Introduction
Colorectal cancer (CRC) is the third most frequently
diagnosed cancer in males and females in the United States
(1). As surgery alone is not
sufficient to cure the majority of patients with CRC, adjuvant
chemotherapy or radiation therapy is also typically administered to
patients (2). A combination of
chemotherapy with folinic acid, 5-fluorouracil and oxaliplatin and
folinic acid, 5-fluorouracil and irinotecan has become the standard
treatment regimen for patients with CRC, providing a higher
response rate compared with conventional chemotherapy (3–6). However,
the response rates of current chemotherapy regimes are <50% and
therefore, alternative molecular targets are required to improve
drug response rates (3–6).
The kinesin superfamily proteins (KIFs) are
microtubule-dependent molecular motors that convert the chemical
energy from ATP hydrolysis to the mechanical action of transporting
cargo along microtubules, suggesting that they have an important
role in intracellular transport and cell division (7). KIFs are classified into 14 distinct
families with varying structural and functional characteristics
(8,9).
Among these, KIF4A is considered to have a role in chromosome
condensation and is involved in the segregation machinery that
functions in mitotic division (10,11).
Alterations in the regulation of KIF4A promote abnormal spindle
separation and lead to aneuploidy in daughter cells (10). Cells with aneuploidy are characterized
by loss or gain of genetic material (11). Therefore, KIF4A expression levels may
be associated with cancer progression.
It has been reported that KIF4A expression is
altered in numerous types of cancer, including cervical (12), lung (13), gastric (14), oral (15) and breast cancer (16). These alterations in cancer cells
suggest that the biological function of KIF4A is associated with
the regulation of the cell cycle and cellular proliferation. A
previous microarray study found that there was increased expression
of KIF4A mRNA in human cervical cancer (12). KIF4A expression was identified as
being upregulated in lung cancer and was significantly associated
with the male gender, non-adenocarcinoma histology and a reduced
survival rate in patients with non-small cell lung cancer (13). In an immunohistochemical (IHC)
evaluation of 106 patients with oral squamous cell cancer, KIF4A
expression levels in cancer tissue were significantly increased
compared with those in normal tissue (15). A previous study demonstrated that
estrogen induces a number of KIFs, including KIF4A, and increased
levels of KIF4A are associated with reduced relapse-free survival
of patients with breast cancer that are positive for the estrogen
receptor (16). These previous
studies indicated that KIF4A may function as an oncogene in a
numerous types of cancer, but the expression and role of KIF4A in
CRC remain to be elucidated. The present study investigated the
biological significance of KIF4A expression levels to clarify the
function of KIF4A in CRC.
Materials and methods
Clinical tissue samples from
patients
A total of 258 surgical specimens were obtained for
analysis from 258 patients with CRC that underwent surgical
resection at Fukushima Medical University Hospital (Fukushima,
Japan) between January 1991 and December 2011. In 63/258 of the
tissues, mRNA was extracted from cancer tissue and adjacent
non-tumor tissue. Information regarding age, gender,
tumor-mode-metastasis (TNM) stage (the 7th classification)
(17,18) and pathological diagnosis, including
lymphatic and venous invasion, were retrospectively collected. At
the time of primary tumor resection, carcinoma tissues were staged
according to the Union for International Cancer Control
classification (17,18). Written informed consent was obtained
from all patients. This study was approved by the Ethics Committee
of Fukushima Medical University (ref. no. 2117).
Cell line culture
The RKO, SW480, Lovo, HCT15, SW48, LS174T, SW620,
LS180 and HCT116 colon cancer cell lines were used in this study
and all were obtained from the American Type Culture Collection
(Manassas, VA, USA). The cells were cultured in the recommended
media (Dulbecco's modified Eagle's medium; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) for RKO, LS174T and LS180 and
Roswell Park Memorial Institute-1640 medium (Sigma-Aldrich; Thermo
Fisher Scientific, Inc.) for SW480, Lovo, HCT15, SW48, SW620 and
HCT116 supplemented with 10% fetal bovine serum (Thermo Fisher
Scientific, Inc.). Cells were maintained in a 37°C incubator in an
atmosphere containing 5% CO2. Cells were regularly
monitored using a light microscope and subcultured once they
reached 80–90% confluency.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from RKO, SW480, Lovo,
HCT15, SW48, LS174T, SW620, LS180 and HCT116 cells using
TRIzol® reagent (Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol as previously described
(19). Complementary DNA (cDNA) was
synthesized from 5 µg of total RNA with a random hexamer using the
SuperScript® III First-Strand Synthesis system (Thermo
Fisher Scientific, Inc.). These cDNAs were used for the measurement
of gene expression with an Applied Biosystems® 7500
Real-time PCR system (Thermo Fisher Scientific, Inc.) using
TaqMan® probes of KIF4A and β-actin which were used as
internal controls (KIF4A, #Hs01020169_m1; β-actin, #Hs99999903_m1;
Thermo Fisher Scientific, Inc.) and experiments were performed in
triplicate with blinded patient information. The thermocycling
conditions were as follows: 50°C for 2 min for AMPerase activation,
95°C for 10 min for Taq activation, 95°C for 15 sec for
denaturation and 60°C for 1 min for annealing and extension.
