Introduction
Chronic lymphocytic leukemia (CLL) is the most
common type of adult leukemia in the Western hemisphere (1). In 2016, ~18 960 new cases of CLL were
diagnosed in the United States (1).
The treatment of CLL has markedly changed in the past years with
the regulatory approval of idelalisib and ibrutinib. In addition,
other antibodies are likely to become widely available in the next
few years (2,3). Over the past decade, the essential role
of the tumor microenvironment in the survival and progression of
CLL has become increasingly clear (4). In the bone marrow and secondary
lymphatic tissues, CLL cells engage in complex, yet not completely
defined, cellular and molecular interactions with stromal cells and
the matrix, which collectively are referred to as the
‘microenvironment’ (5). These
interactions affect CLL cell survival and proliferation, and confer
drug resistance, which may be responsible for residual disease
following conventional therapy (6).
Understanding the association of neoplastic cells with the
microenvironment (i.e., identifying which cells are required for
lymphoma growth and which are involved in regulating lymphoma
growth) will be crucial to developing therapies aimed at targeting
the microenvironment (4,6). Notably, the cross-talk between CLL cells
and bone marrow stromal cells (BMSCs) is bidirectional, causing the
activation of both CLL cells and BMSCs (7). BMSCs attract and affect CLL cells via
G-protein-coupled chemokine receptors, including C-X-C chemokine
receptor type 4, cluster of differentiation (CD) 79a and Notch
homolog 1, translocation-associated (Drosophila) (Notch1),
which are expressed at high levels on CLL cells (8,9). Our group
previously reported that Notch1 constitutively activates the
nuclear factor (NF)-κB signaling pathway through activating its
downstream gene, Hes family BHLH transcription factor 1 (HES-1), in
CLL (10). However, the mechanism by
which stromal cells affect Notch1-driven oncogenic target gene
activation by histone modification is largely unknown. The present
study demonstrates that stromal cells enhance the survival of CLL
cells by regulating HES-1 gene and protein expression, as well as
histone H3K27me3 DNA demethylation. In the present study, several
experiments were designed to determine if a BMSC line, HESS-5,
improves the survival and proliferation of CLL cells in
vitro. The ability of HESS-5 cells to protect CLL cells from
serum deprivation-induced apoptosis and to modify apoptosis
responses to the chemotherapy agent cyclophosphamide (CTX) were
evaluated. In addition, the present study examined whether the
expression of HES-1, one of the key regulators of apoptosis, as
well as H3K27me3, the main histone involved in the gene
transcription of Notch1 (11), are
affected in CLL cells by co-culture with HESS-5 monolayers.
Materials and methods
Cells
The hematopoietic-supportive murine stromal cell
line HESS-5, murine CLL cell line L1210 (The Cell Bank of Type
Culture Collection of Chinese Academy of Sciences, Shanghai,
China), and primary CLL cells from patients with CLL were used in
the present study. The choice of L1210 was due to ease of
accessibility and culturing compared with other human CLL cell
lines in our lab. Samples (10 ml) of human bone marrow were
obtained from a newly diagnosed patient with CLL upon obtaining
informed consent in Fujian Medical University Union Hospital
(Fuzhou, China) between 1st January 2012 and 1st January 2014. The
present study was approved by the Ethics Committee of Fujian
Medical University Union Hospital (document no. 139). The L1210 and
primary CLL cells were cultured in 24-well, flat-bottom plates,
with or without a confluent HESS-5 stromal cell layer in RPMI-1640
with 20% fetal bovine serum, 1% penicillin and streptomycin
[American Type Culture Collection (ATCC), Manassas, VA, USA]. All
cells were cultured at 37°C with 5% CO2. In certain
experiments, cells were grown in serum-free conditions for 24 h,
with or without the presence of 0.5 or 1.0 µM CTX (GE Healthcare
Life Sciences, Uppsala, Sweden).
L1210 cell growth on fixed stromal
cells
To interrupt the metabolic activity of stromal cells
while leaving surface proteins intact, confluent HESS-5 cell layers
were treated with glutaraldehyde as previously described (12). L1210 cells were then incubated on the
fixed stromal cell layers for 48 h in the presence of CTX (0.5 or
1.0 µM). L1210 cells were collected through vigorous pipetting, and
the viability of all samples was evaluated in triplicate by Trypan
Blue exclusion assay as previously described (12).
Evaluation of leukemic cell
viability
L1210 cells were cultured under different
conditions: With or without serum, CTX (0.5 or 1.0 µM), monolayers
or 50% stromal conditioned medium (CM) from HESS-5 cells. Viability
was evaluated by Trypan Blue exclusion assay in triplicate samples
daily. After 48 h of the above treatments, leukemic cells were
transferred to 96-well flat-bottom plates containing fresh
RPMI-1640 medium with 10% fetal calf serum (FCS; ATCC). Viability
was evaluated using an MTT assay as previously described following
an additional 48 h (12).
