Introduction
Ovarian cancer (OC) is an intractable cancer with
the highest mortality rate among all types of female cancers
worldwide (1). Its incidence is
continually increasing along with the prevalence of westernized
lifestyles, the use of hormone replacement therapy and aging
population in Asian regions (2).
Early OC is frequently asymptomatic; accordingly, >70% of
patients are reportedly diagnosed with OC when it has reached an
advanced stage (stage 3 or higher). Furthermore, recurrence or
metastasis occurs in >75% of patients within the first 2 years
following initial treatment (3,4). In the
majority of patients, initial responses can be achieved with
debulking surgery and treatment with taxanes in combination with
platinum-based chemotherapy; however, >75% of those responders
eventually relapse, resulting in chemoresistant and fatal disease
(5,6).
Epigenetic alterations, which are closely associated
with ovarian tumorigenesis, are defined as heritable alterations in
gene expression without changes to the DNA sequence; these include
histone modification, DNA methylation and posttranscriptional gene
regulation by microRNAs (7). Among
the various epigenetic mechanisms that affect gene expression, DNA
methylation is the most extensively studied. Specific DNA
methyltransferases catalyze DNA methylation by transferring a
methyl group, using S-adenosyl methionine as the methyl donor, to
the cytosine residues of CpG dinucleotides (8). CpG methylation in the promoter regions
of specific genes leads to physical obstruction of transcription
factor binding, and recruitment of methyl-CpG-binding domain
proteins and histone deacetylases that are associated with gene
silencing and the formation of inactive heterochromatin (9). DNA methylation is therefore an important
mechanism underlying gene silencing and inactivation, and the
methylation status at promoter CpG sites serves a pivotal role in
the regulation of gene expression (10).
Recent studies have reported that aberrant DNA
methylation may affect the sensitivity of cells to anticancer drugs
by altering the expression of genes that are crucial for drug
response. A number of studies have demonstrated that DNA
hypermethylation is involved in generating drug-resistant
phenotypes by inactivating genes that are required for cytotoxicity
(11–14).
Based on previous studies, we hypothesized that
epigenetically regulated genes involved in drug resistance may
serve as promising novel targets for the effective treatment of
cisplatin-resistant OC. To identify the genes involved in cisplatin
resistance, the cytotoxicities of eight different OC cell lines
were determined, and the cell lines were classified into two groups
(sensitive and resistant). mRNA expression levels were analyzed
with GeneChip Human Gene 1.0 ST Arrays, and DNA methylation
profiles were determined using the Human Methylation450 BeadChip.
Using an integrated approach of analyzing gene expression level and
DNA methylation profiles simultaneously, 26 genes were selected
that were differentially expressed and methylated between the
resistant and sensitive groups. Among these 26 genes, 3-oxoacid CoA
transferase 1 (OXCT1) was selected for further
investigation. OXCT1 protein has been identified as a homodimeric
mitochondrial matrix enzyme involved in ketone body utilization via
the reversible transfer of coenzyme A from succinyl-CoA to
acetoacetate (15); however, the
involvement of this gene in the drug response has not yet been
reported. Epigenetic silencing of OXCT1 via the
hypermethylation of promoter CpGs was revealed in the present
study, and was shown to be associated with cisplatin resistance in
OC. Furthermore, the overexpression of OXCT1 restored
chemosensitivity to cisplatin, indicating that OXCT1 acts as
a suppressor of cisplatin resistance in OC. The results of the
present study offer novel insight into the function of OXCT1
in chemoresistant OC.
Materials and methods
Cell culture
SK-OV-3, PA-1, Caov-3, TOV-21G, TOV-112D, OV-90 and
OVCAR-3 human OC cell lines studied were purchased from the
American Type Culture Collection (Manassas, VA, USA), and the human
OC A2780 cell line was purchased from the European Collection of
Cell Cultures (London, UK). All cell lines were initially cultured
using the medium and supplements recommended by the suppliers.
Table I summarizes components of the
culture media for individual cell lines. All eight cell lines were
grown as monolayers and attached cells were fully disaggregated by
trypsinization between passages. The cell lines were maintained in
a 95% humidified and 5% CO2 atmosphere at 37°C.
 | Table I.Components of culture media for the
human ovarian cancer cell lines studied. |
Table I.
Components of culture media for the
human ovarian cancer cell lines studied.
| Cell line | Components of
culture mediaa |
|---|
| SK-OV-3 | McCoy's 5a + 10%
FBS + 1% P/S |
| PA-1 | MEM α + 10% FBS +
1% P/S |
| Caov-3 | DMEM (1.5 g/l
sodium bicarbonate) + 10% FBS + 1% P/S |
| TOV-21G | MCDB 105 (1.5 g/l
sodium bicarbonate) and Medium 199 (2.2 g/l sodium
bicarbonate) |
|
| 1:1 mix + 10% FBS +
1% P/S |
| TOV-112D | MCDB 105 (1.5 g/l
sodium bicarbonate) and Medium 199 (2.2 g/l sodium
bicarbonate) |
|
| 1:1 mix + 10% FBS +
1% P/S |
| OV-90 | MCDB 105 (1.5 g/l
sodium bicarbonate) and Medium 199 (2.2 g/l sodium
bicarbonate) |
|
| 1:1 mix + 10% FBS +
1% P/S |
| OVCAR-3 | RPMI-1640 (25 mM
HEPES) + 10% FBS + 1% P/S |
| A2780 | RPMI-1640 (25 mM
HEPES) + 10% FBS + 1% P/S |
Cisplatin sensitivity assay
The cisplatin sensitivities of the eight human
ovarian cell lines (SK-OV-3, PA-1, Caov-3, TOV-21G, TOV-112D,
OV-90, OVCAR-3 and A2780) were determined using MTT assays
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). Briefly,
2×104 cells were seeded onto 96-well plates and
incubated at 37°C overnight. The medium was exchanged with fresh
medium supplemented with various cisplatin concentrations (0–100
µM). Following incubation for 48 h, 20 µl 2.5 mg/ml MTT solution
was added to each well and the plates were further incubated for 2
h at 37°C. Dimethyl sulfoxide (100 µl; Sigma-Aldrich; Merck KGaA)
was added to solubilize the MTT formazan product through a 10 min
oscillation at 37°C. Absorbance at 540 nm was determined using a
microplate reader (Molecular Devices LLC, Sunnyvale, CA, USA).