Relative KIF4A gene expression was calculated using the
2−ΔΔCq method (20).
IHC staining and evaluation
IHC staining was performed on paraffin-embedded
histological sections (4-µm thick) using a polymer peroxidase
method, in which colon cancer specimens were fixed in 20%
phosphate-buffered formalin (pH 7.4) at room temperature overnight.
Briefly, following deparaffinization with xylene and rehydration
using alcohol-water mixtures, prior to heat treatment in 10 mM
citric acid (pH 6.0) for antigen retrieval, the sections were
treated with 0.3% hydrogen peroxide in methanol for 30 min to block
endogenous peroxidase activity. Following washing with PBS, the
sections were incubated with rabbit polyclonal anti-KIF4A antibody
(dilution, 1:300; #ab122227; Abcam, Cambridge, UK) and mouse
monoclonal anti-MIB-1 (Ki-67) antibody (dilution, 1:100; Dako;
Agilent Technologies GmbH, Waldbronn, Germany) at 4°C overnight.
Following washing with PBS the slides were treated with a
peroxidase-labeled polymer conjugated to goat anti-rabbit
immunoglobulin (Dako EnVision+ System-HRP Labelled Polymer;
ready-to-use; #K4003; Dako; Agilent Technologies) according to the
manufacturer's protocol, as the secondary antibody for 30 min at
room temperature. The staining was visualized with
diaminobenzidine, followed by counterstaining with hematoxylin.
Colon cancer cell lines were also immunostained for KIF4A and
evaluated for staining intensity.
Expression of these proteins was evaluated using
optical microscopy (BX43; Olympus Corporation, Tokyo, Japan) as
positive when the nucleus of the cancerous tissue and the total
field of view were observed at ×400 magnification. Blinded to the
patients' characteristics and clinical outcomes, the staining of
each specimen was evaluated. The number of stained cells was
counted in per 1,000 cancer cells in the field of cancer tissue by
two investigators. The rate of positively stained cells was
classified as follows: 0, 0–5; 1, 6–20; 2, 21–50; 3, 51–100%. The
staining intensity was scored as 0 (negative), 1 (weak), 2
(moderate) and 3 (intense). The evaluation was expressed as a
product of the score of positive rate and staining intensity.
Positive staining was defined as a score of >2 and negative
staining was scored as 0 or 1.
Western blot analysis
The HCT116 cells were washed twice in ice-cold PBS,
pelleted by centrifugation (200 × g for 5 min) and stored at
−80°C. The pellet was resuspended in radioimmunoprecipitation assay
buffer (Thermo Fisher Scientific, Inc.) with a Halt Protease
Inhibitor Single-Use Cocktail (100X; Thermo Fisher Scientific,
Inc.) and centrifuged at 4°C and 17,400 × g for 20 min.
Total protein concentrations were measured by the Bradford method
using Bradford reagent (Bio-Rad Laboratories, Inc., Hercules, CA,
USA) and a Smart Spec 3000 spectrometer (Bio-Rad Laboratories,
Inc.). Total proteins isolated from cell lines were separated by
4–12% NuPAGE® Bis-Tris Precast Gel (Thermo Fisher
Scientific, Inc.). Tris-Glycine SDS sample buffer (Thermo Fisher
Scientific, Inc.) and 3-Mercapto-1,2-propandiol (Wako Pure Chemical
Industries, Ltd., Osaka, Japan) were added to the total protein
samples and heated at 100°C for 3 min. The 4–12% Tris-Glycine gels
(Thermo Fisher Scientific, Inc.) loaded with the 20 µg protein
samples were electrophoresed in 100 V for 100 min using
Tris-Glycine SDS Running buffer in Invitrogen™ XCell
SureLock™ electrophoresis system (Thermo Fisher
Scientific, Inc.). Following electrophoresis, the gel was
transferred onto the nitrocellulose membrane using an iBlot Gel
Transfer Stacks Nitrocellulose, Mini (Thermo Fisher Scientific,
Inc.). After transfer, the nitrocellulose membrane was washed with
25 ml TBS for 5 min at room temperature and blocked in SuperBlock
blocking buffer in PBS (Thermo Fisher Scientific, Inc.). The
protein blots were incubated with the anti-KIF4A antibody
(dilution, 1:300; catalog no. #ab122227; Abcam) for 1 h at room
temperature and incubated with goat anti-rabbit immunoglobulin
G-horseradish peroxidase (Santa Cruz Biotechnology, Inc., Dallas,
TX, USA) as the secondary antibody for 30 min at room temperature.