Evaluation of leukemic cell
apoptosis
CLL cells or L1210 were cultured under different
conditions: With or without CTX, monolayers or 50% stromal CM of
HESS-5 cells. CLL cells or L1210 were removed from adherent stromal
cells by vigorous pipetting, and then evaluated using flow
cytometry as previously described (12) with an Alexa Fluor488 AnnexinV
apoptosis kit (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA,
USA) according to the manufacturer's protocol. For the terminal
deoxynucleotidyl transferase-mediated 2′-deoxyuridine
5′-triphosphate (dUTP) nick end labeling (TUNEL) assay,
fluorescein-conjugated dUTP incorporated into nucleotide polymers
was detected and quantified using flow cytometry. In the present
study, the In Situ Cell Death Detection kit, Fluorescein
(Roche Diagnostics GmbH, Mannheim, Germany) was used according to
the manufacturer's protocol. Briefly, cells (2×106
cells/well) were cultured on cover glasses in 6-well plates.
Following treatment, cells were washed with PBS, fixed with 4%
paraformaldehyde solution for 1 h at 20–25°C and permeabilized
through incubation for 2 min on ice. Following washing with PBS
cells were incubated with the TUNEL reaction mixture for 60 min at
37°C in the dark. Fluorescence of the stained cells was quantified
using flow cytometry.
RNA isolation and reverse
transcription-polymerase chain reaction (RT-PCR)
After 24 h of CTX treatment, primary CLL cells were
transferred onto a confluent HESS-5 cell layer and cultured for ≤48
h. Leukemic cells were collected by vigorous pipetting. Their RNA
was isolated according to the single-step acid guanidinium
thiocyanate-phenol-chloroform (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) method (13). The
RNA quality and quantity were determined following 1.0% agarose gel
electrophoresis and staining with 1 µg/ml ethidium bromide.
Oligo-(dT) 10 primers (Invitrogen; Thermo Fisher Scientific, Inc.)
were used as for complementary DNA (cDNA) synthesis. Total RNA
template was used per each 10 µl of RT reaction in the presence of
AMV Reverse Transcriptase (Roche Diagnostics GmbH) at 42°C for 1 h.
PCR results were confirmed using 1% agarose gel electrophoresis
then stored at −20°C. PCR amplification reaction mixtures (50 µl)
contained DNA polymerase, dNTPs, cDNA (2 µl), and HES-1, c-MYC
forward/reverse primers (70 pmol/l) and ß2-microglobulin
primers (70 pmol/l; used as control), which were synthesized by
Shanghai Sangong Pharmaceutical Co., Ltd. (Shanghai, China).
Thermal cycler conditions included holding the reactions at 50°C
for 2 min and at 95°C for 10 min, followed by amplification for 40
cycles using 95°C for 15 sec and 60°C for 1 min. The PCR samples
were electrophoresed on 1.0% agarose gels and visualized with 1
µg/ml ethidium bromide. The gel images were digitally captured and
analyzed with AlphaEaseFC software (version 5.0; ProteinSimple;
Bio-Techne, Minneapolis, MN, USA).
Western blot analysis of HES-1
expression
After 24 h of CTX (0.5 or 1.0 µM) treatment, primary
CLL cells were transferred onto the confluent HESS-5 cell layer for
≤48 h. Leukemic cells were collected by vigorous pipetting
following either 24 or 48 h of co-culture, and then lysed in 10 mM
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (pH 7.6), 250 mM
NaCl, 5 mM EDTA and 0.5% Nonidet P-40. The lysates were
fractionated by 12% SDS-PAGE and transferred to nitrocellulose
membranes. The membranes were then blocked in 5% non-fat milk at
room temperature for 1 h and then incubated with mouse anti-human
HES-1 antibody (cat no. RS-2972R; 1:3,000 dilution; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA) and a mouse monoclonal
anti-β-actin antibody (cat no. A5316; control; 1:3,000 dilution;
Sigma-Aldrich; Merck KGaA) overnight at 4°C, and a secondary
horseradish peroxidase-conjugated anti-mouse antibody (cat no.
BL0824; Beijing Zhongshan Jinqiao Biotechnology Co., Ltd., Beijing,
China) for 1 h at room temperature. Detection was performed with
Immobilon Western Detection reagents (Sigma-Aldrich; Merck
KGaA).