Dose-response curves were plotted as the percentage of the control,
which was obtained from the sample with no drug exposure.
Half-maximal inhibitory concentration (IC50) was
evaluated as the concentration of cisplatin that reduces cell
growth by 50% under the experimental conditions. The eight human
ovarian cell lines were classified into two groups: Sensitive and
resistant cell lines.
Total RNA isolation and mRNA
microarray
Total RNA was extracted from the eight human OC cell
lines using the RNeasy Mini kit (Qiagen, Inc., Valencia, CA, USA)
and amplified and labeled according to the Affymetrix GeneChip
Whole Transcript Sense Target Labeling protocol (Thermo Fisher
Scientific, Inc., Waltham, MA, USA). The resulting labeled cDNA was
hybridized to Affymetrix GeneChip Human Gene 1.0 ST Arrays (Thermo
Fisher Scientific, Inc.). The scanned raw expression values were
background-corrected, normalized and summarized using the Robust
Multiarray Averaging approach in the Bioconductor ‘affy’ package
(Bioconductor; http://www.bioconductor.org/). The resulting
log2-transformed data were used for further analyses. To identify
differentially expressed genes (DEGs), moderated t-statistics were
applied based on an empirical Bayesian approach (16). Significantly upregulated and
downregulated DEGs were defined as those with ≥1.5-fold difference
in expression level between the cisplatin-resistant and -sensitive
groups following correction for multiple testing
[Benjamini-Hochberg false discovery rate (BH FDR)-adjusted
P<0.01] (17).
Genomic DNA isolation and CpG
methylation microarray
Genomic DNA was extracted from the eight human OC
cell lines using the QIAmp Mini kit (Qiagen, Inc.), according to
the manufacturer's instructions. For genome-wide screening of DNA
methylation, the Illumina HumanMethylation450 BeadChip (Illumina,
Inc., San Diego, CA, USA) was used, which targets 450,000 specific
CpG sites. DNA methylation values were described by β-values, which
were determined by subtracting the background obtained from
negative controls on the array and calculating the ratio of the
methylated signal intensity to the sum of the methylated and
unmethylated signals. β-values ranged from 0 (completely
unmethylated) to 1 (fully methylated) on a continuous scale for
each CpG site. To identify differentially methylated CpG sites, the
difference in mean β-value (Δβ; mean β-value in resistant
group-mean β-value in sensitive group) was determined. If the
absolute difference in mean β-values (|Δβ|) was >0.3, the sites
were defined as differentially methylated CpG sites. CpG sites or
genes were described as hypermethylated if Δβ>0.3 and as
hypomethylated if Δβ<-0.3.
Integrated analysis of DNA methylation
and gene expression
To identify genes that had expression regulated by
epigenetic alteration in the cisplatin-resistant group, the global
DNA methylation profiling data was integrated with the mRNA
expression profiles using stringent selection criteria
(|Δβ|>0.3, expression fold change >1.5). Expression of
candidate genes was considered to be upregulated (fold change
>1.5) by hypomethylation (Δβ<-0.3) at promoter CpG sites and
downregulated (fold change >1.5) by hypermethylation (Δβ>0.3)
at promoter CpG sites in the cisplatin-resistant group compared
with the cisplatin-sensitive group.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA (1 µg) was converted to cDNA using
Superscript II Reverse Transcriptase and oligo-(dT)12–18
primers (both from Invitrogen; Thermo Fisher Scientific, Inc.),
according to the manufacturer's instructions. RT-qPCR was performed
in a 20 µl reaction mixture containing 1 µl cDNA, 10 µl SYBR Premix
Ex Taq (Takara Bio, Inc., Otsu, Japan), 0.4 µl Rox reference dye
(×50; Takara Bio, Inc.), and 200 nM primers for each gene. The
primer sequences were as follows: OXCT1 forward,
5′-GGGTCCATATCCACGACAACA-3′; OXCT1 reverse,
5′-GACGTGTCCACCTCTAATCATTG-3′; GAPDH forward,
5′-AATCCCATCACCATCTTCCA-3′; and GAPDH reverse,
5′-TGGACTCCACGACGTACTCA-3′. The reactions were run on a 7500 Fast
Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific,
Inc.) at 95°C for 30 sec, followed by 40 cycles of 95°C for 3 sec
and 60°C for 30 sec, and a single dissociation cycle of 95°C for 15
sec, 60°C for 60 sec and 95°C for 15 sec. All reactions were
performed in triplicate, and the specificity of the reaction was
determined using melting curve analysis at the dissociation stage.
Comparative quantification of each target gene was performed based
on the cycle threshold (Cq) normalized to GAPDH using the
2−ΔΔCq method (18).
5-aza-2′-deoxycytidine (5-aza-dc)
treatment
To demethylate methylated CpG sites, the eight human
ovarian cell lines were treated with 10 µM 5-aza-dc (Sigma-Aldrich;
Merck KGaA) for 3 days at 37°C. Each day, the medium was exchanged
with fresh medium supplemented with 10 µM of 5-aza-dc.
Transient transfection
To establish a transient expression system, SK-OV-3
cells were transfected with pCMV-SPORT6-OXCT1 (KRIBB, Daejeon,
Korea) or pEGFP-N3 (Clontech Laboratories, Inc., Mountainview, CA,
USA) plasmids using Lipofectamine 2000® (Invitrogen;
Thermo Fisher Scientific, Inc.). Briefly, the cells were plated at
a density of 6×105 cells/well in 6-well plates and
allowed to grow overnight at 37°C. In total, 2 µg of each plasmid
DNA and 5 µl Lipofectamine 2000 were diluted separately in Opti-MEM
(Gibco; Thermo Fisher Scientific, Inc.) to a total volume of 250
µl. The diluted plasmid DNAs and Lipofectamine 2000 were mixed and
incubated at room temperature for 20 min to generate the
transfection mixtures. The cells were washed with serum-free
McCoy's 5A medium, and subsequently the transfection mixtures were
added to each well of the 6-well plates containing complete growth
medium, and incubated at 37°C for 24 h in a 5% CO2
incubator. The sensitivity to cisplatin of the transfected cells
was determined using the MTT assay, as described for the
aforementioned ‘cisplatin sensitivity assay’.
