Introduction
Recently, oral cavity cancer, including that of the
lip, was found to be the fifteenth most common type of malignancy
across populations worldwide, and the site incidence rate of oral
cancer was 30.5% of all head and neck (H&N) regions (including
the thyroid gland) (1,2), with an incidence of 275,000 cases
annually (3). In the ‘Cancer
Statistics, 2016,’ 31,910 new cases of oral cancer and 6,490 deaths
were estimated in the U.S. (4), and
both of these values are 1.5-fold higher than the values in the
past two decades (5,6). Oral squamous cell carcinoma (OSCC)
represents a majority of oral cancer cases, and the major site of
occurrence is the tongue (7,8). Among the OSCC cases, the rate of
early-stage OSCC has increased gradually, and, to date, 67.4–80% of
all OSCCs have been reported to be early-stage OSCCs (9,10).
Therefore, early-stage tongue squamous cell carcinoma (TSCC) is one
of the most common H&N cancers (1,7,9,10). The
standard treatment of early-stage OSCC, including TSCC remains
surgery, with the addition of adjuvant therapy for advanced
features such as positive surgical margins and detection of venous,
lymphatic, or neural invasion (11),
which has remained relatively unchanged to date (9). Local recurrence and lymph node
metastasis (LNM) are the main causes of death in patients with
early-stage oral cancer (12,13) because the relapse is generally
followed by distant metastasis (14).
The survival rate of the tumor depends on local recurrence, LNM,
and distant metastasis (12). A
relatively higher rate of neck LNM has been reported in OSCC
(4). In particular, TSCC is well
known to be associated with an increased tendency for LNM (15). Early stage cancer has basically good
prognosis, however, the stages of OSCC have relatively poor
prognoses (approximately 40% of patients died within 3 years)
(9), and the primary reason is
delayed LNM (16,17). Delayed LNM is generally considered LNM
that occurs after primary treatment for cancer (neck dissection was
not performed during treatment) (18,19). The
prediction of delayed LNM is important in the treatment of
early-stage oral cancer (20).
Numerous prognostic factors (clinical, pathological, and molecular
features) of LNM have been reported to date (2,9,20–24);
however, this collectively makes daily clinical practice
complex.
MicroRNAs (miRs) are expected to serve as simple
markers for prognosis in cancer patients (25,26). Their
expressions can be easily detected in not only in fresh tissue but
also blood, saliva, and formalin-fixed paraffin-embedded (FFPE)
tissues (27–30). They are small (19–25 nt in length),
non-coding RNAs known to critically regulate various oncogenes or
tumor suppressor gene expressions (31–35). To
date, more than 2,600 human miRs are present in the miRbase
(Release 21), a database that stores miR data, and 60% of human
protein-coding genes have been reported to be regulated by miRs
(36). miRs play important roles in
the regulation of OSCC and may serve as prognostic factors
(15,37–40). To
date, many studies have investigated miR regulation in oral cancer.
As a result, a plethora of miRs have been identified as prognostic
factors for recurrence or metastasis in OSCC patients. There are
also many pathways of cancer progression or metastasis that relate
to miRs are present (41).
Additionally, various kinds of oral cancer-related genes and
pathways are present and these might be associated with tumor
recurrence and metastasis (3,7,25,34,41–43). Among
the miRs, miR-10a, miR-10b, miR-196a, miR-196b, and miR-615, which
reside in Homeobox (HOX) gene clusters, have been the
focus of much attention recently (25,44–46).
Increased regulation of HOX genes is associated with the
proliferation and migration of OSCC cells, which can lead to
recurrence or metastasis in OSCC patients (45). Among the five miRs, miR-615 from
examination has the sparsity of studies (47). The remaining four miRs (miR-10a,
miR-10b, miR-196a, and miR-196b) have been shown to be related to
various target genes and to control many types of malignancies or
other diseases (34,48–51). In
particular, some genes regulated by miR-196, impact recurrence or
metastasis (34,45,46,52,53).
Thus, miR-196 expression and functional have been investigated
(29,46,52,54). Those
four miRs (miR-10a, miR-10b, miR-196a, and miR-196b) have been
shown to be related with OSCC in the previous studies (29,52,55,56).
Therefore, we selected miRs as candidates in this study.
For OSCC patients, it is critically more important
to identify prognostic rather than diagnostic markers. Among all
malignancies, OSCC is relatively easy to detect because the oral
cavity can be directly observed. However, to date, no approaches
can monitor recurrence and metastasis in early-stage TSCC (39). Furthermore, no useful miR marker can
predict recurrence or metastasis, particularly delayed LNM in
patients with early-stage OSCC. Some studies have reported several
miRs as prognostic markers of local recurrence or LNM; however,
their data were relevant to ‘all stages’ and not only to the early
stages of the disease (57,58). Among our candidate miRs, miR-196a
reportedly serves as a useful prognostic marker of ‘locoregional
recurrence’ (local recurrence and/or regional LNM) in OSCC
(29,45,52),
although the cited studies focused on ‘all stages’ of OSCC
throughout the oral cavity and did not distinguish between local
recurrence and LNM. To examine local recurrence or delayed LNM,
only LNM after treatment should be investigated. Moreover, there
are few studies on miR expression in early-stage OSCC (10,59,60). We
hypothesized that the above-mentioned miRs could be useful
prognostic markers of early-stage TSCC. For testing this
hypothesis, we used reverse transcription-quantitative polymerase
chain reaction (RT-qPCR) to analyze the expression of the miRs in
FFPE tissues of patients with early-stage TSCC compared the
findings with those of adjacent normal tissue (ANT) in order to
determine the relationship between miR expressions and disease
features.
Materials and methods
Patients and tissue samples
Informed consent was obtained from all patients. The
Ethics Committee of the University of the Ryukyus (Okinawa, Japan)
approved this study on June 22, 2016 (approval no. 957), and the
study complied with the Declaration of Helsinki. The Conflict of
Interest Committee of the University of the Ryukyus approved this
study on June 22, 2016, and the authors have no conflicts of
interest to disclose. Samples of cancer tissue and ANT were
retrospectively collected from 50 patients with primary early-stage
TSCC (cT1T2N0) clinically (excluding cancer in situ,
dysplasia, pre-cancer diseases, and verrucous carcinoma), who were
aged ≥20 years and diagnosed by our department. All patients
underwent surgical excision between January 2005 and December 2014.
All cancers were located on the mobile tongue. Some cases were from
our previous retrospective study (61), whose aim was to compare patients who
underwent surgery alone to those with neoadjuvant chemotherapy and
surgery. The current study excluded patients who underwent
neoadjuvant therapy. Therefore, no methods or results from the
previous study were included in the current study.
No LNM or distant metastasis was confirmed by
assessing clinical symptoms and radiological lesions before
surgery. All alive patients were followed for ≥2 years after
treatment, which is considered an appropriate timeframe to evaluate
recurrence or metastases (17,62). The
maximum length of follow-up was 5 years (63). We excluded the following patients: i)
who received chemotherapy, radiotherapy, or both before surgery;
ii) who underwent simultaneous neck dissections (in all patients,
no LNM was suspected clinically or radiologically; therefore, LNM
after curative surgery could be defined as ‘delayed’ LNM); iii) two
or more additional resections on the day after the first local
resection because of positive margins; iv) who had coexisting
cancer; and v) patients with a history of H&N cancers or those
who underwent treatment for these tumors.
The surgical margins used were ≥10 mm clinically for
all patients; thus, the resection margins of all tumors were
microscopically free of cancer cells. In three cases with closed
surgical margins, tumors were pathologically found a few days after
surgery, and adjuvant chemotherapy (oral therapy of S-1) was
administered. No patient received adjuvant radiotherapy. All
stained tissues were reviewed by pathologists at the Department of
Pathology, University Hospital of the Ryukyus. Histological grades
were classified by pathologists according to the WHO guidelines
(64) as well, moderate, or poor.
There was no undifferentiated type. The mode of invasion was
divided into four main grades (1, 2, 3, and 4C/4D) by using the
criteria proposed by Yamamoto et al. for hematoxylin and
eosin specimens (65). Lymphatic,
venous, and neural invasions were evaluated with
immunohistochemical stains or special stains, including D2-40 (code
M3619; Dako, Glostrup, Denmark), Victoria blue (no. 4077; Muto Pure
Chemical Co., Ltd., Tokyo, Japan), and S-100 (cat. no. Z0311;
Dako), respectively. Tumor depth was measured vertically between
the adjacent normal mucosa surface and the deepest point of tumor
invasion (66). All stained slides
were 3-µm thick and were obtained from surgical specimens. Patient
history of alcohol use or smoking was determined. No patient chewed
betel quid, which is a high-risk factor for oral cancer. The
definitions of recurrence/metastasis were as follows: i) local
recurrence: lesions appearing in the oral cavity and nearly where
resection for tongue cancer was performed between 6 weeks and 5
years after the first curative surgery; ii) LNM: lesions appearing
only in the neck lymph node region between 6 weeks and 5 years
after the first curative surgery; and iii) distant metastasis:
distant lesions appearing between 6 weeks and 5 years after the
first curative surgery (63). In the
present study, no combination of local recurrence or LNM was found.
At the time of diagnosis, distant metastases, local recurrence, or
LNM was not found. Overall survival (OS) was defined as the time
from the date of surgical excision to the date of death or last
follow-up. Local recurrence-free survival (LRFS), LNM-free survival
(LNMFS), distant metastasis-free survival (DMFS), and disease-free
survival (DFS) were defined as the time from the date of surgical
excision to the date of local recurrence, LNM, distant metastasis,
and any recurrence or metastasis, respectively, or the last
follow-up day, including death without recurrence.
Collection of target samples from FFPE
tissues using laser-capture microdissection (LMD)
FFPE tissues acquired from surgical excisions or
biopsies were collected for analysis. Before performing LMD,
pathologists classified the cancer and ANT areas. ANTs were defined
as epithelial tissues located on the surgical margin that were ≥5
mm away from cancer tissues. A microtome (Leica RM2235; Leica
Biosystems Nussloch GmbH, Nussloch, Germany) was used to prepare
7-µm-thick tissue sections from FFPE tissue blocks, which were
placed on nuclease-free 1.0 PEN Membrane Slides (no.
415190-9081-000; Zeiss GmbH, Jena, Germany). Then, tissue sections
were deparaffinized, stained with cresyl violet, and dried in air
briefly, and the slides were subsequently stored at −20°C. LMD was
performed using a Zeiss PALM Microbeam laser microdissection system
(Carl Zeiss Microscopy, Jena, Germany) and PALM Robo v4.6 software.
For capturing tissue, Adhesive Caps (no. 415190-9201-000; Zeiss
GmbH) were used. Tissue sections (cancer tissue and paired ANT)
were immediately deparaffinized and total RNA was then
extracted.