The protein levels were quantified using a rabbit monoclonal
anti-β-actin antibody (dilution, 1:2,000; catalog no. sc-47778;
Santa Cruz Biotechnology, Inc.) as the internal loading control for
1 h at room temperature. Bound antibodies were detected by enhanced
chemiluminescence detection reagents (Thermo Fisher Scientific,
Inc.) and visualized by autoradiography (ImageQuant™ LAS
4000 IR MultiColor imager; Fujifilm Corporation, Tokyo, Japan).
Small interfering RNA (siRNA)
transfection
Knockdown experiments were performed using siRNA
oligos for KIF4A (#sc-60888; Santa Cruz Biotechnology, Inc.) and
included three target-specific siRNAs and a control siRNA (siRNA-A;
#sc-37,007; Santa Cruz Biotechnology, Inc.) according to the
manufacturer's protocol. The day prior to transfection, the HCT116
CRC cell line was seeded at a density of 15×105
cells/well in a 6-well plate. Transfection using Lipofectamine
RNAiMAX (Thermo Fisher Scientific, Inc.) with a final concentration
of 10 nM siRNA was performed when the cell density was 30–50% in
the 6-well plates and cells were subsequently incubated at 37°C for
48 h.
Cell counting
The cell proliferation rate was assessed using a
Cell Counting kit-8 (CCK-8; Dojindo Molecular Technologies, Inc.,
Kumamoto, Japan) according to the manufacturer's protocol. Briefly,
2×103 HCT116 cells and control cells were plated into
each well in 96-well plates. The absorbance was measured after 24,
48, 72, 96 and 120 h of siRNA transfection. After 1 h of incubation
with 10 µl of CCK-8 reagent at 37°C, the absorbance was measured at
a wavelength of 450 nm using a Benchmark Plus microplate reader
(Bio-Rad Laboratories, Inc.).
Statistical analysis
Mann Whitney test, Fisher's exact test, a
χ2 test and Wilcoxon matched pairs test were performed
by GraphPad Prism 6.0 (GraphPad Software, Inc. La Jolla, CA, USA).
Data of cell viability analysis are shown as the mean ± standard