Methylation-specific PCR (MSP)
analysis of the H3K27me3 gene
Primary CLL cells were seeded onto a confluent
HESS-5 cell layer for ≤48 h. Genomic DNA was extracted from primary
CLL cells following the standard procedure of DNA extraction kit
(GE Healthcare Life Sciences). The genomic DNA was quantified by
Qubit Fluorimeter (Thermo Fisher Scientific, Inc.) and then stored
at −20°C. Approximately 5,000 ng DNA fragments were used for
bisulfite conversion using the Qiagen EpiTect system (Qiagen, Inc.,
Valencia, CA, USA) according to the manufacturer's protocol, and
100 ng of this modified DNA was used as the template for MSP
analysis as previously described (14). Thermal cycler conditions included
holding the reactions at 50°C for 2 min and at 95°C for 10 min per
cycle, for a total of 33 cycles. The PCR samples were
electrophoresed on 1.0% agarose gels and then stained with 1 µg/ml
ethidium bromide. The gel images were digitally captured and
analyzed with AlphaEaseFC software (version 5.0). U266 cells
expressing methylated H3K27me3 were used as a positive control
(15).
Statistical analysis
Data are presented as the mean + the standard error
of the mean. Statistical significance was determined by a Student's
two-tailed t-test using SPSS version 11.0 (SPSS Inc., Chicago, IL,
USA). P<0.05 was considered to indicate a statistically
significant difference.
Results
Stromal cells enhance CLL cell
viability in serum-deprivation conditions and during chemotherapy
exposure
To determine whether co-culture with a stromal cell
monolayer affected leukemic cell survival during chemotherapy drug
exposure and serum-deprivation, L1210 cells were grown on HESS-5
monolayer and compared with L1210 cells maintained in culture
medium alone. In all cases, stromal cell co-culture resulted in
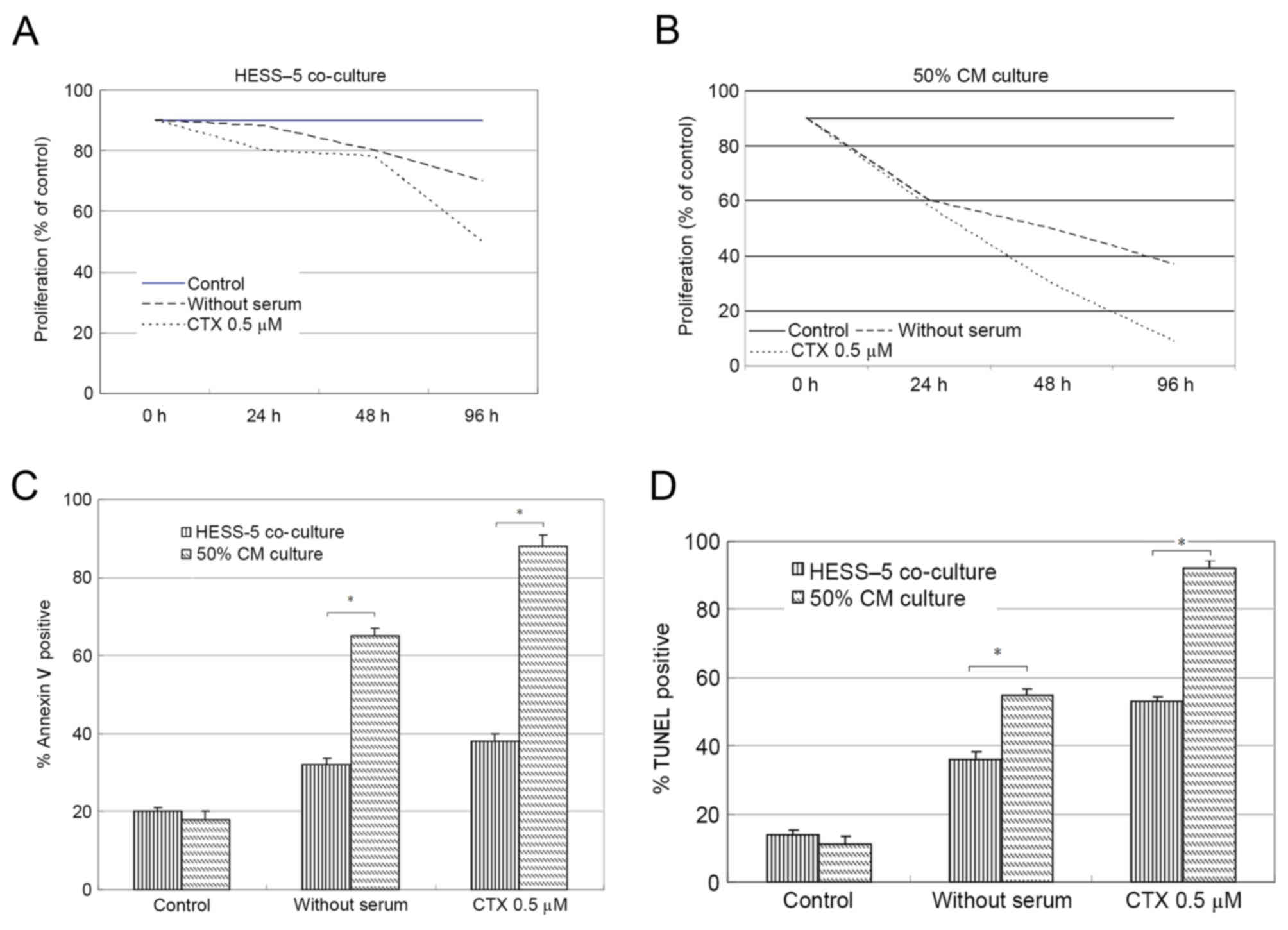
increased L1210 cell viability during treatment (data not shown).