RT-qPCR
Total RNA was extracted from FFPE tissues using an
miRNeasy FFPE kit (Qiagen GmbH, Hilden, Germany) (67), which involved removal of genomic DNA
contamination and RNA purification with RNeasy MinElute. The sample
was stored at −80°C before use for reverse transcription (RT). The
purity of total RNA was examined using BioSpec-nano (Shimadzu,
Kyoto, Japan). RT was performed using a Taqman microRNA RT kit
(Thermo Fisher Scientific, Inc., Waltham, MA, USA) (68) with 5 µl of total RNA, and cDNA samples
were stored at −20°C before use. We selected miR-196a-3p to
evaluate miR-196a-2. TaqMan miRNA assays (Thermo Fisher Scientific,
Waltham, Inc.) were used to assess the relative expressions of the
following five miRs: miR-10a (miR-10a-5p: assay ID 000387, mature
miR sequence: UAC CCU GUA GAU CCG AAU UUG UG), miR-10b (miR-10b-5p:
assay ID 002218, mature miR sequence: UAC CCU GUA GAA CCG AAU UUG
UG), miR-196a-5p (assay ID 241070_mat, mature miR sequence: UAG GUA
GUU UCA UGU UGU UGG G), miR-196a-3p (assay ID 002336, mature miR
sequence: CGG CAA CAA GAA ACU GCC UGAG), and miR-196b (miR-196b-5p:
assay ID 002215, mature miR sequence: UAG GUA GUU UCC UGU UGU UGG
G), according to the manufacturer's instructions. The reactions
were carried out at 50 °C for 2 min and at 95°C for 10 min,
followed by 40 cycles at 95°C for 15 sec and at 60°C for 60 sec.
All experiments were carried out in triplicate. Data were analyzed
with the 2−ΔΔCq method (69). Data were normalized using small
nuclear RNAs 44 and 48 (RNU44, assay ID 001094; RNU48, assay ID
001006; Thermo Fisher Scientific, Inc.) as endogenous controls. In
all 50 patients, RNU44 and 48 could be determined. The undetermined
Cq value of the other five miRs (miR-10a, miR-10b, miR-196a-5p,
miR-196a-3p, and miR-196b) was estimated as 40 Cq (70,71). The
formulas were as follows: ΔCq (cancer) value=(Cq value of cancer
tissue)-(the average Cq value of RNU44 and RNU48); ΔCq (normal)
value=(Cq value of normal tissue)-(the average Cq value of RNU44
and RNU48); and ΔΔCq value=ΔCq (cancer) value-ΔCq (normal) value. A
lower ΔCq value represented higher expression of miR. A lower ΔΔCq
value represented higher expression of miR in cancer tissues than
in paired ANTs of the same patient.
Statistical analysis
The Gaussian distribution of each group was tested
using Shapiro-Wilk test. The homogeneity of variances was confirmed
using the Levene test. For data with a Gaussian distribution, the
difference between two groups was demonstrated using the Student's
t-test or the Welch's t-test, depending on the homogeneity of the
variances. For data that did not conform to a Gaussian
distribution, the Mann-Whitney U test was applied. When
analyzing three or more subgroups, one-way ANOVA or the
Kruskal-Wallis test was used to assess whether the data conformed
to a Gaussian distribution. The association of the ΔCq value
between early-stage TSCC tissue and paired ANT was confirmed using
a paired t-test. The ΔΔCq values and the clinicopathological
characteristics of the patients were evaluated using the tests
mentioned above. Differences in the patient numbers between high
and low ΔΔCq value regulation groups were evaluated using a
two-tailed Fisher's exact test. Survival analyses (OS, LRFS, LNMFS,
DMFS, and DFS) were performed using the Kaplan-Meier method, and
survival curves were compared using the log-rank test. A receiver
operating characteristic (ROC) curve analysis was performed and
area under the ROC curve (AUC) was determined to examine the
feasibility of using the ΔΔCq value as an approach for assessing
the prognosis of delayed LNM and the mode of tumor invasion. Youden
index was calculated for the identification of the best ΔΔCq
cut-off value. P<0.05 was considered to indicate statistical
significance. All statistical analyses were performed using JMP
software (JMP Version Pro 12; SAS Institute Inc., Cary, NC,
USA).
Results
Characteristics
Table I presents the
characteristics of the patients. The numbers of patients with local
recurrence, delayed LNM, and distant metastasis were 4, 17, and 3,
respectively. All distant metastases occurred after treatment and
control of delayed LNM. The clinical T stage was cT1 in 32 patients
and cT2 in 18 patients. The histological grade was G1/well in 31
patients, G2/moderate in 16 patients, and G3/poor in 1 patient. The
mode of tumor invasion was 1–3 in 39 patients and 4C-4D in 11
patients. The depth of the tumor was <5 mm in 42 patient and
between 5 and 10 mm in eight patients. Lymphatic, venous, and
neural invasions were found in six, five, and five patients,
respectively. Nineteen patients were current or past smokers and 22
were current or past alcohol drinkers.
 | Table I.Clinicopathological characteristics
of our patients (n=50). |
Table I.
Clinicopathological characteristics
of our patients (n=50).
| Subgroups | n (%) |
|---|
| Age |
|
|
<60 | 21 (42) |
|
≥60 | 29 (58) |
| Sex |
|
|
Male | 24 (48) |
|
Female | 26 (52) |
| Clinical T
stage |
|
| I | 32 (64) |
| II | 18 (36) |
| Local
recurrence |
|
|
Yes | 4
(8) |
| No | 46 (92) |
| Delayed LNM |
|
|
Yes | 17 (34) |
| No | 33 (66) |
| Distant
metastasis |
|
|
Yes | 3
(6) |
| No | 47 (94) |
| Histological
grade |
|
|
G1/well | 31 (62) |
|
G2/moderate | 16 (32) |
|
G3/poor | 1
(2) |
|
G4/undifferentiated | 0
(0) |
|
Unknown | 2
(4) |
| Mode of tumor
invasion |
|
|
1–3 | 39 (78) |
|
4C-4D | 11 (22) |
| Depth of tumor |
|
| <5
mm | 46 (92) |
| 5 mm ≤
and <10 mm | 4
(8) |
| ≥10
mm | 0
(0) |
| Lymphatic
invation |
|
|
Yes | 6
(12) |
| No | 43 (86) |
|
Unknown | 1
(2) |
| Venous
invasion |
|
|
Yes | 5
(10) |
| No | 45 (90) |
| Neural
infiltration |
|
|
Yes | 5 (10) |
| No | 44 (88) |
|
Unknown | 1
(2) |
| Smoking |
|
| Current
or past | 19 (38) |
|
Never | 31 (62) |
| Alcohol intake |
|
| Current
or past | 22 (44) |
|
Never | 25 (50) |
|
Unknown | 3 (6) |
All 5 miRs were significantly
expressed in early-stage TSCC tissues
The levels of all five candidate miRs showed
significant differences between early-stage TSCC tissues and ANTs,
and P-values for miR-196a-5p, miR-196b, miR-10a, miR-10b, and
miR-196a-3p were <0.001, <0.001, <0.001, <0.001, and
0.003, respectively (Fig. 1A-E).
miR-196a-5p and miR-196b were upregulated (Fig. 1A and B) while miR-196a-3p, miR-10a,
and miR-10b were downregulated in early-stage TSCC tumors (Fig. 1C-E). Thus, all five miRs were viable
biomarkers.
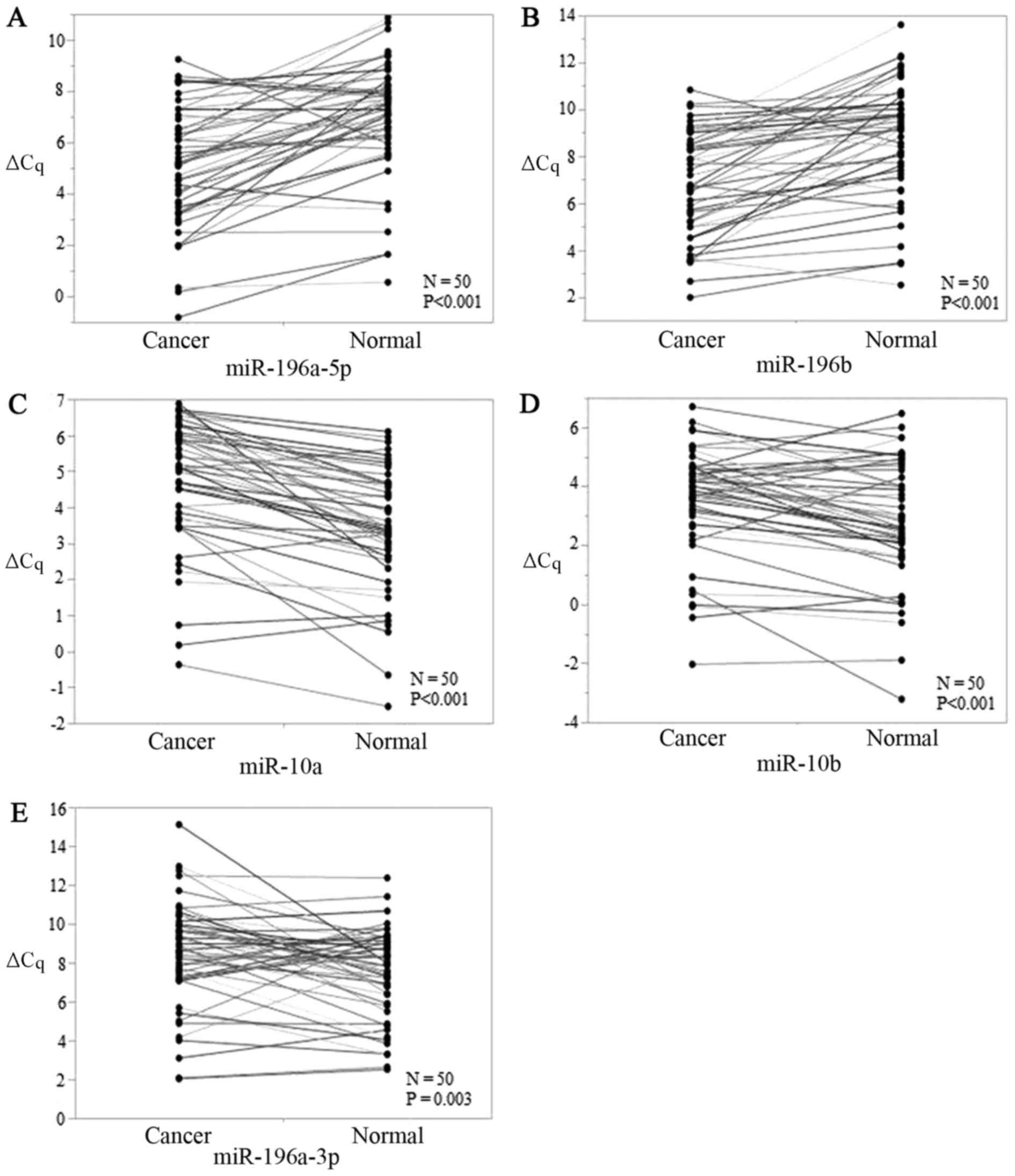 | Figure 1.ΔCq value expressions of (A-E) 5
candidate miRs [(A) miR-196a-5p, (B) miR-196b, (C) miR-10a, (D)
miR-10b, and (E) miR-196a-3p] in early-stage TSCC tissues and
paired adjacent normal tissues. P-values of miR-196a-5p, miR-196b,
miR-10a, miR-10b, and miR-196a-3p were <0.001, <0.001,
<0.001, <0.001, and 0.003 respectively. miR, microRNA; TSCC,
tongue squamous cell carcinoma. |
Clinicopathological significance of
miR levels in early-stage TSCC
We analyzed the association between miR expression
(using ΔΔCq values) and clinicopathological parameters (Table II). For several characteristics, the
P-values were significant. The ΔΔCq values of miR-196a-5p and
miR-196a-3p were significantly different between the local
recurrence and no recurrence groups. The ΔΔCq values of miR-196a-5p
and miR-196a-3p were also significantly different between the
delayed LNM and no LNM groups. In the distant metastasis subgroup,
the ΔΔCq values of miR-196b were significantly different.