deviation. Survival rate curves were generated using the
Kaplan-Meier method and compared by the log-rank test. P<0.05
was considered to indicate a statistically significant
difference.
Results
KIF4A is upregulated in CRC
samples
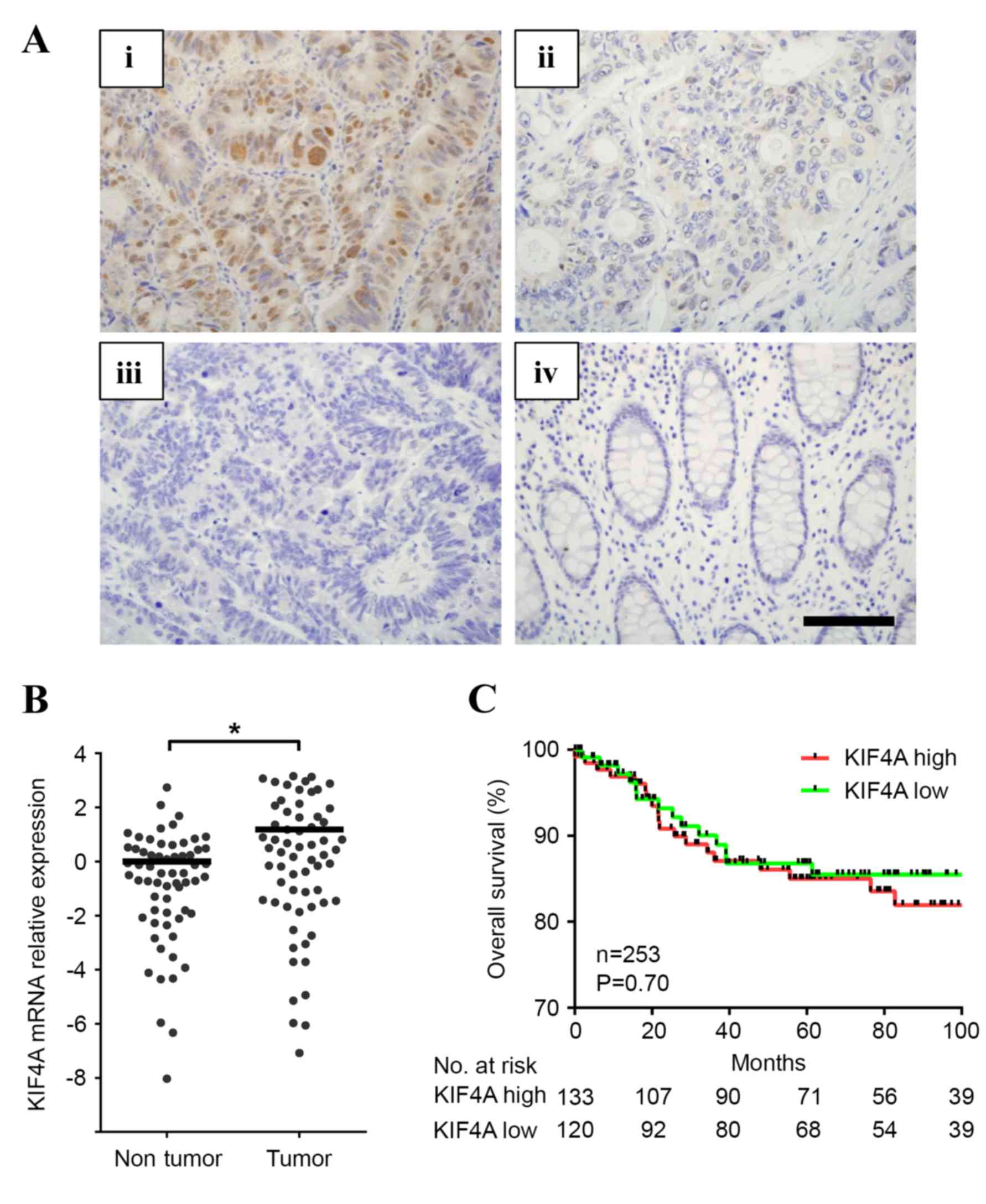
KIF4A expression was evaluated using IHC staining in
258 patients with CRC (Fig. 1A).
KIF4A expression was positive in 132 cases (51.1% with scores of 2
or 3) and negative in 126 cases (48.9% with scores of 0 or 1).
KIF4A expression levels were observed in the nucleus and cytoplasm
of cancer cells, whereas no expression was observed in the normal
mucosa. mRNA expression levels of KIF4A were analyzed and compared
between tumor and adjacent non-cancerous tissues. KIF4A mRNA
expression was elevated in tumor tissues in 63 of the 258 available
cases (2.26-fold increase; Wilcoxon matched pairs test, P=0.0013)
suggesting KIF4A may have an oncogenic role in CRC (Fig. 1B).
KIF4A expression levels and the clinicopathological
characteristics were evaluated in patients with CRC (Table I). The positive expression of KIF4A
was significantly associated with positive lymph node metastasis
(P<0.05). However, KIF4A expression levels were not associated
with age, gender, tumor location, TNM stage, depth of invasion,
venous invasion or liver metastasis. Notably, as KIF4A is located
on chromosome Xq13.1, the clinical significance of KIF4A expression
in each gender was investigated. However, KIF4Aexpression levels
did not associate with any clinicopathological characteristics in
male or female patients. Additionally, Kaplan-Meier analysis
demonstrated that there was no association between the increase in
KIF4A levels and overall survival rate (P=0.70; Fig. 1C).
 | Table I.Clinicopathological characteristics
and KIF4A IHC expression. |
Table I.
Clinicopathological characteristics
and KIF4A IHC expression.
|
|
| KIF4A IHC |
|
|---|
|
|
|
|
|
|---|