The viability of CLL cells cultured on stromal cell layers
increased by ~3-fold when compared with that of cells cultured in
medium alone. Upon treatment with 0.5 µM CTX for 48 h, the CLL cell
number decreased by 30%, but stromal cell co-culture significantly
rescued the viability of these CLL cells. After 96 h of
serum-deprivation, the number of CLL cells cultured in medium alone
was reduced by 40% compared with 70% when serum-deprived cells were
co-cultured with HESS-5 cells (Fig.
1A).
Following serum-free culture or CTX exposure, CLL
cells were co-cultured with stromal cells for 48 h, prior to being
collected and plated on 96-well flat-bottom plates with fresh
medium (10% FCS). Soluble factors derived from stromal cells (50%
CM) did not alter the response of leukemic cells to serum
withdrawal or chemotherapy treatment (Fig. 1B).
Stromal cells inhibit leukemic cell
apoptosis induced by CTX
To determine whether CTX induced apoptosis, L1210
cells were evaluated by flow cytometry, expression following
chemotherapy drug exposure in the presence or absence of HESS-5
monolayer. The number of apoptotic cells increased from 20% in the
control group to 90% in the experimental group after 48 h of CTX
treatment. However, co-culture with stromal cells following CTX
exposure resulted in significantly reduced numbers of apoptotic
leukemic cells (40%; P<0.01; Fig.
1C). The same results were also attained by TUNEL assay
(Fig. 1D).
BMSCs enhance the expression of HES-1
and c-MYC in CLL cells following treatment with CTX
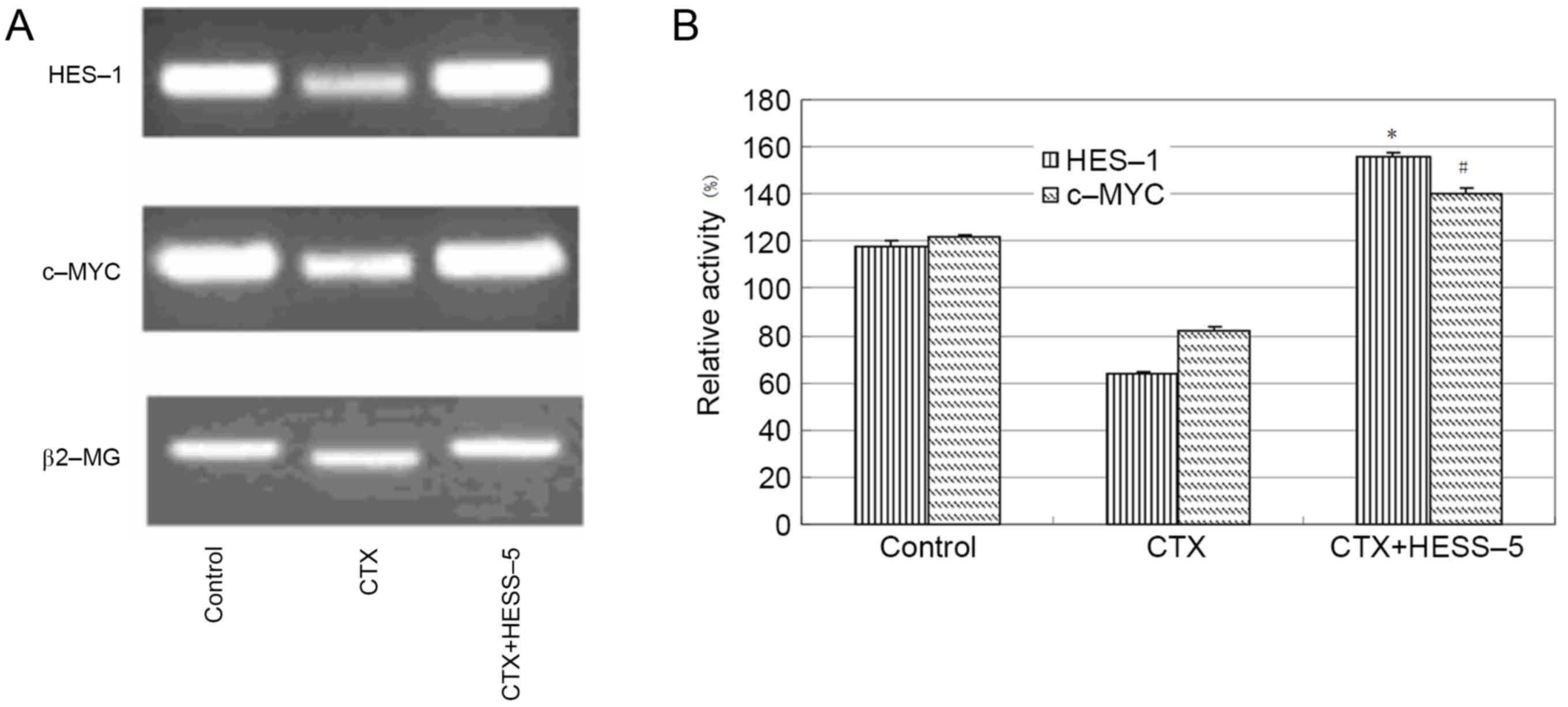
To further explore the mechanism of stromal cell
protection of leukemic cells from apoptosis, HES-1 and c-MYC
messenger RNA expression, as well as HES-1 protein expression, were
measured in primary CLL cells treated with CTX in the presence or
absence of a HESS-5 monolayer. HES-1 and c-MYC gene expression was
higher in CLL cells supported by stromal cells than in CTX-treated