Furthermore, in the sex, clinical stage, mode of tumor invasion,
depth of tumor, and smoking subgroups, the ΔΔCq values of miR-10b,
miR-196b, miR-196a-5p, miR-196a-3p, and miR-10b, respectively, were
significantly different. In contrast, there was no significant
relationship between miRs levels and age, alcohol intake, and other
pathological features such as histological grade, venous/lymphatic
invasion, or neural infiltration (Table
II). We assessed the most appropriate biomarker candidates. As
described above, miR-196a-5p and miR-196b were upregulated
(Fig. 1A and B) while miR-196a-3p,
miR-10a, and miR-10b were downregulated in early-stage TSCC tissues
than in ANTs (Fig. 1C-E). We thought
that miR-196a-5p (local recurrence), miR-196a-3p (LNM), miR-196b
(distant metastasis), and miR-10b (smoking) were not appropriate
markers because some combinations of the subgroups and ΔΔCq values
were controversial. Further, miR-10b (gender) and miR-196b
(clinical, stage) were not appropriate markers because these can be
confirmed clinically. On the other hand, we thought that
miR-196a-5p (LNM and mode of tumor invasion) and miR-196a-3p (local
recurrence) were appropriate candidates because the expressions
were consistent.
 | Table II.Association of miR levels and
clinicopathological characteristics of our patients (n=50). |
Table II.
Association of miR levels and
clinicopathological characteristics of our patients (n=50).
| Subgroup | miR-10a | miR-10b | miR-196a-5p | miR-196a-3p | miR-196b |
|---|
| Age |
|
|
|
|
|
<60 | 1.420±1.330 | 0.542
(−2.02–3.695) | −2.027±1.819 | 0.445±2.144 | −2.091±1.744 |
|
≥60 | 1.21±0.993 | 0.985
(−2.135–2.245) | −1.845±1.999 | 1.512±2.541 | −1.696±2.149 |
|
P-value | 0.5250 | 0.1628 | 0.7423 | 0.1247 | 0.4921 |
| Gender |
|
|
|
|
|
|
Male | 1.181±1.303 | 0.345±1.253 | −2.101±2.043 | 1.128±2.545 | −2.102±1.628 |
|
Female | 1.406±0.976 | 1.058±1.146 | −1.756±1.799 | 1.005±2.345 | −1.640±2.267 |
|
P-value | 0.4904 | 0.0410 | 0.5288 | 0.8603 | 0.4665 |
| Clinical stage |
|
|
|
|
|
| I | 1.301±1.245 | 0.678±1.255 | −1.714±1.690 | 1.032±2.603 | −1.443±1.647 |
| II | 1.292±0.955 | 0.784±1.246 | −2.290±2.249 | 1.121±2.121 | −2.606±2.331 |
|
P-value | 0.9793 | 0.7737 | 0.3112 | 0.9017 | 0.0451 |
| Local
recurrence |
|
|
|
|
|
|
Yes | 0.813±0.760 | 0.711±1.092 | 0.265±0.554 | 3.783±3.902 | −0.271±1.147 |
| No | 1.340±1.162 | 0.716±1.263 | −2.112±1.866 | 0.828±2.154 | −2.000±1.984 |
|
P-value | 0.3805 | 0.9931 | 0.0153 | 0.0175 | 0.0942 |
| Delayed LNM |
|
|
|
|
|
|
Yes | 1.199±1.164 | 0.545±1.514 | −2.995±1.452 | 0.011±1.668 | −2.224±1.979 |
| No | 1.349±1.140 | 0.804±1.088 | −1.369±1.896 | 1.606±2.583 | −1.666±1.982 |
|
P-value | 0.6636 | 0.4897 | 0.0033 | 0.0114 | 0.3349 |
| Distant
metastasis |
|
|
|
|
|
|
Yes | 2.065
(−0.677–2.69) | 0.905±0.587 | −2.254±0.287 | 0.882±1.055 | −0.645±0.420 |
| No | 1.165
(−0.655–4.58) | 0.704±1.273 | −1.900±1.967 | 1.076±2.486 | −1.939±2.016 |
|
P-value | 0.7131 | 0.7882 | 0.2976 | 0.8946 | 0.0059 |
| Histological
grade |
|
|
|
|
|
|
G1/well | 1.178±1.190 | 0.642±1.063 | −1.529±1.854 | 1.359±2.821 | −0.96
(−7.03–2.27) |
|
G2/moderate | 1.455±1.103 | 0.95±1.461 | −2.479±1.983 | 0.495±1.602 | −2.825
(−5.155–0.81) |
|
G3/poor | 1.165 | 1.29 | −2.195 | 0.755 | −3.21 |
|
Unknown | 1.962±1.028 | −0.285±2.453 | −3.402±1.446 | 1.202±1.156 | −2.805
(−4.6–1.01) |
|
P-value | 0.7321 | 0.5453 | 0.2774 | 0.7249 | 0.5092 |
| Mode of tumor
invasion |
|
|
|
|
|
|
1–3 | 1.283±1.175 | 0.92
(−2.02–3.695) | −1.584±1.838 | 1.285±2.619 | −1.697±2.059 |
|
4C-4D | 1.349±1.051 | 0.745
(−2.135–1.45) | −3.12±1.727 | 0.282±1.319 | −2.447±1.615 |
|
P-value | 0.8679 | 0.2511 | 0.0168 | 0.0918 | 0.2716 |
| Depth of tumor |
|
|
|
|
|
| <5
mm | 1.289±1.081 | 0.785±1.069 | −1.832±1.941 | −1.831±2.018 | 1.081±2.483 |
| 5 mm ≤
and <10 mm | 1.406±1.905 | −0.078±2.658 | −2.955±1.209 | −2.211±1.676 | 0.876±1.751 |
|
P-value | 0.8457 | 0.5638 | 0.2633 | 0.7173 | 0.8730 |
| Lymphatic
invasion |
|
|
|
|
|
|
Yes | 1.364±1.347 | 0.823±1.051 | −1.039±1.308 | 1.343±3.713 | −1.643±1.413 |
| No | 1.315±1.125 | 0.672±1.273 | −2.028±1.977 | 0.955±2.224 | −1.914±2.075 |
|
Unknown | 0.175 | 1.955 | −2.63 | 4.07 | −0.935 |
|
P-value | 0.6153 | 0.5878 | 0.4685 | 0.4339 | 0.8563 |
| Venous
invasion |
|
|
|
|
|
|
Yes | 1.199±0.569 | 0.066±1.303 | −2.725±1.534 | 0.354±0.720 | −2.802±2.230 |
| No | 1.309±1.189 | 0.788±1.226 | −1.832±1.939 | 1.143±2.530 | −1.757±1.950 |
|
P-value | 0.8396 | 0.2199 | 0.3264 | 0.1278 | 0.2677 |
| Neural
infiltration |
|
|
|
|
|
|
Yes | 0.67±0.630 | −0.002±1.318 | −2.995
(−6.485–3.285) | 1.005±2.367 | −1.651±2.596 |
| No | 1.395±1.164 | 0.769±1.221 | −2.025
(−4.735–0.725) | 1.003±2.435 | −1.907±1.953 |
|
Unknown | 0.175 | 1.955 | −2.63 | 4.07 | −0.935 |