| Characteristics | Total (n=258) | Positive (n=132) | Negative (n=126) | P-value |
|---|
| Age |
|
|
| 0.64 |
| ≥65 | 142 | 75 | 69 |
|
|
<65 | 116 | 57 | 59 |
|
| Gender |
|
|
| 0.49 |
| Male | 149 | 79 | 70 |
|
|
Female | 109 | 53 | 59 |
|
| Stage |
|
|
| 0.12 |
|
0 | 10 |
2 |
8 |
|
| I | 40 | 23 | 17 |
|
| II | 93 | 43 | 50 |
|
| III | 75 | 48 | 27 |
|
| IV | 40 | 16 | 24 |
|
| Tumor location |
|
|
| 0.66 |
|
Proximal | 86 | 42 | 44 |
|
|
Distant | 89 | 49 | 40 |
|
|
Rectum | 83 | 41 | 42 |
|
|
Histologya |
|
|
| 0.24 |
| Tub1 | 120 | 59 | 61 |
|
|
Tub2 | 106 | 63 | 43 |
|
|
Por | 10 |
3 |
7 |
|
|
Muc | 19 |
5 | 14 |
|
|
Other |
3 |
0 |
3 |
|
| Depth |
|
|
| 0.22 |
|
Tis | 10 |
2 |
8 |
|
| T1 | 26 | 13 | 13 |
|
| T2 | 32 | 22 | 10 |
|
| T3 | 172 | 95 | 82 |
|
| T4 | 18 |
5 | 13 |
|
| Lymphatic
invasion |
|
|
| 0.35 |
|
Absent | 155 | 71 | 84 |
|
|
Present | 95 | 59 | 36 |
|
| Venous
invasion |
|
|
| 0.54 |
|
Absent | 59 | 28 | 31 |
|
|
Present | 199 | 104 | 95 |
|
| Lymph node
metastasis |
|
|
| <0.01 |
|
Negative | 155 | 71 | 84 |
|
|
Positive | 95 | 59 | 36 |
|
| Liver
metastasis |
|
|
| 0.59 |
|
Negative | 226 | 117 | 109 |
|
|
Positive | 32 | 15 | 17 |
|
Knockdown of KIF4A inhibition cell
growth in colon cancer cells
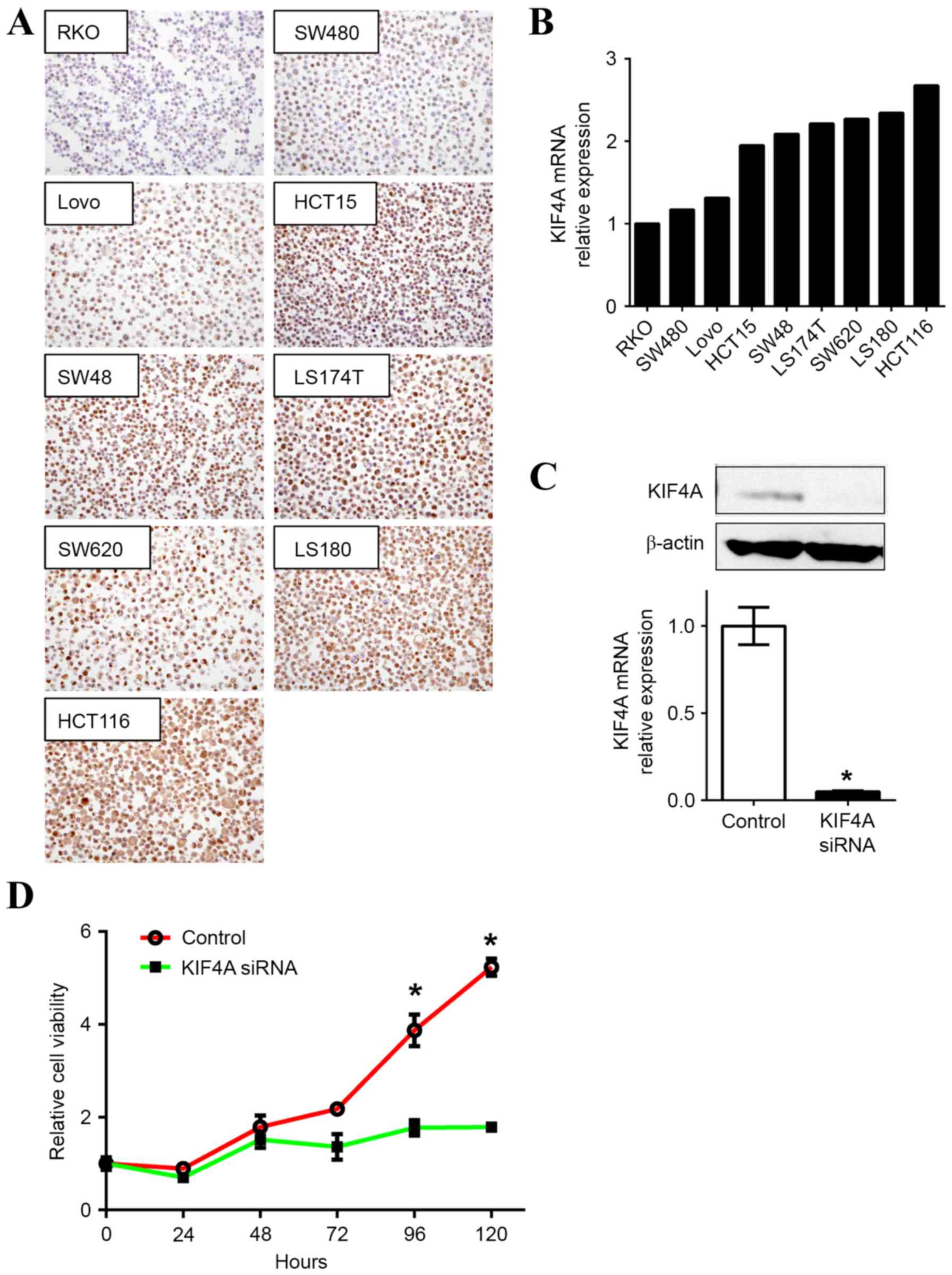
To evaluate the role of KIF4A in colon cancer
progression, gene knockdown technology was used to investigate cell
proliferation. KIF4A expression levels in RKO, SW480, Lovo, HCT15,
SW48, LS174T, SW620, LS180 and HCT116 colon cancer cell lines were
evaluated to select appropriate cells for further experiments in
the current study. KIF4A protein expression levels were examined
using IHC staining (Fig. 2A) and
KIF4A mRNA expression was evaluated using RT-qPCR (Fig. 2B). Concordant with the IHC results,
KIF4A mRNA expression was highest in the HCT116 cells compared with
the eight other colon cancer cell lines and therefore was used for
further experiments.