Statistical analysis
Data are expressed as the mean ± standard deviation
of ≥3 independent experiments. Statistical analyses were performed
using GraphPad Prism 5.0 software (GraphPad Software, Inc., La
Jolla, CA, USA). The unpaired Student's t-test was used to perform
statistical analysis between two groups. P<0.05 was considered
to indicate a statistically significant difference.
Results
Determination of cisplatin resistance
in eight human OC cell lines
The sensitivity to cisplatin of the eight human
ovarian cell lines (SK-OV-3, PA-1, Caov-3, TOV-21 G, TOV-112D,
OV-90, OVCAR-3 and A2780) was determined using the MTT cytotoxicity
assay. The highest IC50 value for cisplatin was observed
in SK-OV-3 cells (69.8 µM), and the lowest IC50 value
was observed in PA-1 cells (4.1 µM). The IC50 values of
the eight human OC cell lines increased in the order PA-1, TOV-21
G, TOV-112D, Caov-3, A2780, OVCAR-3, OV-90 and SK-OV-3, as
presented in Fig. 1. Based on the
IC50 values for cisplatin, the cell lines were
classified into two groups as sensitive (PA-1, TOV-21G, TOV-112D,
Caov-3 and A2780) and resistant (OVCAR-3, OV-90, and SK-OV-3).
Identification of differentially
expressed genes between cisplatin-resistant and cisplatin-sensitive
groups
To identify DEGs, the present study applied
moderated t-statistics based on an empirical Bayesian approach
(16). Significantly upregulated and
downregulated DEGs were defined as genes with ≥1.5-fold difference
in expression level between cisplatin-resistant and -sensitive
groups based on the microarray, following correction for multiple
testing (BH FDR-adjusted P<0.01) (17). Using this criterion, the expression
levels of 376 genes were altered in the cisplatin-resistant group
compared with in the cisplatin-sensitive group.
Identification of differentially
methylated CpG sites between cisplatin-resistant and
cisplatin-sensitive groups
To identify differentially methylated CpG sites, the
difference in Δβ was used. If |Δβ|>0.3, the sites were defined
as differentially methylated CpG sites. CpG sites or genes were
described as hypermethylated if Δβ>0.3 and as hypomethylated if
Δβ<-0.3. By these criteria, the promoter methylation of 5,384
genes (12,293 CpGs) was altered in the cisplatin-resistant group
compared with the cisplatin-sensitive group.
Selection of cisplatin
resistance-associated genes using integrated analysis
To identify genes whose expression was regulated by
DNA methylation during the development of cisplatin resistance, the
global DNA methylation profiling data was integrated with the mRNA
expression profiles using stringent selection criteria (Δβ>0.3;
expression fold change >1.5). Expression of candidate genes was
considered to be upregulated (fold change >1.5) by
hypomethylation (Δβ<-0.3) at promoter CpG sites and
downregulated (fold change >1.5) by hypermethylation (Δβ>0.3)
at promoter CpG sites in the cisplatin-resistant group compared
with the cisplatin-sensitive group. Using these criteria, 26
candidate genes were selected. The candidate genes are presented in
Table II.
 | Table II.Candidate genes with expression
levels regulated by DNA methylation during cisplatin-resistance
development. |
Table II.
Candidate genes with expression
levels regulated by DNA methylation during cisplatin-resistance
development.
|
| DNA
methylation | Gene
expression |
|---|
|
|
|
|
|---|
| Gene symbol | Difference in β
valuea | P-value | Chromosome | CpG site | Fold change
(log2)b | P-value |
|---|
| MFSD2A | 0.795151465 | 0.000000402 | 1 | 40420603 | 0.94105296 | 0.015471851 |
