines should be performed. The
abilities of growth and motility in gain- and loss-function of
CREB5 in ovarian cell lines should be tested. These results may
provide a strong evidence to support the clinical data.
Acknowledgements
This study was supported by grants from the Science
and Technology Planning Project of Guangdong Province, China (grand
no. 2016A030313820); the Science and Technology Planning Project of
Huangpu Guangdong Province, China (grand no. 201544-03); and the
Science and Technology Planning Project of Guangzhou City, China
(grand no. 201704020163).
References
|
1
|
Gloss BS and Samimi G: Epigenetic
biomarkers in epithelial ovarian cancer. Cancer Lett. 342:257–263.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Matulonis UA, Sood AK, Fallowfield L,
Howitt BE, Sehouli J and Karlan BY: Ovarian cancer. Nat Rev Dis
Primers. 2:160612016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Aletti GD, Gallenberg MM, Cliby WA, Jatoi
A and Hartmann LC: Current management strategies for ovarian
cancer. Mayo Clin Proc. 82:751–770. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang X, Liu H, Xie Z, Deng W, Wu C, Qin
B, Hou J and Lu M: Epigenetically regulated miR-449a enhances
hepatitis B virus replication by targeting cAMP-responsive element
binding protein 5 and modulating hepatocytes phenotype. Sci Rep.
6:253892016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zu YL, Maekawa T, Nomura N, Nakata T and
Ishii S: Regulation of trans-activating capacity of CRE-BPa by
phorbol ester tumor promoter TPA. Oncogene. 8:2749–2758.
1993.PubMed/NCBI
|
|
6
|
Qi L and Ding Y: Involvement of the CREB5
regulatory network in colorectal cancer metastasis. Yi Chuan.
36:679–684. 2014.PubMed/NCBI
|
|
7
|
Seo HS, Liu DD, Bekele BN, Kim MK, Pisters
K, Lippman SM, Wistuba II and Koo JS: Cyclic AMP response
element-binding protein overexpression: A feature associated with
negative prognosis in never smokers with Non-small cell lung
cancer. Cancer Res. 68:6065–6073. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jarboe EA, Folkins AK, Drapkin R, Ince TA,
Agoston ES and Crum CP: Tubal and ovarian pathways to pelvic
epithelial cancer: A pathological perspective. Histopathology.
53:127–138. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hennessy BT, Coleman RL and Markman M:
Ovarian cancer. Lancet. 374:137–1382. 2009. View Article : Google Scholar
|
|
10
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Grann AF, Nørgaard M, Blaakær J,
Søgaard-Andersen E and Jacobsen JB: Survival of patients with
ovarian cancer in central and northern Denmark, 1998–2009. Clin
Epidemiol. 3 Suppl 1:S59–S64. 2011. View Article : Google Scholar
|
|
12
|
Bender E: Trials show delayed recurrence
in ovarian cancer. Cancer Discov. 3:OF82013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Amate P, Huchon C, Dessapt AL, Bensaid C,
Medioni J, Le Frère Belda MA, Bats AS and Lécuru FR: Ovarian
cancer: Sites of recurrence. Int J Gynecol Cancer. 23:1590–1596.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wen AY, Sakamoto KM and Miller LS: The
role of the transcription factor CREB in immune function. J
Immunol. 185:6413–6419. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Carlezon WA Jr, Duman RS and Nestler EJ:
The many faces of CREB. Trends Neurosci. 28:436–445. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Silva AJ, Kogan JH, Frankland PW and Kida
S: CREB and memory. Annu Rev Neurosci. 21:127–148. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mayr B and Montminy M: Transcriptional
regulation by the phosphorylation-dependent factor CREB. Nat Rev
Mol Cell Biol. 2:599–609. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Arnould T, Vankoningsloo S, Renard P,
Houbion A, Ninane N, Demazy C, Remacle J and Raes M: CREB
activation induced by mitochondrial dysfunction is a new signaling
pathway that Impairs cell proliferation. EMBO J. 21:53–63. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Vaudry D, Stork PJ, Lazarovici P and Eiden
LE: Signaling pathways for PC12 cell differentiation: Making the
right connections. Science. 296:1648–1649. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bender RA, Lauterborn JC, Gall CM, Cariaga
W and Baram TZ: Enhanced CREB phosphorylation in immature dentate
gyrus granule cells precedes neurotrophin expression and indicates
a specific role of CREB in granule cell differentiation. Eur J
Neurosci. 13:679–686. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kinjo K, Sandoval S, Sakamoto KM and
Shankar DB: The role of CREB as a proto-oncogene in hematopoiesis.
Cell Cycle. 4:1134–1135. 2005. View Article : Google Scholar : PubMed/NCBI
|