The knockdown of KIF4A using siRNA oligonucleotides
in the HCT116 cells (KIF4A-siRNA) confirmed that the downregulation
of KIF4A affected the mRNA and protein levels (Fig. 2C). Although no morphological changes
were observed in the KIF4A-depleted cells, cell proliferation was
attenuated. There was a significant decrease after 96 h (P<0.05)
in the cell growth of the KIF4A-siRNA treated cells compared with
the control cells (Fig. 2D).
KIF4A does not associate with positive
Ki-67 labeling index results in CRC
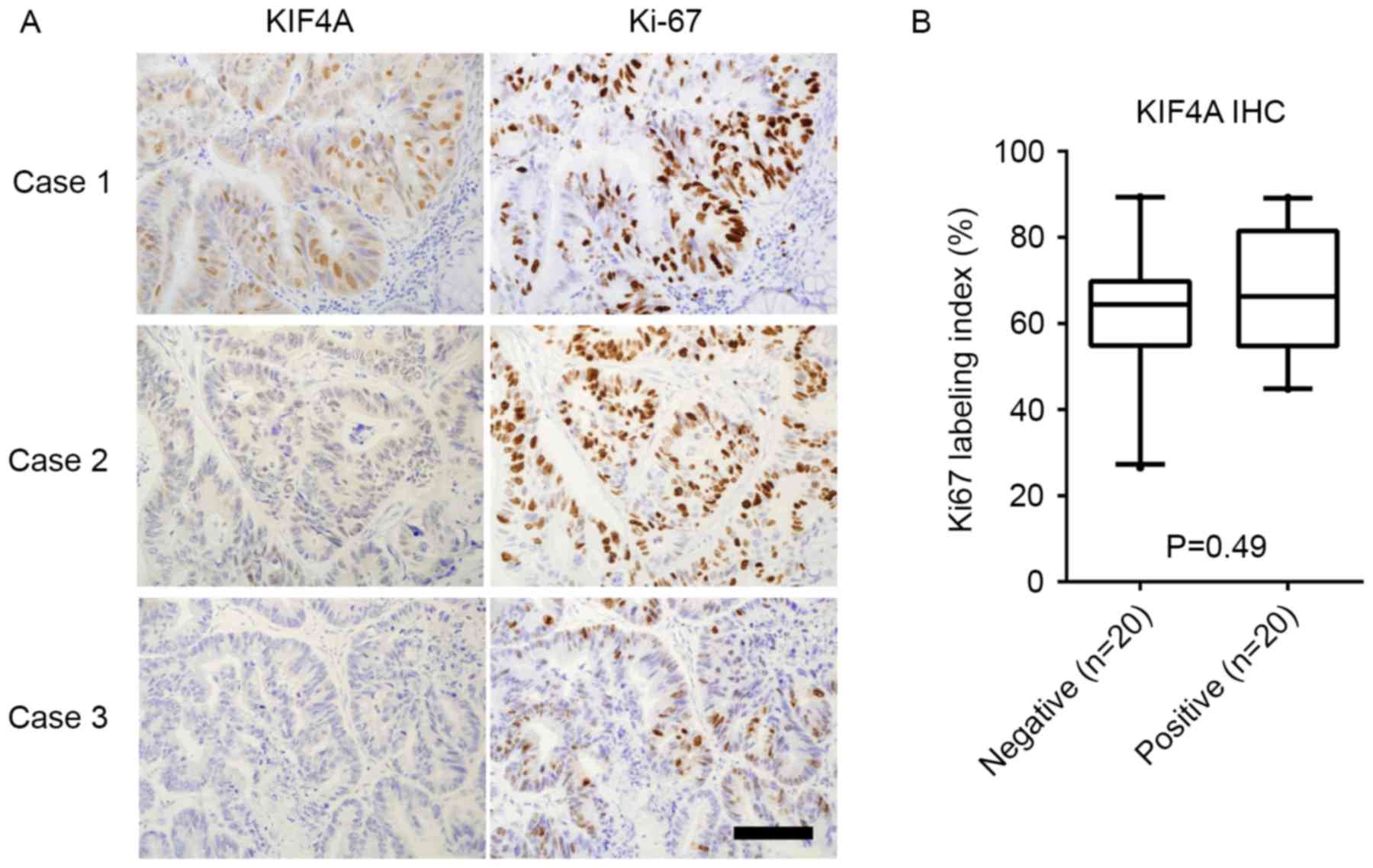
To further evaluate whether KIF4A accelerates
cell proliferation in CRC, Ki-67 IHC staining was performed on 40
patient tissue samples (Fig. 3). The
association between the KIF4A staining results and the Ki-67
labeling index was also investigated; however, no significant
association was observed (Mann Whitney test; P=0.49; Fig. 3).