|
| 0.528977905 | 0.000119729 | 1 | 40420537 |
|
|
|
| 0.638894305 | 0.000139608 | 1 | 40420635 |
|
|
| FKBP10 | 0.392134207 | 0.0000166 | 17 | 39968802 | 3.577717953 | 0.000795819 |
|
| 0.635697744 | 0.000000435 | 17 | 39968772 |
|
|
|
| 0.450164388 | 0.00000461 | 17 | 39968804 |
|
|
|
| 0.418122533 | 0.000429913 | 17 | 39968600 |
|
|
|
MARVELD1 | 0.470414467 | 0.000134843 | 10 | 99474521 | 1.346310667 | 0.000609707 |
|
HIST1H2BF | 0.539404884 | 0.000000755 | 6 | 26199465 | 1.109350157 | 0.005487531 |
| HIVEP2 | −0.649083953 | 0.00000458 | 6 | 143249236 | −2.267853853 | 0.032947151 |
| ZNF257 | −0.439560377 | 0.000513184 | 19 | 22235199 | −2.515197001 | 0.001691734 |
|
| −0.556456643 | 0.000598785 | 19 | 22235022 |
|
|
|
| −0.7204497 | 0.00000623 | 19 | 22234992 |
|
|
|
| −0.588986337 | 0.000100124 | 19 | 22235281 |
|
|
| ZFP3 | −0.699854973 | 0.0000295 | 17 | 4981598 | −1.681825862 | 0.0000449 |
|
| −0.756465323 | 0.000142409 | 17 | 4981610 |
|
|
|
| −0.614927003 | 0.00043569 | 17 | 4981403 |
|
|
|
| −0.824812407 | 0.0000127 | 17 | 4981603 |
|
|
| INA | 0.649938177 | 0.00020484 | 10 | 105036701 | 4.355589328 | 0.0000111 |
|
HIST1H3D | 0.397435269 | 0.0000184 | 6 | 26199702 | 0.6066328 | 0.04004395 |
| LEPREL2 | 0.31302834 | 0.00022931 | 12 | 6938638 | 0.920190572 | 0.003181991 |
|
| 0.433342418 | 0.0000252 | 12 | 6938635 |
|
|
| OXCT1 | 0.550269195 | 0.000511071 | 5 | 41870875 | 1.716584541 | 0.013127834 |
|
| 0.497255367 | 0.000143461 | 5 | 41870856 |
|
|
|
| 0.511680656 | 0.000544145 | 5 | 41870860 |
|
|
|
| 0.377481771 | 0.0000288 | 5 | 41869963 |
|
|
| NME4 | 0.725727483 | 0.0000517 | 16 | 446668 | 1.482274313 | 0.009042391 |
| MAP3K12 | 0.342133404 | 0.0000789 | 12 | 53893000 | 1.81515751 | 0.008388664 |
| DLG4 | −0.56680592 | 0.000631474 | 17 | 7108468 | 1.410099319 | 0.04186927 |
|
| −0.522567633 | 0.000105786 | 17 | 7108653 |
|
|
| TMEM180 | 0.420509153 | 0.000114655 | 10 | 104221598 | 0.789638858 | 0.047135907 |
| DCBLD1 | −0.469961667 | 0.000118409 | 6 | 117869857 | −1.955294821 | 0.002032113 |
| DNAJC15 | −0.545150123 | 0.000684898 | 13 | 43597565 | −3.261718049 | 0.014808162 |
| MAPRE3 | 0.499877566 | 0.000140963 | 2 | 27194315 | −0.808403845 | 0.021186922 |
| VSIG10L | 0.564933039 | 0.000167731 | 19 | 51843854 | 0.813198578 | 0.007985285 |
|
HIST1H2BB | −0.65683174 | 0.000172726 | 6 | 26044274 | −1.511985487 | 0.031374454 |
|
| −0.575591967 | 0.000262479 | 6 | 26043990 |
|
|
|
| −0.6231011 | 0.00030871 | 6 | 26044220 |
|
|
| FAM188B | 0.474266434 | 0.000703141 | 7 | 30810858 | 0.755956576 | 0.00432309 |
|
| 0.533877189 | 0.000787152 | 7 | 30810882 |
|
|
|
| 0.417810231 | 0.000671844 | 7 | 30810864 |
|
|
|
| 0.504869284 | 0.000261883 | 7 | 30810870 |
|
|
| BAMBI | 0.53068002 | 0.000322864 | 10 | 28965584 | −2.435784283 | 0.016972961 |
| NAGA | 0.545561447 | 0.000456257 | 22 | 42466345 | 0.637024627 | 0.01300139 |
| ST3GAL2 | 0.327738228 | 0.000518788 | 16 | 70473447 | −0.880289271 | 0.024322723 |
| RUNX1 | −0.484414007 | 0.000602711 | 21 | 36421955 | −1.735856425 | 0.019877857 |
|
ZC3HAV1L | 0.661703861 | 0.000765107 | 7 | 138720989 | 1.317571487 | 0.048052422 |
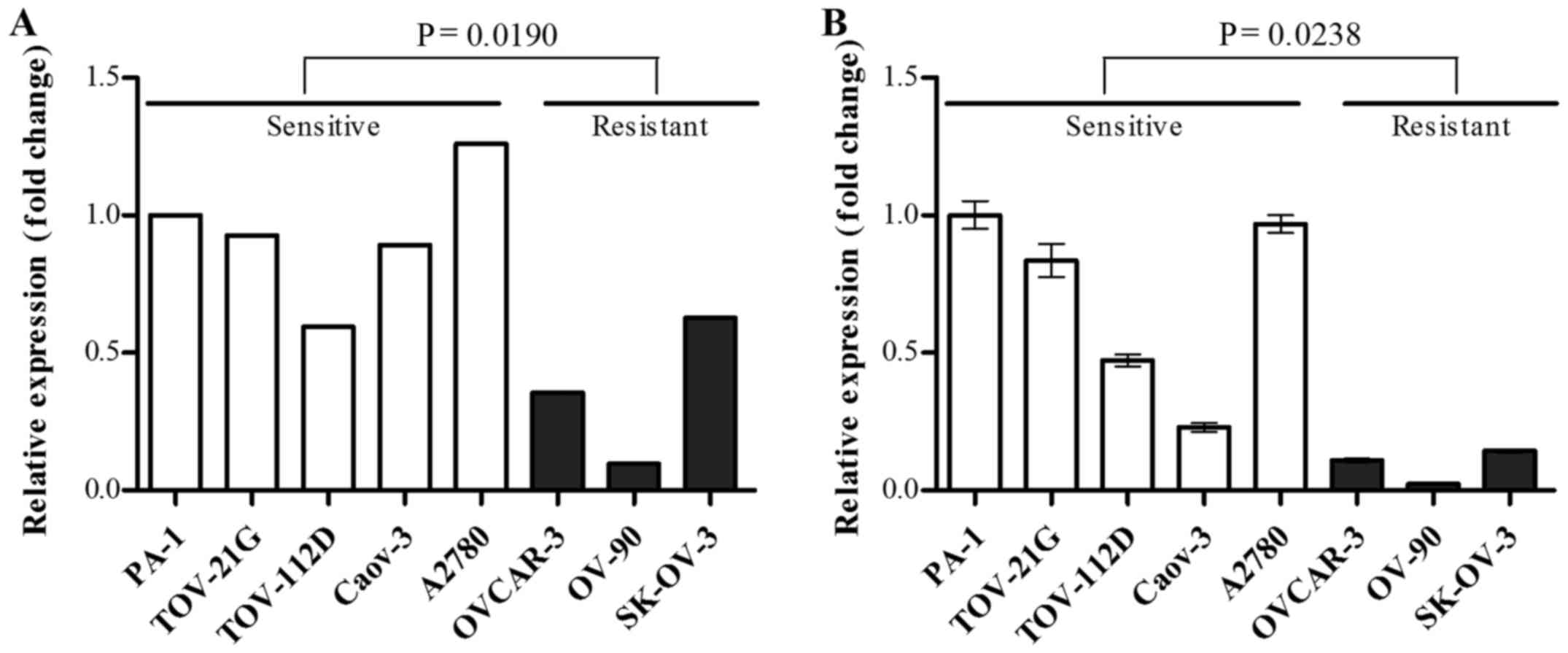
mRNA expression level of OXCT1 was
downregulated in the cisplatin-resistant group
Among the 26 cisplatin resistance-associated genes,
OXCT1 was selected as, to the best of our knowledge, its
association with chemosensitivity has not been reported in previous
cancer studies, and its expression level was confirmed by RT-qPCR.