CLL cells alone (Fig. 2A and B).
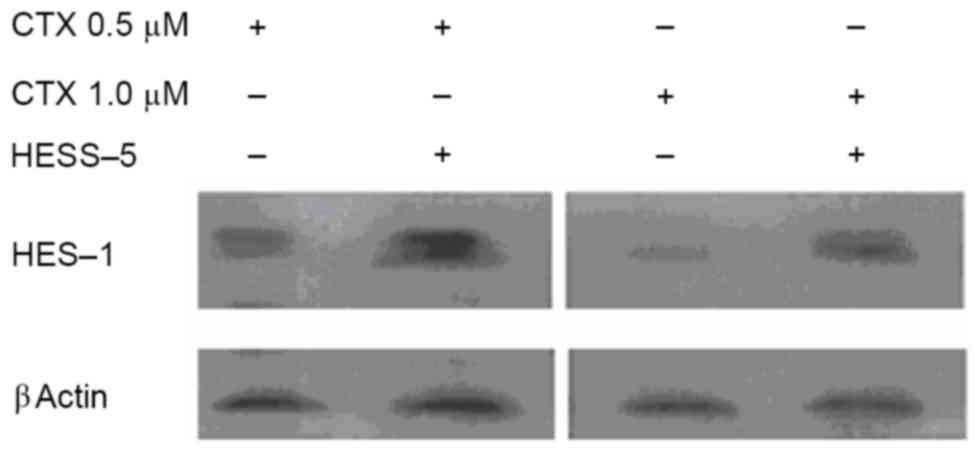
Western blot analysis revealed that HES-1 protein expression was
increased in primary CLL cells treated with two concentrations of
CTX (0.5 and 1.0 µM) for 24 h and then cultured for additional 48 h
with the HESS-5 cell monolayer (Fig.
3).
Co-culture of primary CLL cells with
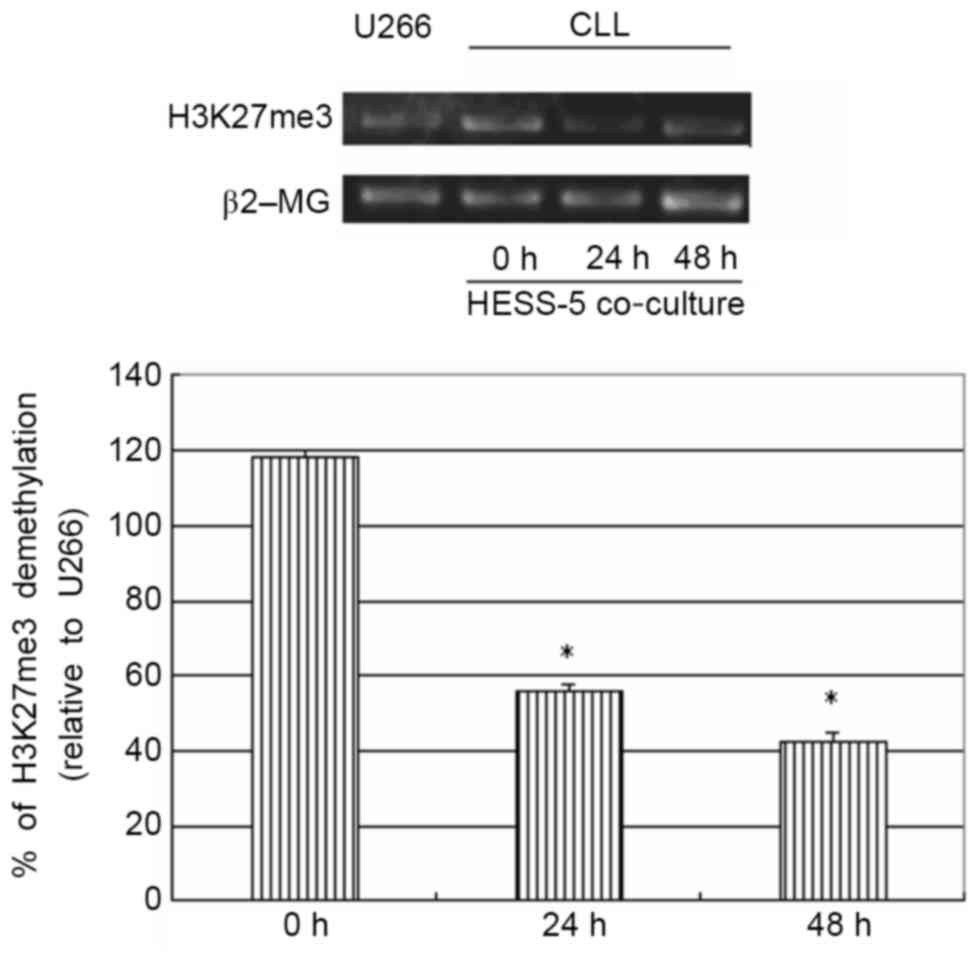
stromal cells results in demethylation of H3K27me3
To further explore the possible mechanism of stromal
cell protection of leukemic cells from apoptosis, H3K27me3 gene
expression was measured in primary CLL cells treated with CTX in
co-culture with a HESS-5 stromal cell layer. The H3K27me3 gene has
been shown to be methylated in primary CLL cells (16). However, in the present study,
co-culture with HESS-5 cells led to a significant demethylation of
this gene in primary CLL cells (P<0.05; Fig. 4).
Discussion
Growth and differentiation in vivo of
hematopoietic cells, including normal and leukemic cells, require
direct contact of hematopoietic cells with stromal cells (4). CLL cells display enhanced survival in
vivo, but they die spontaneously and are difficult to maintain
alive in vitro (5,7). It has been reported that BMSCs
synthesize several cytokines, including colony stimulating factors,
interleukin (IL)-6, IL-7, IL-10, transforming growth factor-β and
stem cell factor, which constitute a complex regulatory network
together with extracellular matrix proteins (17). Contact with the bone marrow stroma
induces the proliferation and survival of acute myeloid leukemia,
acute lymphoblastic leukemia and CLL cells in culture (17,18).
HESS-5 cells, a murine stromal cell line capable of supporting
early human progenitors in vitro (19), was used in the present study as a
model to evaluate hematopoietic-stromal cell interactions.
The present study observed that stromal cell CM
conferred negligible protection for leukemic cells during serum
deprivation or CTX-induced apoptosis. Therefore, the role of