Discussion
The present study identified that KIF4A expression
levels are increased in tumor samples from patients with CRC and
this may be associated with positive lymph node metastasis. The
tumor mRNA and protein expression levels of KIF4A were upregulated
in patients with CRC. Previous studies have established that KIF4A
expression levels are increased in a number of malignant tumors and
are associated with poor patient prognosis (12–16).
Concordant with these previous studies, the results of the present
study indicate a potential oncogenic role for KIF4A in CRC.
However, no significant associations between KIF4A protein
expression levels and patient survival rate were observed and
further investigation is required to identify if KIF4A may be an
effective prognostic biomarker for CRC.
In addition, the present study found that the
downregulation of KIF4A suppressed the cell proliferation of colon
cancer cells, further suggesting that KIF4A is associated with CRC
progression and metastasis. Similarly, it was reported that mutated
KIF4A in colon cancer cells lengthens the duration of mitosis and
reduces the speed of cell proliferation compared with wild-type
cells (10). The knockdown of KIF4A
expression levels has also been demonstrated to suppress cellular
proliferation in the SBC-5 lung squamous cell carcinoma cell line
(13). The downregulation of KIF4A
may induce cell cycle arrest in oral squamous cell carcinoma
(15). As KIF4A is involved in
regulating the M phase of the cell cycle and controls cellular
proliferation via activation of the spindle assembly checkpoint
(10), the level of cell cycle arrest
identified in cells with KIF4A knockdown was similar to the level
observed following the use of microtubule inhibitors (15). A previous study also reported that the
downregulation of KIF4A is associated with chemosensitivity in
breast cancer cells and demonstrated that KIF4A is directly
mediated by doxorubicin-induced cell apoptosis via the upregulation
of poly(ADP-ribose) polymerase-1 (21). Therefore, KIF4A expression levels are
hypothesized to promote cell malignancy and lymph node metastasis
in CRC. However, the association between KIF4A function and the
chemosensitivity of CRC cells identified in the current study
requires further investigation.
Several studies have reported a tumor suppressor
role for KIF4A (14,22). In total 13/23 (56.6%) gastric
carcinoma cases identified reduced expression levels of KIF4
compared with corresponding adjacent normal tissues (14). These 13 cases demonstrated a
significant association with poorer differentiation of gastric
cancer cells. In addition, in vitro experiments found that
KIF4A expression inhibits gastric cancer cell proliferation
(14). Another study reported that
the loss of KIF4 may trigger tumor formation; embryonic stem cells
in the KIF4 knockout mouse model had mitotic defects associated
with the loss of a molecular motor function and generation of
aneuploidy (22). Notably, the loss
of KIF4A expression may result in the onset of carcinogenesis in a
number of cell lines from the NCI-60 Human Tumor Cell Line Screen
(22). In the present study, the
effect of KIF4A expression levels on cell proliferation was
investigated in patients with CRC. When comparing KIF4A expression
levels and the Ki-67 index, no significant association was
observed. Therefore, the functional role of KIF4A remains to be
determined and further experimental and mouse model studies (KIF4A
transgenic or knockout mice) are required.
In conclusion, to the best of our knowledge the
present study is the first to demonstrate an association between
the KIF4A expression status and a functional role in CRC. These
results suggest that KIF4A may be a potential therapeutic target
and this may improve clinical outcomes of patients with CRC.
Acknowledgements
The present study was supported by JSPS KAKENHI
(grant no. 15k10143).
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Venook AP, Weiser MR and Tepper JE:
Colorectal cancer: All hands on deck. Am Soc Clin Oncol Educ Book.
1–89. 2014.
|
|
3
|
de Gramont A, Figer A, Seymour M, Homerin
M, Hmissi A, Cassidy J, Boni C, Cortes-Funes H, Cervantes A, Freyer
G, et al: Leucovorin and fluorouracil with or without oxaliplatin
as first-line treatment in advanced colorectal cancer. J Clin
Oncol. 18:2938–2947. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Douillard JY, Cunningham D, Roth AD,
Navarro M, James RD, Karasek P, Jandik P, Iveson T, Carmichael J,
Alakl M, et al: Irinotecan combined with fluorouracil compared with
fluorouracil alone as first-line treatment for metastatic
colorectal cancer: A multicentre randomised trial. Lancet.