OXCT1 mRNA expression level was significantly decreased in
the cisplatin-resistant group compared with the cisplatin-sensitive
group, in agreement with the results of gene expression microarray
(Fig. 2).
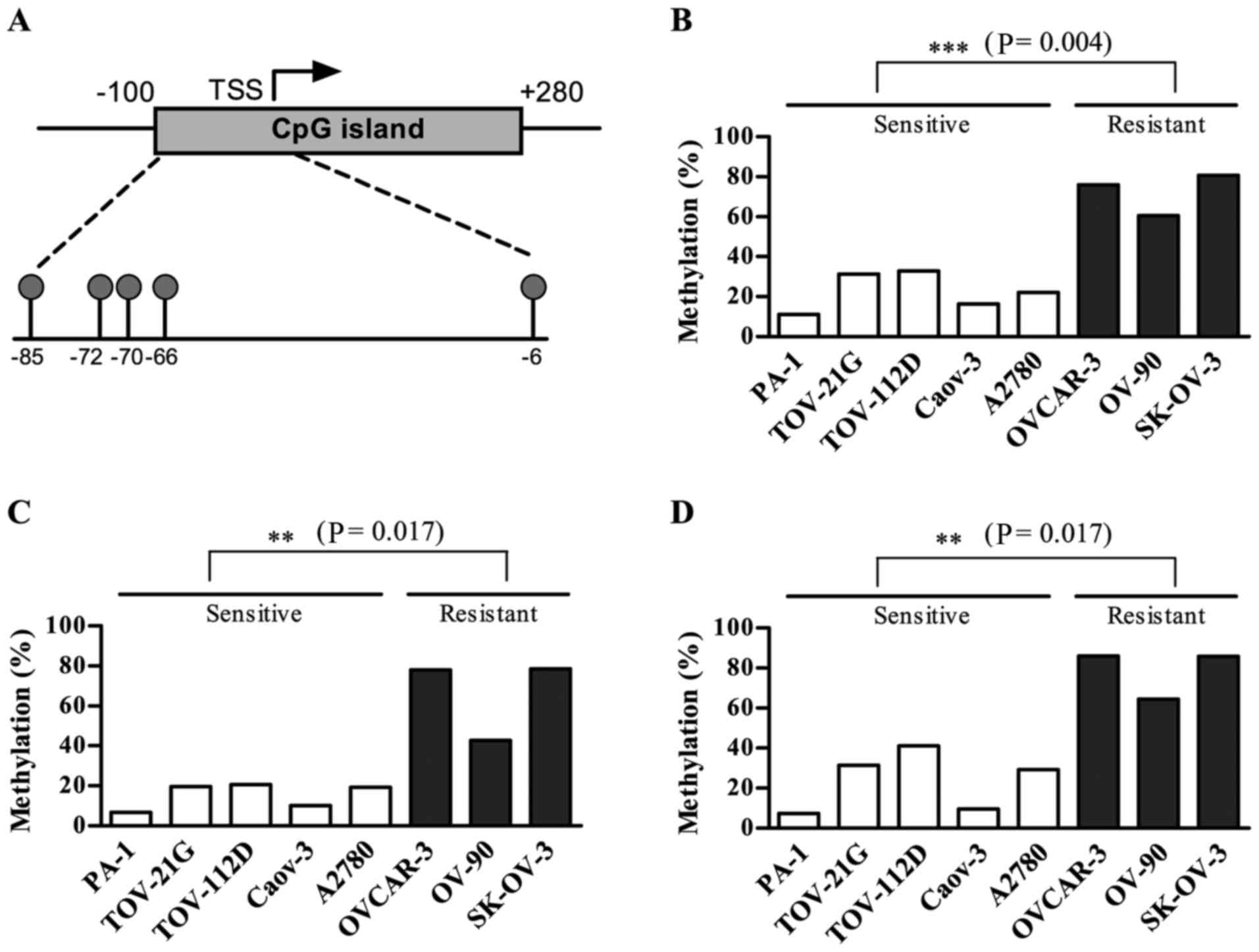
OXCT1 expression was suppressed by DNA
methylation in the cisplatin-resistant group
Substantial DNA methylation changes during the
acquisition of chemoresistance have been widely reported in various
types of cancer (19). In particular,
DNA hypermethylation of CpG islands (CGIs) within promoter regions
serves a prominent role in the development of drug resistance by
silencing genes that are required for cytotoxicity (11). Thus, the present study investigated
the DNA methylation status of CGIs within the promoter region of
the OXCT1 gene using the Illumina HumanMethylation450
BeadChip, which included five CpG sites within the CGI promoter
region of the OXCT1 gene, located between positions
41,870,792 and 41,870,890 of chromosome 5 (human GRCh37/hg19). The
five CpGs were at positions −85, −72, −70, −66 and −6, relative to
the transcription start site (TSS) as presented in Fig. 3A. Among the five CGI promoter CpGs,
the three CpG sites located at −85, −70 and −66 from the TSS were
significantly hypermethylated in the cisplatin-resistant group
compared with the cisplatin-sensitive group (Fig. 3).