|
P-value | 0.2493 | 0.2564 | 0.6001 | 0.4640 | 0.8660 |
| Smoking |
|
|
|
|
|
| Current
or past | 1.120±1.008 | 0.235±1.027 | −2.463±1.949 | 1.599±3.009 | −1.575
(−5.195–0.185) |
|
Never | 1.407±1.215 | 1.011±1.281 | −1.590±1.836 | 0.736±1.956 | −1.36
(−7.03–2.27) |
|
P-value | 0.3931 | 0.0303 | 0.1175 | 0.2749 | 0.5622 |
| Alcohol intake |
|
|
|
|
|
| Current
or past | 1,353±1.340 | 0.383±1.270 | −2.071±1.944 | 1.306±2.819 | −1.23
(−5.195–0.185) |
|
Never | 1.310±0.998 | 1.050±1.130 | −1.728±1.851 | 0.851±2.068 | −1.49
(−7.03–2.27) |
|
Unknown | 0.794±0.741 | 0.371±1.611 | −2.434±2.704 | 1.067±2.707 | −4.385
(−4.51–0.4325) |
|
P-value | 0.7335 | 0.1635 | 0.7459 | 0.8194 | 0.5670 |
|
Association between miR-196a-5p levels
and LNMFS in patients with early-stage TSCC
We divided patients using the median ΔΔCq value as
the cut-off value. On comparing the low miR-196a-5p ΔΔCq value
regulation (i.e., high expression of miR-196a-5p) group and the
high regulation group, no bias was found except with regard to
delayed LNM (Table III). Thus,
miR-196a-5p could be a prognostic marker. In contrast, on comparing
the low miR-196a-3p ΔΔCq value regulation (i.e., high expression of
miR-196a-3p) group and the high regulation group, bias was found
for delayed LNM and depth of tumor (data not shown). Thus,
miR-196a-3p could not be considered as a prognostic marker of
‘local recurrence’ because of the biases. The Kaplan-Meier analysis
of each survival curve [OS, LRFS, LNMFS, DMFS, and DFS (Fig. 2A-E, respectively)] indicated that
LNMFS was shorter in the low miR-196a-5p ΔΔCq value regulation
(i.e., higher regulation in TSCC tissues than in ANTs) group than
in the high regulation group (P=0.0079) (Fig. 2C). In contrast, OS, LRFS, DMFS, and
DFS showed no significant differences (P=0.6205, 0.0502, 0.5382,
and 0.1049, respectively) (Fig. 2A, B, D
and E). In subsequent ROC analysis, ΔΔCq values revealed that
miR-196a-5p levels yielded P=0.0025, an AUC of 0.740 and a cut-off
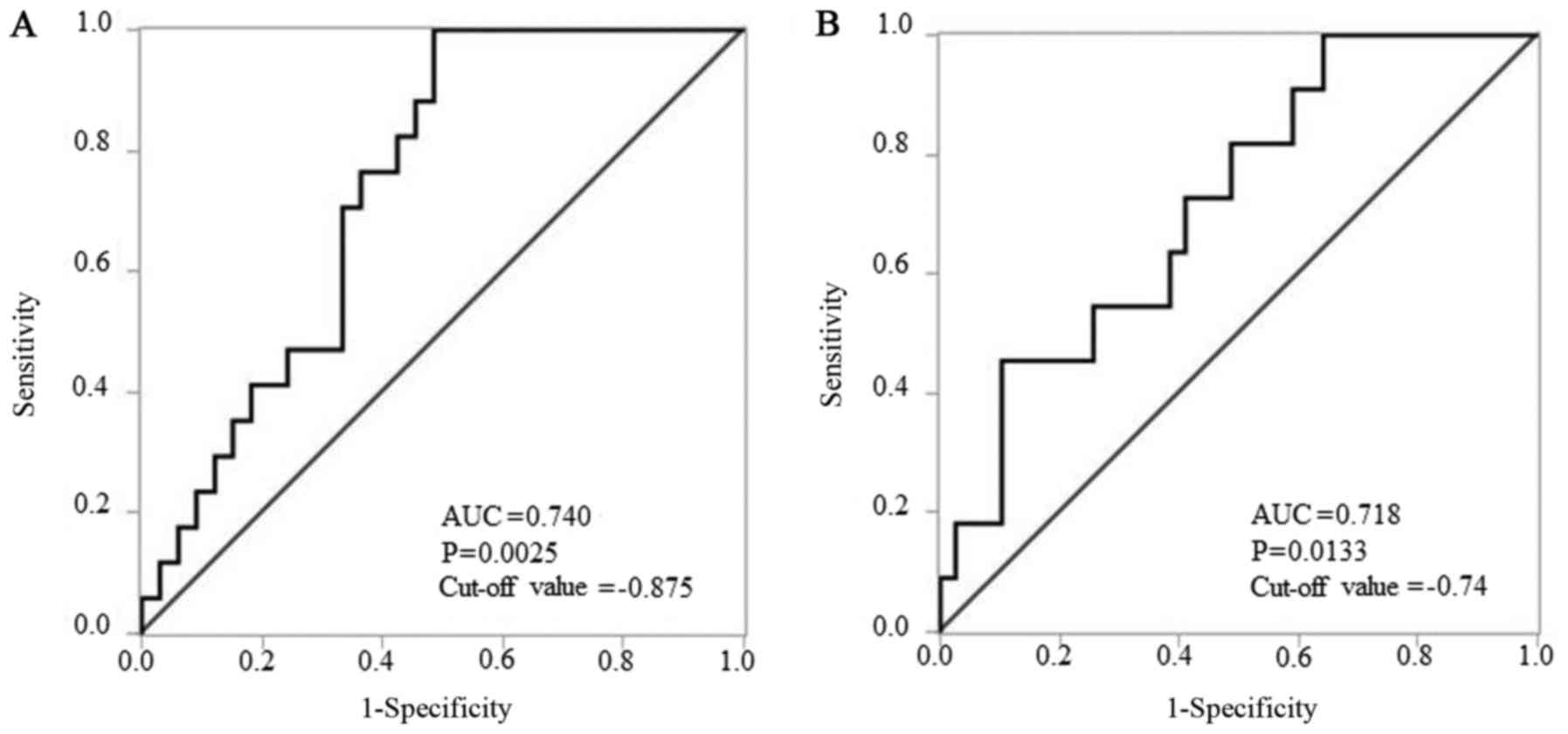
value of −0.875 to distinguish delayed LNM (Fig. 3A). According to the result presented
in Table II, the mode of tumor
invasion was also evaluated by ROC analysis. ΔΔCq values revealed
that miR-196a-5p levels yielded P=0.0133, an AUC of 0.718 and a
cut-off value of −0.74 to distinguish YK grade 4C-4D from YK grade
1–3 (Fig. 3B).
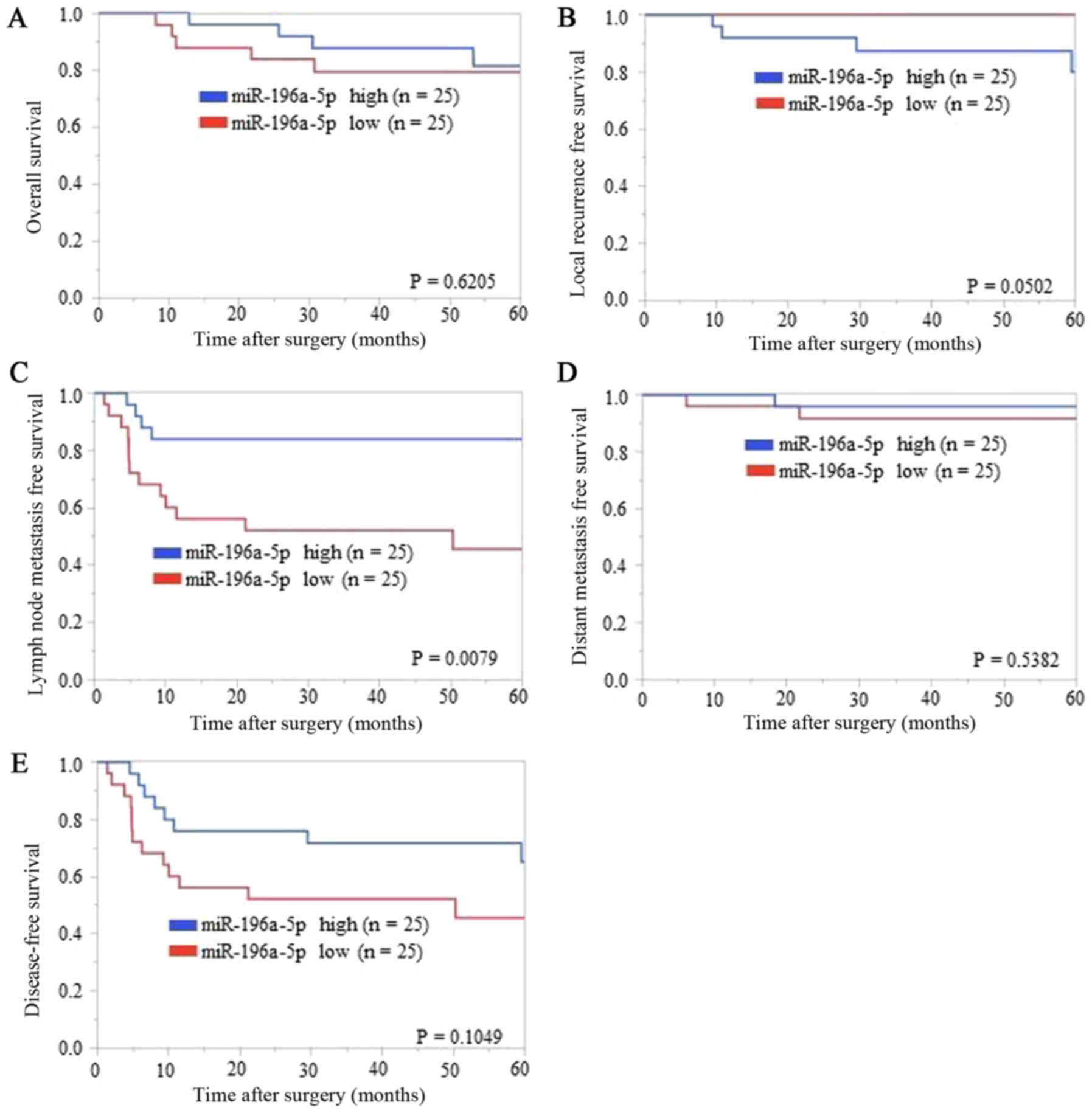 | Figure 2.Association of ΔΔCq value regulation
of miR-196a-5p expression with (A) overall survival, (B) local
recurrence-free survival, (C) lymph node metastasis-free survival,
(D) distant metastasis-free survival, and (E) disease-free
survival. Kaplan-Meier analysis (cut-off: The median ΔΔCq value)
indicated that patients with relatively low ΔΔCq value regulation
levels of miR-196a-5p experienced significantly shorter lymph node
metastasis-free survival than those with high ΔΔCq value regulation
levels (P=0.0079, log-rank test). Overall survival, local
recurrence-free survival, distant metastasis-free survival, and
disease-free survival showed no significant differences between low
and high groups of miR-196a-5p regulations (P=0.6205, P=0.0502,
P=0.5382, and P=0.1049, respectively, log-rank test). |
 | Table III.Association between microRNA-196a-5p
ΔΔCq value regulation and subgroups (n=50). |
Table III.
Association between microRNA-196a-5p
ΔΔCq value regulation and subgroups (n=50).
|
| Case number of
groups |
|
|---|
|
|
|
|
|---|
| Subgroup | High regulation
(n=25) | Low regulation
(n=25) | P-value |
|---|
| Age |
|
|
|
|
<60 | 10 | 11 | 1.000 |
|
≥60 | 15 | 14 |
|
| Gender |
|
|
|
|
Male | 12 | 12 | 1.000 |
|
Female | 13 | 13 |
|
| Clinical stage |
|
|
|
| I | 18 | 14 | 0.377 |
| II | 7 | 11 |
|
| Local
recurrence |
|
|
|
|
Yes | 4 | 0 | 0.110 |
| No | 21 | 25 |
|
| Delayed LNM |
|
|
|
|
Yes | 4 | 13 | 0.016 |
| No | 21 | 12 |
|
| Distant
metastasis |
|
|
|
|
Yes | 1 | 2 | 1.000 |
| No | 24 | 23 |
|
| Histological
grade |
|
|
|
|
G1/well | 17 | 14 | 0.653 |
|
G2/moderate | 8 | 8 |
|
|
G3/poor | 0 | 1 |
|
|
Unknown | 0 | 2 |
|
| Mode of tumor
invasion |
|
|
|
|
1–3 | 22 | 17 | 0.171 |
|
4C-4D | 3 | 8 |
|
| Depth of tumor |
|
|
|
| <5
mm | 24 | 22 | 0.609 |
| 5 mm ≤
and <10 mm | 1 | 3 |
|
| Lymphatic
invasion |
|
|
|
|
Yes | 4 | 2 | 0.667 |
| No | 21 | 22 |
|
|
Unknown | 0 | 1 |
|
| Venous
invasion |
|
|
|
|
Yes | 2 | 3 | 1.000 |
| No | 23 | 22 |
|
| Neural
infiltration |
|
|
|
|
Yes | 2 | 3 | 0.667 |
| No | 23 | 21 |
|
|
Unknown | 0 | 1 |
|
| Smoking |
|
|
|
| Current
or past | 9 | 10 | 1.000 |
|
Never | 16 | 15 |
|
| Alcohol intake |
|
|
|
| Current
or past | 11 | 11 | 1.000 |
|
Never | 13 | 12 |
|
|
Unknown | 1 | 2 |
|
Discussion
Our study illustrates two important clinical issues.