proteins on the stromal cell surface was investigated using
glutaraldehyde-fixed stromal cells. Previous reports have shown
that this fixation process disrupts the metabolism of adherent 3T3
cells, thus allowing to investigate the role of signaling through
ligand-receptor interactions in the absence of cytokine production
(20). It has also been suggested
that signals between stromal cells and hematopoietic progenitors
can be bidirectional, with the adhesion of hematopoietic cells to
stromal cells altering stromal cell function (21). In the present study, fixed stromal
cells protected leukemic cells from CTX-induced cell death,
similarly to viable stromal cells. These observations support the
conclusion that the signals protecting leukemic cell survival
primarily result from cell-surface interactions, but not from
soluble factors in stromal CM.
The present study also demonstrated that HESS-5
cells mediated the inhibition of apoptosis that was associated with
increased HES-1 and c-MYC expression. Notch1 is known to be
involved in the pathogenesis of leukemia. Unlike normal B cells,
CLL cells express both Notch1 and 2, and their ligands (10). Co-expression of the Notch receptors
and their ligands results in the constitutive activation of the
Notch target molecule HES-1, which is a potential biomarker in
leukemia cells (22,23). In CLL, upon Notch1 activation, the
cleaved intracellular portion of the Notch1 receptor translocates
into the nucleus, where it recruits a transcription complex to
modify the expression of several target genes, including HES-1,
c-MYC and NF-κB (10). Notch1
mutations in CLL have been reported to cause disruptions to HES-1
expression, resulting in the stabilization of active intracellular
Notch1 and deregulated Notch signaling (24). In fact, Notch1 mutations are
significantly more common in CLL harboring +12 trisomy with
intermediate-risk disease compared with low-risk CLL (25,26).