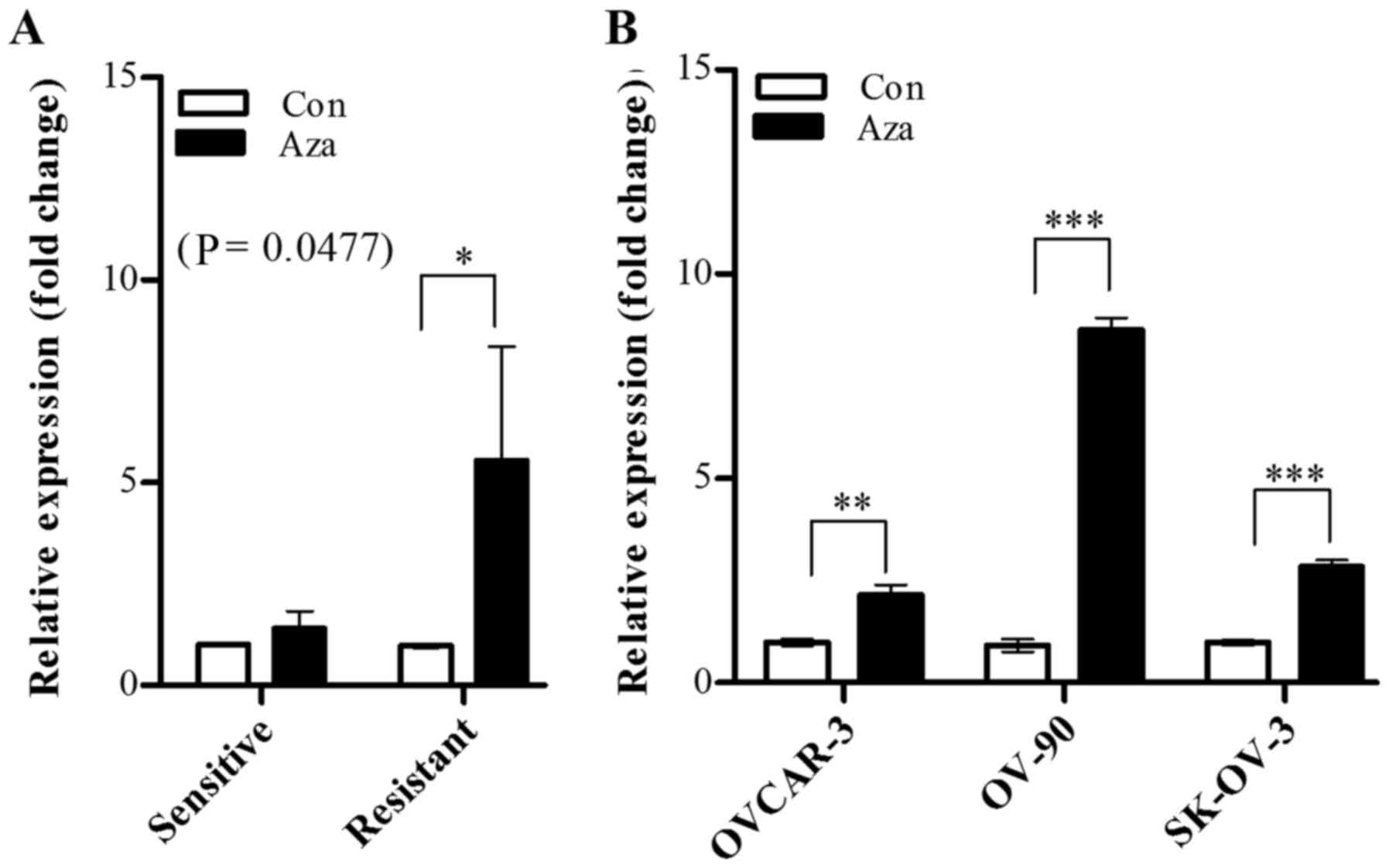
Subsequently, whether OXCT1 expression is
regulated by epigenetic modification was investigated using a DNA
methyltransferase inhibitor. The cisplatin-sensitive and
cisplatin-resistant groups were treated with 5-aza-dc, and
OXCT1 expression was evaluated by RT-qPCR. The results
demonstrated that OXCT1 expression level was significantly
increased in the cisplatin-resistant group, whereas there was no
significant increase detected the in cisplatin-sensitive group
(Fig. 4A). In all the
cisplatin-resistant cell lines (OVCAR-3, OV-90 and SK-OV-3), the
OXCT1 expression level of 5-aza-dc-treated cells was
restored, in the range of 2.1–6.8-fold, compared with that of
5-aza-dc-untreated cells, which indicated that the OXCT1
expression level was suppressed by hypermethylation in
cisplatin-resistant cell lines (Fig.
4B).
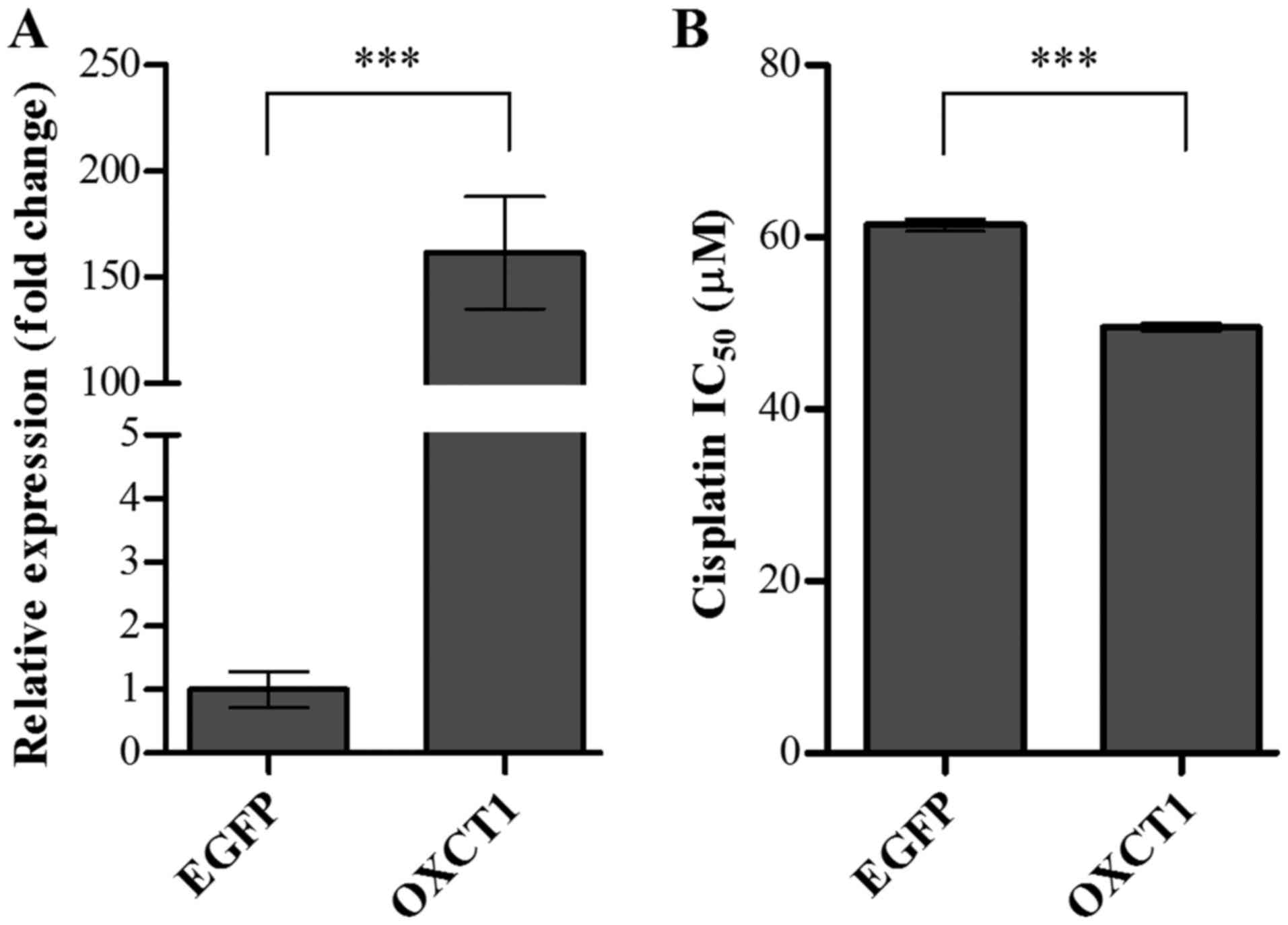
Overexpression of OXCT1 enhanced
sensitivity to cisplatin in the SK-OV-3 OC cell line
To determine whether OXCT1 overexpression in
a cisplatin-resistant cell line improved sensitivity to cisplatin,
SK-OV-3 cells were transiently transfected with OXCT1 or
EGFP expression plasmid constructs. Following a 24-h
transfection, the expression levels of OXCT1 were determined
by RT-qPCR. Compared with the EGFP-transfected control
cells, the level of OXCT1 expression was increased to
161.5-fold in OXCT1-transfected cells (Fig. 5A). The sensitivity to cisplatin was
also determined in EGFP- or OXCT1-transfected cells
using an MTT assay. IC50 was evaluated in SK-OV-3 cells
transfected with EGFP- or OXCT1-overexpressing
constructs following treatment with cisplatin at various
concentrations. Overexpression of OXCT1 significantly
decreased the IC50 for cisplatin by ~21% compared with
EGFP-transfected control cells (Fig. 5B). These results indicated that
overexpression of OXCT1 in the cisplatin-resistant cell line
significantly attenuated resistance to cisplatin.
Discussion
The present study provides evidence that
epigenetically regulated genes are involved in cisplatin resistance
of OC cells. mRNA expression profiles were integrated with DNA
methylation profiles to identify candidate genes for drug
resistance, and the OXCT1 gene was revealed to be a
promising target for modulating cisplatin resistance.
Cisplatin is one of the most effective
broad-spectrum anticancer drugs, and this platinum-based anticancer
drug activates the intrinsic apoptosis pathway via formation of
platinum-DNA adducts, resulting in DNA strand breaks during mitotic
cell division, which induce apoptosis (20). The DNA strand break initiates multiple