First, to the best of our knowledge, this is the first study to
examine LNM-related miR in early-stage TSCC and is the first miR
study of delayed LNM of H&N cancer. Second, miR-196a-5p was
significantly upregulated in early-stage TSCC. In addition,
miR-196a-5p upregulation increased the risk of delayed LNM and was
associated with a shorter LNMFS rate in early-stage TSCC.
We searched the literature from 1993 (72) to 2017 using PubMed and Google Scholar;
however, no reports on LNM-related miRs in early-stage TSCC or miR
studies on delayed LNM of H&N cancer were found. Identification
of a useful prognostic marker of LNM is an urgent issue in cancer
patients who are not only in the advanced stage but also in the
early stage.
We focused on delayed LNM for three reasons. First,
in general, early-stage OSCC patients have relatively good
prognosis when compared to that in advanced-stage patients
(73). Fundamentally, early-stage
tongue cancer is less likely to result in the distant metastasis
(14), which is consistent with our
finding; however, delayed LNM worsens the prognosis (16) because LNM is generally followed by
distant metastasis (14), which is
similar to the findings in our cases. In our study, all distant
metastasis cases occurred after delayed LNM. Second, the treatment
of early-stage OSCC is still controversial with regard to whether
elective neck dissections (ENDs) can be performed (11,74,75), even
after a report by D'cruz et al (76) that described the importance of ENDs in
early-stage OSCC to improve the survival rate. However, the current
guideline of The National Comprehensive Cancer Network (11) does not strongly define END as the
standard therapy for early-stage oral cancer. Finally, to evaluate
the treatment results without performing END, it is important to
clearly distinguish ‘delayed’ LNM from ‘simultaneous’ LNM (20).
miR can be reliable markers in daily cancer
treatment. Many markers of cancer recurrence or metastasis have
been reported (including those described in the new guidelines of
the Union for International Cancer Control (2)) and are in use currently. This tends to
make the clinician rather uncertain because the markers are too
many to use in daily practice. In a recent review, Yu et al
reported that no current clinical or pathological tools that are
useful for monitoring recurrence or metastasis in early-stage OSCC
patients (39). On the other hand,
Irani recently suggested that ‘the mode of tumor invasion’ is
important as a prognostic factor for LNM (77). Based on our results (Table I), we conclude that pathological
properties such as mode of tumor invasion, depth of tumor,
venous/lymphatic invasion, and neural infiltration can be rarely
observed in such early-stage cases (16,78).
Additionally, a relatively high rate of well-differentiated tumors
was found among our patients, which is not rare and is consistent
with a previous finding in a study by Yanamoto et al
(63). Only ‘the mode of tumor
invasion’ appeared to be a prognostic factor of delayed LNM
according to our results (Table
III, Fig. 3B). In contrast,
targeted miRs could be evaluated by RT-qPCR and were significantly
regulated in cancers from patients with delayed LNM when compared
to those from patients without LNM in our study. As mentioned
above, in the daily practice of OSCC treatment, a potential marker
to predict delayed LNM before or immediately after treatment is
strongly desired. miR is a useful approach that can easily detect
the presence of OSCC not only in fresh tissues but also in blood,
saliva, and FFPE tissues (27–30).
When selecting a miR, we suggest that it is
important to confirm how many oncogenes or tumor suppressor genes
with solid evidence are targeted by that miR. Three reasons for the
above suggestion have been presented. First, miRs are often
differentially expressed in different cancer conditions such as
cancer cells in vitro, tissues in vivo, and human
tissues. The functions of miRs are associated with their up- or
downregulation (39) and their
expression affect several target genes. Many miRs have been
reported to be expressed in OSCC (15,39);
however, controversially, one study reported that certain miRs were
significantly more upregulated in cancer tissues or cells than in
normal tissues or cells (39). In
contrast, it was reported that these miRs were downregulated in
other cancer tissues or cells (39).
For example, miR-148b is upregulated in TSCC cell lines (79) but downregulated in Syrian hamster OSCC
(80). Similarly, miR-197 was found
to be strongly upregulated in LMD of human TSCC (81) but downregulated in human TSCC cell
lines (79). Second, one particular
miR targets thousands of messenger RNAs (mRNAs); in contrast, one
mRNA can be targeted by hundreds of different miRs (31), which contiguously affects several
pathways of tumor processes through mRNA (30). Thus, complicated pathways of cancer
initiation or progression exist (25,82).
Moreover, cancer progression itself has a multistep process
associated with multiple alterations of oncogenes and tumor
suppressor genes involving miR functions (31). Thus, many miRs are reported as
candidates of potential clinical utility, including LNM with OSCC
(39,83–86).
Third, there are a plethora of miRs related to LNM in OSCC of human
patients, in vivo or in vitro, including
let-7d (77), let-7g
(87), miR-17 and miR-20a (88), miR-21 (89), miR-26b (90), miR-29b (91), miR-31 (92), miR-34a (93), miR-93 (94), miR-134 (95), miR-138 (84), miR-153 (96), miR-155-5p (97), miR-181 (98), miR-196a/b (52), miR-203 (99), miR-205 (83), miR-211 (100), miR-214-3p (10), miR-222 (101), miR-363 (102), miR-372 and miR-373 (85), miR-375 (10), miR-376c-3p (86), miR-491-5p (103), miR-1246 (104), miR-1275 (105). However, no reports have evaluated
only ‘delayed’ LNM of OSCC. In addition, no report has described
LNM of early-stage TSCC. Therefore, when choosing miR for a study,
we suggest that it is important to confirm how many oncogenes or
tumor suppressor genes with solid evidence are targeted by the miR.
Among so many miRs related to LNM, we focused on the following
HOX gene-related miRs (44–46,52):
miR-10a, miR-10b, miR-196a, miR-196b, and miR-615. Those miRs are
within the HOX gene clusters that are related to functions
of many types of H&N cancers (44–46).
HOX genes are cancer development genes and
tend to differ with regard to their up- or downregulation depending
on the cancer site or type (106).
HOX genes are suggested to be dysregulated in human OSCC
(107). In oral cancer cells or
tissues, many HOX genes are upregulated (45). HOX genes are reported as
oncogenes that regulate epithelial-mesenchymal transition, cancer
invasion, and apoptotic pathways (45). Upregulation of particular HOX
genes is associated with upregulation of miRs within each
HOX gene. The miRs: miR-10, miR-196, and miR-615 are present
within four types of HOX gene clusters: miR-10a and
miR-196a-1 are located in HOXB on chromosome 17, miR-10b is
located in HOXD on chromosome 2, miR-615 and miR-196a-2 are
located in HOXC on chromosome 12, and miR-196b is located in
HOXA on chromosome 7 (45,46,108).
miR-196a generally means miR-196a-5p. miR-196a-1 and miR-196a-2
have the same sequence; however, they are located on different
chromosomes (34). We selected
miR-196a-3p (miR passenger strand of miR-196a-2; not present in
miR-196a-1) because we attempted to compare the functions of
miR-196a-1 and miR-196a-2 (29,34).
miR-615 is not reported to be related to cancer (46); therefore, we excluded miR-615 from
examination as a prognostic marker because of the sparsity of
studies. miR-10a and miR-10b have not been reported to be related
to LNM of OSCC to date. However, miR-10 s have been shown to be
associated with cancer metastasis. miR-10a was reported to regulate
metastasis in various types of cancer (109) and to contribute to LNM of gastric
cancer (110), while miR-10b was
reported to be significantly upregulated in OSCC cell lines and to
promote cell migration and invasion (56). We eventually selected five miRs
(miR-10a, miR-10b, miR-196a-5p, miR-196a-3p, and miR-196b) as
candidate prognostic markers. In our study, the levels of all five
candidate miRs were significantly regulated in cancer tissues when
compared to ANTs. This confirmed their possible utility as markers
of early-stage TSCC. In our subsequent analyses, we found that
miR-196a-5p was a possible prognostic marker of delayed LNM in
early-stage TSCC.
In a recent study, significant miR-196a-5p
upregulation was observed in early-stage TSCC tissues. Furthermore,
miR-196a-5p upregulation increased the risk of delayed LNM and was
especially associated with a shorter LNMFS rate in early-stage
TSCC. Indeed, our study is not the first to report that miR-196a-5p
upregulation is associated with OSCC metastasis (29,52), and
we needed previous evidence to rationalize our study of
miR-196a-5p. Liu et al reported that miR-196a was
significantly upregulated in tumor tissues and plasma in ‘all
stages’ of OSCC (29), suggesting
that it might serve as a diagnostic and prognostic marker of LNM.
In contrast, we investigated only prognostic markers because
diagnostic markers are less important as OSCC is particularly easy
to detect clinically. We also limited the study to patients with
early-stage TSCC for four reasons. First, miR expression in cancer
tissue is specific to site or cancer type. miRs expressed in the
tongue and in other sites of the oral cavity differ (26). Furthermore, Harris et al
reported that the expression of most miRs was different in three
sites of the H&N region via microarray analysis of human SCC
tissue (111). Moreover, miRs
expressions of H&N SCC were different compared to those of
H&N ACC using microarray analysis (112). Second, miR expression can be
different between early-stage and advanced-stage cancer, and this
has been reported previously in microarray studies (58,89).
Despite being the same type of cancer, early-stage and
advanced-stage OSCCs showed significantly different miR-31
expression levels (92). Other
studies reported distinct expression modes in oral cancer and
premalignant disease (such as dysplasia and leukoplakia) (113,114).
The expressions of some genes are distinct between oral carcinoma
in situ and early-stage OSCC (115). Therefore, OSCC appears and
progresses by complicated multi-stage processes (39,116).