355:1041–1047. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Goldberg RM, Sargent DJ, Morton RF, Fuchs
CS, Ramanathan RK, Williamson SK, Findlay BP, Pitot HC and Alberts
SR: A randomized controlled trial of fluorouracil plus leucovorin,
irinotecan and oxaliplatin combinations in patients with previously
untreated metastatic colorectal cancer. J Clin Oncol. 22:23–30.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tournigand C, André T, Achille E, Lledo G,
Flesh M, Mery-Mignard D, Quinaux E, Couteau C, Buyse M, Ganem G, et
al: FOLFIRI followed by FOLFOX6 or the reverse sequence in advanced
colorectal cancer: A randomized GERCOR study. J Clin Oncol.
22:229–237. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nakagawa T, Tanaka Y, Matsuoka E, Kondo S,
Okada Y, Noda Y, Kanai Y and Hirokawa N: Identification and
classification of 16 new kinesin superfamily (KIF) proteins in
mouse genome. Proc Natl Acad Sci USA. 94:pp. 9654–9659. 1997;
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lawrence CJ, Dawe RK, Christie KR,
Cleveland DW, Dawson SC, Endow SA, Goldstein LS, Goodson HV,
Hirokawa N, Howard J, et al: A standardized kinesin nomenclature. J
Cell Biol. 167:19–22. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Miki H, Okada Y and Hirokawa N: Analysis
of the kinesin superfamily: Insights into structure and function.
Trends Cell Biol. 15:467–476. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wandke C, Barisic M, Sigl R, Rauch V, Wolf
F, Amaro AC, Tan CH, Pereira AJ, Kutay U, Maiato H, et al: Human
chromokinesins promote chromosome congression and spindle
microtubule dynamics during mitosis. J Cell Biol. 198:847–863.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mazumdar M, Sundareshan S and Misteli T:
Human chromokinesin KIF4A functions in chromosome condensation and
segregation. J Cell Biol. 166:613–620. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Narayan G, Bourdon V, Chaganti S,
Arias-Pulido H, Nandula SV, Rao PH, Gissmann L, Dürst M, Schneider
A, Pothuri B, et al: Gene dosage alterations revealed by cDNA
microarray analysis in cervical cancer: Identification of candidate
amplified and overexpressed genes. Genes Chromosomes Cancer.
46:373–384. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Taniwaki M, Takano A, Ishikawa N, Yasui W,
Inai K, Nishimura H, Tsuchiya E, Kohno N, Nakamura Y and Daigo Y:
Activation of KIF4A as a prognostic biomarker and therapeutic
target for lung cancer. Clin Cancer Res. 13:6624–6631. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gao J, Sai N, Wang C, Sheng X, Shao Q,
Zhou C, Shi Y, Sun S, Qu X and Zhu C: Overexpression of
chromokinesin KIF4 inhibits proliferation of human gastric
carcinoma cells both in vitro and in vivo. Tumour Biol. 32:53–61.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Minakawa Y, Kasamatsu A, Koike H, Higo M,
Nakashima D, Kouzu Y, Sakamoto Y, Ogawara K, Shiiba M, Tanzawa H
and Uzawa K: Kinesin family member 4A: A potential predictor for
progression of human oral cancer. PLoS One. 8:e859512013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zou JX, Duan Z, Wang J, Sokolov A, Xu J,
Chen CZ, Li JJ and Chen HW: Kinesin family deregulation coordinated
by bromodomain protein ANCCA and histone methyltransferase MLL for
breast cancer cell growth, survival and tamoxifen resistance. Mol
Cancer Res. 12:539–549. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sobin LH GM and Wittekind CH:
International Union Against Cancer (UICC) TNM Classification of
Malignant Tumors. Oxford, UK: Wiley-Blackwell; 2009
|
|
18
|
Sobin LH and Compton CC: TNM seventh
edition: What's new, what's changed: Communication from the
international union against cancer and the american joint committee
on cancer. Cancer. 116:5336–5339. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Okano M, Kumamoto K, Saito M, Onozawa H,
Saito K, Abe N, Ohtake T and Takenoshita S: Upregulated annexin A1
promotes cellular invasion in triple-negative breast cancer. Oncol
Rep. 33:1064–1070. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang H, Lu C, Li Q, Xie J, Chen T, Tan Y,
Wu C and Jiang J: The role of Kif4A in doxorubicin-induced
apoptosis in breast cancer cells. Mol Cells. 37:812–818. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mazumdar M, Lee JH, Sengupta K, Ried T,
Rane S and Misteli T: Tumor formation via loss of a molecular motor
protein. Curr Biol. 16:1559–1564. 2006. View Article : Google Scholar : PubMed/NCBI
|