Although stromal cells were demonstrated to stimulate the
expression of HES-1 and c-MYC in CLL cells in the present study,
further studies are warranted to evaluate whether stromal cells can
activate mutated Notch1 in a similar manner.
CLL hypermethylation has been reported to frequently
affect DNA repeats (27). In general,
hypermethylation frequently targets genes already silenced in
non-tumor cells by repressive histone modifications such as
H3K27me3 (28). Thus, although
certain tumor-suppressor genes become de novo methylated and
silenced in cancer, hypermethylation affects mostly genes already
silenced in normal cells (28,29). The
H3K27me3 methylation mark is associated with chromatin condensation
and transcriptional repression (16).
The results of the present study confirmed that stromal cells in
direct contact with CLL cells lead to the demethylation of
H3K27me3, which is associated with transcriptional activation of
the target gene HES-1.
As the present study was performed using the mouse
CLL line, further studies are required to validate the present
results in human CLL cell lines. In summary, the present study has
shown that cell-cell contact with BMSCs protects CLL cells from
apoptosis by upregulating HES-1 and c-MYC expression, which is
associated with H3K27me3 demethylation. Further studies will be
necessary to identify other factors and mechanisms involved in the
interaction between stromal and leukemic cells.
Acknowledgements
The present study was supported by the Fujian
Natural Science Fund (grant no. 2016J01458) and the New Century
Talent in Fujian (grant no. JA10128) (both received by Zhenshu Xu),
sponsored by National and Fujian Provincial Key Clinical Specialty
Discipline Construction Program, P.R.C and Construction Project of
Fujian Medical Center of Hematology (Min201704).
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Suresh T, Lee L, Joshi J and Barta S: New
antibody approaches to lymphoma therapy. J Hematol Oncol. 7:582014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wu J, Fu J, Zhang M and Liu D:
Blinatumomab: A bispecific T cell engager (BiTE) antibody against
CD19/CD3 for refractory acute lymphoid leukemia. J Hematol Oncol.
8:1042015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nwabo Kamdje AH and Krampera M: Notch
signaling in acute lymphoblastic leukemia: Any role for stromal
microenvironment. Blood. 118:6506–6514. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Burger JA, Ghia P, Rosenwald A and
Caligaris-Cappio F: The microenvironment in mature B-cell
malignancies: A target for new treatment strategies. Blood.
114:3367–3375. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sun Z, Wang S and Zhao RC: The roles of
mesenchymal stem cells in tumor inflammatory microenvironment. J
Hematol Oncol. 7:142014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ding W, Nowakowski GS, Knox TR, Boysen JC,
Maas ML, Schwager SM, Wu W, Wellik LE, Dietz AB, Ghosh AK, et al:
Bi-directional activation between mesenchymal stem cells and CLL
B-cells: Implication for CLL disease progression. Br J Haematol.
147:471–483. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Vlad A, Deglesne PA, Letestu R,
Saint-Georges S, Chevallier N, Baran-Marszak F, Varin-Blank N,
Ajchenbaum-Cymbalista F and Ledoux D: Down-regulation of CXCR4 and
CD62L in chronic lymphocytic leukemia cells is triggered by B-cell
receptor ligation and associated with progressive disease. Cancer
Res. 69:6387–6395. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang L, Lawrence MS, Wan Y, Stojanov P,
Sougnez C, Stevenson K, Werner L, Sivachenko A, DeLuca DS, Zhang L,
et al: SF3B1 and other novel cancer genes in chronic lymphocytic
leukemia. N Engl J Med. 365:2497–2506. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xu ZS, Zhang JS, Zhang JY, Wu SQ, Xiong
DL, Chen HJ, Chen ZZ and Zhan R: Constitutive activation of NF-kB
signaling by NOTCH1 mutation in chronic lymphocytic leukemia. Oncol
Rep. 33:1609–1614. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ross J, Mavoungou L, Bresenick EH and
Milot E: GATA-1 utilizes lkaros and polycomb repressive complex 2
to suppress Hes-1 and to promote erythropoiesis. Mol Cell Biol.
32:3624–3638. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hasslen SR, Burns AR, Simon SL, Smith CW,
Starr K, Barclay AN, Michie SA, Nelson RD and Erlandsen SL:
Preservation of spatial organization and antigenicity of leukocyte
surface molecules by aldehyde fixation: Flow cytometry and
high-resolution FESEM studies of CD62L, CD11b and Thy-1. J
Histochem Cytochem. 44:1115–1122. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chomozynski P and Sacchi N: Single step
method of RNA isolation by acid guanidinium thiocyanate phenol
chloroform extraction. Anal Biochem. 162:156–159. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cai MY, Hou JH, Rao HL, Luo RZ, Li M, Pei
XQ, Lin MC, Guan XY, Kung HF, Zeng YX and Xie D: High expression of
H3K27me3 in human hepatocellular carcinomas correlates closely with
vascular invasion and predicts worse worse prognosis in patients.