cellular self-defense systems, including DNA damage repair,
exocytosis of toxic metal compounds and alterations in gene
expression, and these responses result in chemoresistance to
cisplatin (21). Therefore, effector
genes responsible for cisplatin resistance may be due to a
defective influx route (reduced endocytosis of cisplatin), changes
to other putative proteins for cisplatin uptake, or altered
expression of detoxifying enzymes. Additionally, aberrant promoter
hypermethylation-mediated gene silencing is an epigenetic hallmark
of drug resistance; cisplatin treatment induces promoter
hypermethylation, alters gene expression profiles and renders cells
cisplatin-resistant (11).
OXCT1 encodes 3-oxoacid-conenzyme A
transferase 1, which is a homodimeric mitochondrial matrix enzyme
that serves a central role in extrahepatic ketone body catabolism
(ketone bodies to acetyl-CoA to mitochondrial tricarboxylic acid
cycle entrance) (15). There is
currently no direct evidence linking OXCT1 to cisplatin
sensitivity; however, a previous study demonstrated that
OXCT1 may be involved in autophagy-mediated apoptosis in
epithelial cell cancer cells (22).
In autophagy-mediated apoptosis, c-Jun N-terminal kinase
phosphorylates B-cell lymphoma-2 (Bcl-2) and B-cell lymphoma-extra
large (Bcl-xL), disrupting the Beclin/Bcl-2 and Beclin1/Bcl-xL
complexes; this process results in necroptosis in cancer cells
(23).
The nature of autophagy in drug resistance is
paradoxical and its role in carcinogenesis is context-dependent.
Regarding tumor-suppressive mechanisms, autophagy can inhibit
inflammation and genomic stability at an early stage (24). A previous study demonstrated that the
loss of the autophagy-regulating Beclin gene results in poor
recovery from ischemic stress with accumulation of cellular
aggregates and denatured proteins. Consequently, essential cellular
processes, including mitosis and centrosome functions. Are damaged
leading to chromosomal instability (24). Conversely, inhibition of autophagy can
sensitize cancer cells to ionizing radiation and chemotherapeutic
drugs (25). Further studies are
required to elucidate the precise mechanism underlying OXCT1
in cisplatin resistance.
In conclusion, the present study demonstrated that
OXCT1 acts as a suppressor of cisplatin resistance, and its
gene silencing by hypermethylation of CGI within the promoter
region is associated with cisplatin resistance in OC. The results
of the present study provide evidence of a potential novel
therapeutic target for the treatment of chemoresistant OC.