Thus, we suggest that early-stage and advanced-stage TSCC have
different miRs impacting and regulating pathways. Furthermore, we
excluded premalignant disease and carcinoma in situ in the
current study. Third, as mentioned above, early-stage TSCC is one
of the most common H&N cancers, and TSCC is the most aggressive
type of OSCC (21). Fourth, it is
relatively easy in early-stage OSCC to distinguish neck LNM from
local recurrence. In contrast, it is often difficult to classify
advanced cancer as one or another relapse, especially after neck
dissection (117). We speculate that
limiting our study to early-stage TSCC helped to effectively
evaluate only delayed LNM or local recurrence and tumor-specific
miRs.
miR-196a-5p targets several HOX genes
(35,118) and directly targets many other genes
(35). Lately, it has been reported
that miR-196-5p is aberrantly expressed in many kinds of cancers
and targets many genes, including several genes of the HOX
family, and functions, such as cancer proliferation, metastasis,
and invasion (34,46). miR-196a-5p is upregulated in OSCC
tissues and cells, and its inhibition of mRNA translation
contributes to many cancer processes (cell proliferation,
migration, and invasion), as an oncogene (29,35,52). A
certain HOX gene upregulation indicates miRs located in the
HOX gene. Therefore, cancer progression by HOX gene
upregulation as an oncogene is related to miR-196a-5p upregulation
in H&N SCC (45). Furthermore,
HOX genes are related to HOTAIR (119,120),
which exists upstream of miR-196a-2 and has been reported to be
highly regulated in OSCC and significantly upregulated in cancers
with LNM when compared to the regulation in cancers without
metastasis (35,121). As an oncogene of H&N cancer and
oral cancer, miR-196-5p targets many genes, as described above
(34,35). Annexin A1 is targeted by
miR-196a-5p over-regulation and is related to metastasis as well as
proliferation, invasion, and radio resistance of H&N cancer
(53). NME4 was directly
inhibited by miR-196a-5p with significant upregulation in cancer
tissues and was correlated with neck LNM of oral cancer (52). MAMDC2 has been reported as a
novel direct target of miR-196a-5p in H&N SCC (46). All the above-mentioned genes are
regulated and caused LNM by upregulation of miR-196a-5p, which is
similar to our results. Based on the results of our study,
miR-196a-5p can be a potential prognostic marker of delayed LNM. In
addition, future studies are required to confirm whether miR-196-5p
can be used as a therapeutic marker for OSCC metastasis, as
reported by Chen et al (35).
The use of a biomarker for diagnosis or prognosis
of cancer is a general concept (3).
Several molecular markers (e.g., protein, mRNA, and miR) have been
studied by using tissue, blood, and saliva (113). However, a reliable biomarker that
can detect cancer early, provide a more accurate diagnosis, predict
prognosis, and allow the patient to receive the best treatment is
strongly desired (113). As
described above, for OSCC patients, it is critically more important
to identify prognostic markers than diagnostic markers. To
establish a miR as a biomarker with a highly reliable assay system
for routine clinical purposes, four conditions are needed. One, the
examination of the biomarker should be easy and the method should
be reproducible for clinical use (122). Two, many examination cases are
needed (10,54). Three, noninvasive methods should be
preferred (123). Four, tissues
should be accurately collected (122). Moreover, as described above, we
should pay attention to the characteristics of the miRs. The
biomarker should be specific for early-stage TSCC and not for other
H&N cancers because miR expression is stage- and site-specific,
as well as cancer-specific (26,92,111,112).
In the current study, the tissues of patients were accurately
collected, and there were epithelial tumor tissues and epithelial
ANTs collected by LMD (81). The FFPE
examination was noninvasive, and it is relatively easy to increase
the number of cases than other type of samples. On the other hand,
fresh tissue was collected with an invasive (scalpel) method and,
sometimes, epithelial tissues were collected along with stroma
tissues, making the examination results uncertain (122). In the future, we plan to confirm the
current results in a prospective study. Recently, Adami et
al reported the advantages of brush biopsy (noninvasive) of
oral cancer (122). This method
allows easy assessment of the difference in OSCC tissue and
epithelial ANT for pretreatment timing (124), and accumulation of cases is strongly
expected in the future (123).
ROC analysis is a standard approach to identify a
detection cut-off for a disease biomarker (125). In ROC analysis, the AUC can be
calculated to evaluate the power of an assay system to accurately
distinguish between true and false results for diseases, especially
cancer (125,126). As used by many researchers, the AUC
scale was defined by Hosmer et al (127): if AUC=0.5, no discrimination; if
0.5<AUC<0.7, poor discrimination; if 0.7≤AUC<0.8,
acceptable discrimination; if 0.8≤AUC<0.9, excellent
discrimination; and if AUC≥0.9, discrimination is considered. Based
on our results, miR-196-5p can be a potential prognostic marker. In
our ROC analysis, our results were statistically significant
according to the findings of previous studies on biomarkers
(126,128,129)
(P<0.05 and AUC>0.7).
miR is a useful marker that can easily detect the
expression of OSCC not only in fresh tissues but also blood,
saliva, and FFPE tissues (27–30). Among
these tissues, FFPE tissues are useful to evaluate miR expressions
(57,58,130).
Very old FFPE tissues, such as those obtained 10–19 years
previously, have been used in miR study (28,131,132).
Therefore, we could use old tissue for evaluation, with the oldest
sample being 12 years old. Certainly, our total RNA amount and
density were low by using BioSpec-nano; however, in all patients,
the Cq values of RNU44 and RNU48 could be determined (Cq
values=23.4–36.58) for use. mRNA in FFPE tissues is difficult to
examine using the above method because formalin fixation reduces
the recovery and quality of RNA (131). In contrast, miR in FFPE tissues can
be examined because miRs are small and are protected by the RISC
complex (133). miRs in FFPE tissues
are reportedly robust and well-regulated in RT-qPCR and frozen
tissue sample (131). FFPE tissues
are valuable for conducting retrospective studies of human cancer
(131). Therefore, many miR studies
using FFPE can be accumulated in the future.
The present study has some limitations. First, the
results of the present study might be limited because of the
retrospective study design and inclusion of only 50 patients.
Additionally, fresh tissues were not analyzed. However, the number
of patients appeared to be sufficient according to previous studies
(83–85,134,135).
Further studies that assess fresh cancer tissues and include a
higher number of patients should be performed. Second, the study
was biased as it investigated only miR-10a, miR-10b, miR-196a-5p,
miR-196a-3p, and miR-196b. Third, differential expression did not
allow identification of regulatory mechanisms, particularly when
the activities/expressions of the targets/pathway output were not
investigated. We suggest that one miR repressed many target genes;
thus, studies are needed to evaluate miR with solid evidence from
past studies rather than those involving unknown new target genes
and pathways. In contrast, the main strengths of this study are
that it provides data to indicate miR-196a as an acceptable marker
and that it can be considered as a pilot study for similar studies
in the future. FFPE is considered useful to evaluate the expression
levels of miRs.
In conclusion, to the best of our knowledge, this
is the first study to examine LNM-related miR in early-stage TSCC
and also the first miR study of ‘delayed’ LNM of H&N cancer. In
addition, our findings revealed that miR-196a-5p is a potential new
biomarker for the prognosis of TSCC and our results serve as a
foundation for further studies (not only fresh tissues and FFPE but
also blood and saliva of preoperative and postoperative samples) to
evaluate the utility of this miR in the prediction of delayed LNM
during the treatment of patients with early-stage TSCC.
Furthermore, the results enhanced efforts to prevent metastasis
when combined with close follow-up and aggressive adjuvant therapy
or END. Moreover, our data will serve as a foundation for future
studies to evaluate whether miR-196a-5p can serve as a therapeutic
marker for preventing metastasis.
Acknowledgements
The authors would like to thank Reika Takamatsu and
Toshiyuki Nishihira (Department of Pathology and Oncology, Graduate
School of Medicine, University of the Ryukyus) for the technical
support.
Glossary
Abbreviations
Abbreviations:
|
miR
|
microRNA
|
|
TSCC
|
tongue squamous cell carcinoma
|
|
Cq
|
quantification cycle
|
|
LNM
|
lymph node metastasis
|
|
H&N
|
head and neck
|
|
OSCC
|
oral squamous cell carcinoma
|
|
FFPE
|
formalin-fixed paraffin-embedded
|
|
HOX
|
homeobox
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
|
ANT
|
adjacent normal tissue
|
|
OS
|
overall survival
|
|
LRFS
|
local recurrence-free survival
|
|
LNMFS
|
lymph node metastasis-free
survival
|
|
DMFS
|
distant metastasis-free survival
|
|
DFS
|
disease-free survival
|
|
LMD
|
laser-capture microdissection
|
|
RT
|
reverse transcription
|
|
ROC
|
receiver operating characteristic
|
|
AUC
|
area under the curve
|
|
END
|
elective neck dissection
|
|
mRNA
|
messenger RNA
|
References
|
1
|
IARC: GLOBOCAN 2012: Estimated Cancer
Incidence, Mortality and Prevalence Worldwide in 2012. http://globocan.iarc.fr/Pages/fact_sheets_population.aspxFebruary
22–2017
|
|
2
|
Brierley JD, Gospodarowicz M and Wittekind
C: Lip and Oral CavityTNM Classification of Malignant Tumours. 8th
edition. Wiley Blackwell; New York, NY: pp. 18–21. 2016
|
|
3
|
Sinevici N and O'sullivan J: Oral cancer:
Deregulated molecular events and their use as biomarkers. Oral
Oncol. 61:12–18. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 660:7–30. 2016. View Article : Google Scholar
|
|
5
|
Jemal A, Murray T, Ward E, Samuels A,
Tiwari RC, Ghafoor A, Feuer EJ and Thun MJ: Cancer statistics,
2005. CA Cancer J Clin. 55:2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Parker SL, Tong T, Bolden S and Wingo PA:
Cancer statistics, 1996. CA Cancer J Clin. 46:5–27. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li Z, Wang Y, Qiu J, Li Q, Yuan C, Zhang
W, Wang D, Ye J, Jiang H, Yang J and Cheng J: The polycomb group
protein EZH2 is a novel therapeutic target in tongue cancer.
Oncotarget. 4:2532–2549. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang C, Huang H, Huang Z, Wang A, Chen X,
Huang L, Zhou X and Liu X: Tumor budding correlates with poor
prognosis and epithelial-mesenchymal transition in tongue squamous
cell carcinoma. J Oral Pathol Med. 40:545–551. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Schwam ZG and Judson BL: Improved
prognosis for patients with oral cavity squamous cell carcinoma:
Analysis of the national cancer database 1998–2006. Oral Oncol.
52:45–51. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yoon AJ, Wang S, Shen J, Robine N,
Philipone E, Oster MW, Nam A and Santella RM: Prognostic value of
miR-375 and miR-214-3p in early stage oral squamous cell carcinoma.
Am J Transl Res. 6:580–592. 2014.PubMed/NCBI
|
|
11
|
National ComprehensiveCancer Network, .
NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines):
Head and Neck Cancers 2017. https://www.nccn.org/professionals/physician_gls/pdf/head-and-neck.pdfFebruary
24–2017
|
|
12
|
Rogers SN, Brown JS, Woolgar JA, Lowe D,
Magennis P, Shaw RJ, Sutton D, Errington D and Vaughan D: Survival
following primary surgery for oral cancer. Oral Oncol. 45:201–211.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kelner N, Vartanian JG, Pinto CA,
Coutinho-Camillo CM and Kowalski LP: Does elective neck dissection
in T1/T2 carcinoma of the oral tongue and floor of the mouth
influence recurrence and survival rates? Br J Oral Maxillofac Surg.
52:590–597. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ong HS, Gokavarapu S, Wang LZ, Tian Z and
Zhang CP: Low pretreatment lymphocyte-monocyte ratio and high
platelet-lymphocyte ratio indicate poor cancer outcome in early
tongue cancer. J Oral Maxillofac Surg. 75:1762–1774. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yu X and Li Z: MicroRNA expression and its
implications for diagnosis and therapy of tongue squamous cell
carcinoma. J Cell Mol Med. 20:10–16. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Seki M, Sano T, Yokoo S and Oyama T:
Tumour budding evaluated in biopsy specimens is a useful predictor
of prognosis in patients with cN0 early stage oral squamous cell
carcinoma. Histopathology. 70:869–879. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ariji Y, Goto M, Fukano H, Sugita Y, Izumi
M and Ariji E: Role of intraoral color Doppler sonography in
predicting delayed cervical lymph node metastasis in patients with
early-stage tongue cancer: A pilot study. Oral Surg Oral Med Oral
Pathol Oral Radiol. 119:246–253. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Habu N, Imanishi Y, Kameyama K, et al:
Expression of Oct3/4 and Nanog in the head and neck squamous
carcinoma cells and its clinical implications for delayed neck
metastasis in stage I/II oral tongue squamous cell carcinoma. BMC
Cancer. 15:7302015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Goto M, Hanai N, Ozawa T, Hirakawa H,
Suzuki H, Hyodo I, Kodaira T, Ogawa T, Fujimoto Y, Terada A, et al:
Prognostic factors and outcomes for salvage surgery in patients
with recurrent squamous cell carcinoma of the tongue. Asia Pac J
Clin Oncol. 12:e141–e148. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Luksic I and Suton P: Predictive markers
for delayed lymph node metastases and survival in early-stage oral
squamous cell carcinoma. Head Neck. 39:694–701. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Almangush A, Coletta RD, Bello IO, Bitu C,
Keski-Säntti H, Mäkinen LK, Kauppila JH, Pukkila M, Hagström J,
Laranne J, et al: A simple novel prognostic model for early stage
oral tongue cancer. Int J Oral Maxillofac Surg. 44:143–150. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Montebugnoli L, Gissi DB, Flamminio F,
Gentile L, Dallera V, Leonardi E, Beccarini T and Foschini MP:
Clinicopathologic parameters related to recurrence and locoregional
metastasis in 180 oral squamous cell carcinomas. Int J Surg Pathol.
22:55–62. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Dunkel J, Vaittinen S, Grénman R, Kinnunen
I and Irjala H: Prognostic markers in stage I oral cavity squamous
cell carcinoma. Laryngoscope. 123:2435–2441. 2013.PubMed/NCBI
|
|
24
|
Harada Y, Izumi H, Noguchi H, Kuma A,
Kawatsu Y, Kimura T, Kitada S, Uramoto H, Wang KY, Sasaguri Y, et
al: Erratum to: Strong expression of polypeptide
N-acetylgalactosaminyltransferase 3 independently predicts
shortened disease-free survival in patients with early stage oral
squamous cell carcinoma. Tumour Biol. 36:10003–10004. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Villegas-Ruiz V, Juárez-Méndez S,
Pérez-González OA, Arreola H, Paniagua-García L, Parra-Melquiadez
M, Peralta-Rodríguez R, López-Romero R, Monroy-García A,
Mantilla-Morales A, et al: Heterogeneity of microRNAs expression in
cervical cancer cells: Over-expression of miR-196a. Int J Clin Exp
Pathol. 7:1389–1401. 2014.PubMed/NCBI
|
|
26
|
Boldrup L, Coates PJ, Laurell G, Wilms T,
Fahraeus R and Nylander K: Downregulation of miRNA-424: A sign of
field cancerisation in clinically normal tongue adjacent to
squamous cell carcinoma. Br J Cancer. 112:1760–1765. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu X, Wang A, Heidbreder CE, Jiang L, Yu
J, Kolokythas A, Huang L, Dai Y and Zhou X: MicroRNA-24 targeting
RNA-binding protein DND1 in tongue squamous cell carcinoma. FEBS
Lett. 584:4115–4120. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Rosenfeld N, Aharonov R, Meiri E,
Rosenwald S, Spector Y, Zepeniuk M, Benjamin H, Shabes N, Tabak S,
Levy A, et al: MicroRNAs accurately identify cancer tissue origin.
Nat Biotechnol. 26:462–469. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu CJ, Tsai MM, Tu HF, Lui MT, Cheng HW
and Lin SC: miR-196a overexpression and miR-196a2 gene polymorphism
are prognostic predictors of oral carcinomas. Ann Surg Oncol.
Suppl. 3 20 Suppl:S406–S414. 2013.
|
|
30
|
Liu CJ, Lin SC, Yang CC, Cheng HW and
Chang KW: Exploiting salivary miR-31 as a clinical biomarker of
oral squamous cell carcinoma. Head Neck. 34:219–224. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Di Leva G, Garofalo M and Croce CM:
MicroRNAs in cancer. Annu Rev Pathol. 9:287–314. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Reddy KB: MicroRNA (miRNA) in cancer.
Cancer Cell Int. 15:382015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lin S and Gregory RI: MicroRNA biogenesis
pathways in cancer. Nat Rev Cancer. 15:321–333. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lu YC, Chang JT, Chan EC, Chao YK, Yeh TS,
Chen JS and Cheng AJ: miR-196, an emerging cancer biomarker for
digestive tract cancers. J Cancer. 7:650–655. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chen ZY, Chen X and Wang ZX: The role of
microRNA-196a in tumorigenesis, tumor progression, and prognosis.
Tumour Biol. Oct 18–2016.(Epub ahead of print). View Article : Google Scholar
|
|
36
|
Friedman RC, Farh KK, Burge CB and Bartel
DP: Most mammalian mRNAs are conserved targets of microRNAs. Genome
Res. 19:92–105. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Nohata N, Hanazawa T, Kinoshita T, Okamoto
Y and Seki N: MicroRNAs function as tumor suppressors or oncogenes:
Aberrant expression of microRNAs in head and neck squamous cell
carcinoma. Auris Nasus Larynx. 40:143–149. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lamouille S, Subramanyam D, Blelloch R and
Derynck R: Regulation of epithelial-mesenchymal and
mesenchymal-epithelial transitions by microRNAs. Curr Opin Cell
Biol. 25:200–207. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yu T, Li C, Wang Z, Liu K, Xu C, Yang Q,
Tang Y and Wu Y: Non-coding RNAs deregulation in oral squamous cell
carcinoma: Advances and challenges. Clin Transl Oncol. 18:427–436.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Tu HF, Lin SC and Chang KW: MicroRNA
aberrances in head and neck cancer: Pathogenetic and clinical
significance. Curr Opin Otolaryngol Head Neck Surg. 21:104–111.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang H, Li Y and Lai M: The microRNA
network and tumor metastasis. Oncogene. 29:937–948. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Bracken CP, Gregory PA, Khew-Goodall Y and
Goodall GJ: The role of microRNAs in metastasis and
epithelial-mesenchymal transition. Cell Mol Life Sci. 66:1682–1699.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kim KY, Lee GY and Cha IH: Biomarker
detection for the diagnosis of lymph node metastasis from oral
squamous cell carcinoma. Oral Oncol. 48:311–319. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Severino P, Brüggemann H, Andreghetto FM,
Camps C, Mde F Klingbeil, de Pereira WO, Soares RM, Moyses R,
Wünsch-Filho V, Mathor MB, et al: MicroRNA expression profile in
head and neck cancer: HOX-cluster embedded microRNA-196a and
microRNA-10b dysregulation implicated in cell proliferation. BMC
Cancer. 13:5332013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Platais C, Hakami F, Darda L, Lambert DW,
Morgan R and Hunter KD: The role of HOX genes in head and neck
squamous cell carcinoma. J Oral Pathol Med. 45:239–247. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Darda L, Hakami F, Morgan R, Murdoch C,
Lambert DW and Hunter KD: The role of HOXB9 and miR-196a in head
and neck squamous cell carcinoma. PLoS One. 10:e01222852015.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Jiang Y, Zhang Y, Li F, Du X and Zhang J:
CDX2 inhibits pancreatic adenocarcinoma cell proliferation via
promoting tumor suppressor miR-615-5p. Tumour Biol. 37:1041–1049.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Pearson CE: Co-opting endogenous microRNAs
for therapy. Nat Med. 18:1011–1012. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Hoss AG, Kartha VK, Dong X, Latourelle JC,
Dumitriu A, Hadzi TC, Macdonald ME, Gusella JF, Akbarian S, Chen
JF, et al: MicroRNAs located in the Hox gene clusters are
implicated in huntington's disease pathogenesis. PLoS Genet.
10:e10041882014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Shan Q, Zheng G, Zhu A, Cao L, Lu J, Wu D,
Zhang Z, Fan S, Sun C, Hu B, et al: Epigenetic modification of
miR-10a regulates renal damage by targeting CREB1 in type 2
diabetes mellitus. Toxicol Appl Pharmacol. 306:134–143. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Bourguignon LY, Wong G and Shiina M:
Up-regulation of histone methyltransferase, DOT1L, by matrix
hyaluronan promotes MicroRNA-10 expression leading to tumor cell
invasion and chemoresistance in cancer stem cells from head and
neck squamous cell carcinoma. J Biol Chem. 291:10571–10585. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Lu YC, Chang JT, Liao CT, Kang CJ, Huang
SF, Chen IH, Huang CC, Huang YC, Chen WH, Tsai CY, et al:
OncomiR-196 promotes an invasive phenotype in oral cancer through
the NME4-JNK-TIMP1-MMP signaling pathway. Mol Cancer. 13:2182014.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Suh YE, Raulf N, Gäken J, Lawler K, Urbano
TG, Bullenkamp J, Gobeil S, Huot J, Odell E and Tavassoli M:
MicroRNA-196a promotes an oncogenic effect in head and neck cancer
cells by suppressing annexin A1 and enhancing radioresistance. Int
J Cancer. 137:1021–1034. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Saito K, Inagaki K, Kamimoto T, Ito Y,
Sugita T, Nakajo S, Hirasawa A, Iwamaru A, Ishikura T, Hanaoka H,
et al: MicroRNA-196a is a putative diagnostic biomarker and
therapeutic target for laryngeal cancer. PLoS One. 8:e714802013.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Kozaki K, Imoto I, Mogi S, Omura K and
Inazawa J: Exploration of tumor-suppressive microRNAs silenced by
DNA hypermethylation in oral cancer. Cancer Res. 68:2094–2105.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Lu YC, Chen YJ, Wang HM, Tsai CY, Chen WH,
Huang YC, Fan KH, Tsai CN, Huang SF, Kang CJ, et al: Oncogenic
function and early detection potential of miRNA-10b in oral cancer
as identified by microRNA profiling. Cancer Prev Res (Phila).