Mol Med. 17:12–20. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kikuchi J, Koyama D, Wada T, Izumi T,
Hofgaard PO, Bogen B and Furukawa Y: Phosphorylation-mediated EZH2
inactivation promotes drug resistance in multiple myeloma. J Clin
Invest. 125:4375–4390. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Amin S, Walsh M, Wilson C, Parker AE,
Oscier D, Willmore E, Mann D and Mann J: Cross-talk between DNA
methylation and active histone modifications regulates aberrant
expression of ZAP70 in CLL. J Cell Mol Med. 16:2074–2084. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Moshaver B, van der Pol MA, Westra AH,
Ossenkoppele GJ, Zweegman S and Schuurhuis GJ: Chemotherapeutic
treatment of bone marrow stromal cells strongly affects their
protective effect acute myeloid leukemia cell survival. Leuk
Lymphoma. 49:134–148. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lagneaux L, Delforge A, Bron D, De Bruyn C
and Stryckmans P: Chronic lymphocytic leukemic B cells are rescued
from apoptosis by contact with normal bone marrow stromal cells.
Blood. 91:2387–2396. 1998.PubMed/NCBI
|
|
19
|
Shimakura Y, Kawada H, Ando K, Sato T,
Nakamura Y, Tsuji T, Kato S and Hotta T: Murine stromal cell line
HESS-5 maintains reconstituting ability of ex vivo-generated
hematopoietic stem cells from human bone marrow and
cytokine-mobilized peripheral blood. Stem Cells. 18:183–189. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Roberts RA, Spooncer E, Parkinson EK, Lord
BI, Allen TD and Dexter TM: Metabolically inactive 3T3 cells can
substitute for marrow stromal cells to promote the proliferation
and development of multipotent hematopoietic stem cells. J Cell
Physiol. 132:203–214. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jiang H, Sugimoto K, Sawada H, Takashita
E, Tohma M, Gonda H and Mori KJ: Mutual education between
hematopoietic cells and bone marrow stromal cells through direct
cell-to-cell contact: Factors that determine the growth of bone
marrow stroma-dependent leukemic (HB-1) cells. Blood. 92:834–841.
1998.PubMed/NCBI
|
|
22
|
Smith AD, Roda D and Yap TA: Strategies
for modern biomarker and drug development in oncology. J Hematol
Oncol. 7:702014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ma S, Shi Y, Pang Y, Dong F, Cheng H, Hao
S, Xu J, Zhu X, Yuan W, Cheng T and Zheng G: Notch1-induced T cell
leukemia can be potentiated by microenvironmental cues in the
spleen. J Hematol Oncol. 7:712014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fabbri G, Rasi S, Rossi D, Trifonov V,
Khiabanian H, Ma J, Grunn A, Fangazio M, Capello D, Monti S, et al:
Analysis of the chronic lymphocytic leukemia coding genome: Role of
NOTCH1 mutational activation. J Exp Med. 208:1389–1401. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Balatti V, Bottoni A, Palamarchuk A, Alder
H, Rassenti LZ, Kipps TJ, Pekarsky Y and Croce CM: NOTCH1 mutations
in CLL associated with trisomy 12. Blood. 119:329–331. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xu Z, Zhang J, Wu S, Zheng Z, Chen Z and
Zhan R: Younger patients with chronic lymphocytic leukemia benefit
from rituximab treatment: A single center study in China. Oncol
Lett. 5:1266–1272. 2013.PubMed/NCBI
|
|
27
|
Fabris S, Bollati V, Agnelli L, Morabito
F, Motta V, Cutrona G, Matis S, Grazia Recchia A, Gigliotti V,
Gentile M, et al: Biological and clinical relevance of quantitative
global methylation of repetitive DNA sequences in chronic
lymphocytic leukemia. Epigenetics. 6:188–194. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Martín-Subero JI, Kreuz M, Bibikova M,
Bentink S, Ammerpohl O, Wickham-Garcia E, Rosolowski M, Richter J,
Lopez-Serra L, Ballestar E, et al: New insights into the biology
and origin of mature aggressive B-cell lymphomas by combined
epigenomic, genomic, and transcriptional profiling. Blood.
113:2488–2497. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lu Q, Lin X, Feng J, Zhao X, Gallagher R,
Lee YM, Chiao JW and Liu DL: Phenylhexyl isothiocyanate has dual
function as histone deacetylase inhibitor and hypomethylating agent
and can inhibit myeloma cell growth by targeting critical pathways.
J Hematol Oncol. 1:62008. View Article : Google Scholar : PubMed/NCBI
|