Acknowledgements
The present study was supported by the Korea Health
Technology R&D Project through the Korea Health Industry
Development Institute), funded by the Ministry of Health &
Welfare, Republic of Korea (grant no. HI13C2148).
Glossary
Abbreviations
Abbreviations:
|
OC
|
ovarian cancer
|
|
OXCT1
|
3-oxoacid CoA transferase 1
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
|
5-aza-dC
|
5-aza-2′-deoxycytidine
|
|
FBS
|
fetal bovine serum
|
|
P/S
|
penicillin/streptomycin
|
|
DEGs
|
differentially expressed genes
|
|
CGI
|
CpG islands
|
|
TSS
|
transcription start site
|
References
|
1
|
Lowe KA, Chia VM, Taylor A, O'Malley C,
Kelsh M, Mohamed M, Mowat FS and Goff B: An international
assessment of ovarian cancer incidence and mortality. Gynecol
Oncol. 130:107–114. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Notani PN: Global variation in cancer
incidence and mortality. Curr Sci-Bangalore. 81:465–474. 2001.
|
|
3
|
Salani R, Backes FJ, Fung MF, Holschneider
CH, Parker LP, Bristow RE and Goff BA: Posttreatment surveillance
and diagnosis of recurrence in women with gynecologic malignancies:
Society of gynecologic oncologists recommendations. Am J Obstet
Gynecol. 204:466–478. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ruan Z, Liu J and Kuang Y: Isolation and
characterization of side population cells from the human ovarian
cancer cell line SK-OV-3. Exp Ther Med. 10:2071–2078. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Borley J, Wilhelm-Benartzi C, Brown R and
Ghaem-Maghami S: Does tumour biology determine surgical success in
the treatment of epithelial ovarian cancer? A systematic literature
review. Br J Cancer. 107:1069–1074. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mantia-Smaldone GM, Edwards RP and Vlad
AM: Targeted treatment of recurrent platinum-resistant ovarian
cancer: Current and emerging therapies. Cancer Manag Res. 3:25–38.
2011.PubMed/NCBI
|
|
7
|
Lopez J, Percharde M, Coley HM, Webb A and
Crook T: The context and potential of epigenetics in oncology. Br J
Cancer. 100:571–577. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bird A: Does DNA methylation control
transposition of selfish elements in the germline? Trends Genet.
13:469–472. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Thiel G, Lietz M and Hohl M: How mammalian
transcriptional repressors work. Eur J Biochem. 271:2855–2862.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Suzuki MM and Bird A: DNA methylation
landscapes: Provocative insights from epigenomics. Nat Rev Genet.
9:465–476. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chang X, Monitto CL, Demokan S, Kim MS,
Chang SS, Zhong X, Califano JA and Sidransky D: Identification of
hypermethylated genes associated with cisplatin resistance in human
cancers. Cancer Res. 70:2870–2879. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen CC, Lee KD, Pai MY, Chu PY, Hsu CC,
Chiu CC, Chen LT, Chang JY, Hsiao SH and Leu YW: Changes in DNA
methylation are associated with the development of drug resistance
in cervical cancer cells. Cancer Cell Int. 15:982015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Su HY, Lai HC, Lin YW, Liu CY, Chen CK,
Chou YC, Lin SP, Lin WC, Lee HY and Yu MH: Epigenetic silencing of
SFRP5 is related to malignant phenotype and chemoresistance of
ovarian cancer through Wnt signaling pathway. Int J Cancer.
127:555–567. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li M, Balch C, Montgomery JS, Jeong M,
Chung JH, Yan P, Huang TH, Kim S and Nephew KP: Integrated analysis
of DNA methylation and gene expression reveals specific signaling
pathways associated with platinum resistance in ovarian cancer. BMC
Med Genomics. 2:342009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nam GH, Ahn K, Bae JH, Cho BW, Park KD,
Lee HK, Yang YM, Kim TH, Seong HH, Han K and Kim HS: Identification
of ORF sequences and exercise-induced expression change in
thoroughbred horse OXCT1 gene. Gene. 496:45–48. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Smyth GK: Linear models and empirical
bayes methods for assessing differential expression in microarray
experiments. Stat Appl Genet Mol Biol. 3:Article32004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: A practical and powerful approach to
multiple testing. J R Statist Soc B. 57:289–300. 1995.
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang YW, Zheng Y, Wang JZ, Lu XX, Wang Z,
Chen LB, Guan XX and Tong JD: Integrated analysis of DNA
methylation and mRNA expression profiling reveals candidate genes
associated with cisplatin resistance in non-small cell lung cancer.
Epigenetics. 9:896–909. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang G, Reed E and Li QQ: Molecular basis
of cellular response to cisplatin chemotherapy in non-small cell
lung cancer (Review). Oncol Rep. 12:955–965. 2004.PubMed/NCBI
|
|
21
|
Shen DW, Pouliot LM, Hall MD and Gottesman
MM: Cisplatin resistance: A cellular self-defense mechanism
resulting from multiple epigenetic and genetic changes. Pharmacol
Rev. 64:706–721. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sanchez-Alvarez R, Martinez-Outschoorn UE,
Lin Z, Lamb R, Hulit J, Howell A, Sotgia F, Rubin E and Lisanti MP:
Ethanol exposure induces the cancer-associated fibroblast phenotype
and lethal tumor metabolism: Implications for breast cancer
prevention. Cell Cycle. 12:289–301. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shi S, Wang Q, Xu J, Jang JH, Padilla MT,
Nyunoya T, Xing C, Zhang L and Lin Y: Synergistic anticancer effect
of cisplatin and Chal-24 combination through IAP and c-FLIPL
degradation, Ripoptosome formation and autophagy-mediated
apoptosis. Oncotarget. 6:1640–1651. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kimmelman AC: The dynamic nature of
autophagy in cancer. Genes Dev. 25:1999–2010. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wu WK, Coffelt SB, Cho CH, Wang XJ, Lee
CW, Chan FK, Yu J and Sung JJ: The autophagic paradox in cancer
therapy. Oncogene. 31:939–953. 2012. View Article : Google Scholar : PubMed/NCBI
|