5:665–674. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Ganci F, Sacconi A, Manciocco V, Sperduti
I, Battaglia P, Covello R, Muti P, Strano S, Spriano G, Fontemaggi
G, et al: MicroRNA expression as predictor of local recurrence risk
in oral squamous cell carcinoma. Head Neck. 38:E189–E197. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Sasahira T, Kurihara M, Bhawal UK, Ueda N,
Shimomoto T, Yamamoto K, Kirita T and Kuniyasu H: Downregulation of
miR-126 induces angiogenesis and lymphangiogenesis by activation of
VEGF-A in oral cancer. Br J Cancer. 107:700–706. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Zhang X, Yang H, Lee JJ, Kim E, Lippman
SM, Khuri FR, Spitz MR, Lotan R, Hong WK and Wu X: MicroRNA-related
genetic variations as predictors for risk of second primary tumor
and/or recurrence in patients with early-stage head and neck
cancer. Carcinogenesis. 31:2118–2123. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Gombos K, Horváth R, Szele E, Juhász K,
Gocze K, Somlai K, Pajkos G, Ember I and Olasz L: miRNA expression
profiles of oral squamous cell carcinomas. Anticancer Res.
33:1511–1517. 2013.PubMed/NCBI
|
|
61
|
Kina S, Nakasone T, Kinjo T, Maruyama T,
Kawano T and Arasaki A: Impact of metronomic neoadjuvant
chemotherapy on early tongue cancer. Cancer Chemother Pharmacol.
78:833–840. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Peisker A, Raschke GF, Guentsch A, Luepke
P, Roshanghias K and Schultze-Mosgau S: Evaluation of a
post-treatment follow-up program in patients with oral squamous
cell carcinoma. Clin Oral Investig. 21:135–141. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Yanamoto S, Yamada S, Takahashi H,
Kawasaki G, Ikeda H, Shiraishi T, Fujita S, Ikeda T, Asahina I and
Umeda M: Predictors of locoregional recurrence in T1-2N0 tongue
cancer patients. Pathol Oncol Res. 19:795–803. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Johnson N, Franceschi S, Ferlay J, Ramadas
K, Schmid S, MacDonald DG, Bouquot JE and Slootweg PJ: Squamous
cell carcinoma. In: World Health Organization Classification of
TumoursPathology & Genetics Head and Neck Tumours. Barnes C,
Eveson JW, Reichart P and Sidransky D: IARC Press; Lyon: pp.
168–175. 2005
|
|
65
|
Yamamoto E, Kohama G, Sunakawa H, Iwai M
and Hiratsuka H: Mode of invasion, bleomycin sensitivity and
clinical course in squamous cell carcinoma of the oral cavity.
Cancer. 51:2175–2180. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Huang SH, Hwang D, Lockwood G, Goldstein
DP and O'Sullivan B: Predictive value of tumor thickness for
cervical lymph-node involvement in squamous cell carcinoma of the
oral cavity: A meta-analysis of reported studies. Cancer.
115:1489–1497. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Zeka F, Vanderheyden K, De Smet E,
Cuvelier CA, Mestdagh P and Vandesompele J: Straightforward and
sensitive RT-qPCR based gene expression analysis of FFPE samples.
Sci Rep. 6:214182016. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Nagao Y, Hisaoka M, Matsuyama A, Kanemitsu
S, Hamada T, Fukuyama T, Nakano R, Uchiyama A, Kawamoto M,
Yamaguchi K, et al: Association of microRNA-21 expression with its
targets, PDCD4 and TIMP3, in pancreatic ductal adenocarcinoma. Mod
Pathol. 25:112–121. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Fanale D, Amodeo V, Bazan V, Insalaco L,
Incorvaia L, Barraco N, Castiglia M, Rizzo S, Santini D, Giordano
A, et al: Can the microRNA expression profile help to identify
novel targets for zoledronic acid in breast cancer? Oncotarget.
7:29321–29332. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
McCall MN, McMurray HR, Land H and
Almudevar A: On non-detects in qPCR data. Bioinformatics.
30:2310–2316. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Lee RC, Feinbaum RL and Ambros V: The C.
elegans heterochronic gene lin-4 encodes small RNAs with antisense
complementarity to lin-14. Cell. 75:843–854. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Scott SE, Grunfeld EA and McGurk M: The
idiosyncratic relationship between diagnostic delay and stage of
oral squamous cell carcinoma. Oral Oncol. 41:396–403. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Mitani S, Tomioka T, Hayashi R, Ugumori T,
Hato N and Fujii S: Anatomic invasive depth predicts delayed
cervical lymph node metastasis of tongue squamous cell carcinoma.
Am J Surg Pathol. 40:934–942. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Orabona GD, Bonavolontà P, Maglitto F,
Friscia M, Iaconetta G and Califano L: Neck dissection versus
‘watchful-waiting’ in early squamous cell carcinoma of the tongue
our experience on 127 cases. Surg Oncol. 25:401–404. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
D'Cruz AK, Vaish R, Kapre N, Dandekar M,
Gupta S, Hawaldar R, Agarwal JP, Pantvaidya G, Chaukar D, Deshmukh
A, et al: Elective versus therapeutic neck dissection in
node-negative oral cancer. N Engl J Med. 373:521–529. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Irani S: miRNAs signature in head and neck
squamous cell carcinoma metastasis: A literature review. J Dent
(Shiraz). 17:71–83. 2016.PubMed/NCBI
|
|
78
|
Ganly I, Goldstein D, Carlson DL, Patel
SG, O'Sullivan B, Lee N, Gullane P and Shah JP: Long-term regional
control and survival in patients with ‘low-risk,’ early stage oral
tongue cancer managed by partial glossectomy and neck dissection
without postoperative radiation: The importance of tumor thickness.
Cancer. 119:1168–1176. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Hebert C, Norris K, Scheper MA, Nikitakis
N and Sauk JJ: High mobility group A2 is a target for miRNA-98 in
head and neck squamous cell carcinoma. Mol Cancer. 6:52007.
View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Yu T, Wang XY, Gong RG, Li A, Yang S, Cao
YT, Wen YM, Wang CM and Yi XZ: The expression profile of microRNAs
in a model of 7,12-dimethyl-benz [a]anthrance-induced oral
carcinogenesis in Syrian hamster. J Exp Clin Cancer Res. 28:642009.
View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Wong TS, Liu XB, Chung-Wai Ho A, Po-Wing
Yuen A, Wai-Man Ng R and Ignace Wei W: Identification of pyruvate
kinase type M2 as potential oncoprotein in squamous cell carcinoma
of tongue through microRNA profiling. Int J Cancer. 123:251–257.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Grizzi F, Di Ieva A, Russo C, Frezza EE,
Cobos E, Muzzio PC and Chiriva-Internati M: Cancer initiation and
progression: An unsimplifiable complexity. Theor Biol Med Model.
3:372006. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Brito BL, Lourenço SV, Damascena AS,
Kowalski LP, Soares FA and Coutinho-Camillo CM: Expression of stem
cell-regulating miRNAs in oral cavity and oropharynx squamous cell
carcinoma. J Oral Pathol Med. 45:647–654. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Manikandan M, Magendhra Deva Rao AK,
Rajkumar KS, Rajaraman R and Munirajan AK: Altered levels of
miR-21, miR-125b-2*, miR-138, miR-155, miR-184 and miR-205 in oral
squamous cell carcinoma and association with clinicopathological
characteristics. J Oral Pathol Med. 44:792–800. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Tu HF, Chang KW, Cheng HW and Liu CJ:
Upregulation of miR-372 and −373 associates with lymph node
metastasis and poor prognosis of oral carcinomas. Laryngoscope.
125:E365–E370. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Chang WM, Lin YF, Su CY, Peng HY, Chang
YC, Lai TC, Wu GH, Hsu YM, Chi LH, Hsiao JR, et al: Dysregulation
of RUNX2/activin-A axis upon miR-376c downregulation promotes lymph
node metastasis in head and neck squamous cell carcinoma. Cancer
Res. 76:7140–7150. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Peng SC, Liao CT, Peng CH, Cheng AJ, Chen
SJ, Huang CG, Hsieh WP and Yen TC: MicroRNAs MiR-218, MiR-125b, and
Let-7g predict prognosis in patients with oral cavity squamous cell
carcinoma. PLoS One. 9:e1024032014. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Chang CC, Yang YJ, Li YJ, Chen ST, Lin BR,
Wu TS, Lin SK, Kuo MY and Tan CT: MicroRNA-17/20a functions to
inhibit cell migration and can be used a prognostic marker in oral
squamous cell carcinoma. Oral Oncol. 49:923–931. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Li J, Huang H, Sun L, Yang M, Pan C, Chen
W, Wu D, Lin Z, Zeng C, Yao Y, et al: MiR-21 indicates poor
prognosis in tongue squamous cell carcinomas as an apoptosis
inhibitor. Clin Cancer Res. 15:3998–4008. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Cao J, Guo T, Dong Q, Zhang J and Li Y:
miR-26b is downregulated in human tongue squamous cell carcinoma
and regulates cell proliferation and metastasis through a
COX-2-dependent mechanism. Oncol Rep. 33:974–980. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Yang CN, Deng YT, Tang JY, Cheng SJ, Chen
ST, Li YJ, Wu TS, Yang MH, Lin BR, Kuo MY, et al: MicroRNA-29b
regulates migration in oral squamous cell carcinoma and its
clinical significance. Oral Oncol. 51:170–177. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Siow MY, Ng LP, Vincent-Chong VK,
Jamaludin M, Abraham MT, Rahman Abdul ZA, Kallarakkal TG, Yang YH,
Cheong SC and Zain RB: Dysregulation of miR-31 and miR-375
expression is associated with clinical outcomes in oral carcinoma.
Oral Dis. 20:345–351. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Jia LF, Wei SB, Mitchelson K, Gao Y, Zheng
YF, Meng Z, Gan YH and Yu GY: miR-34a inhibits migration and
invasion of tongue squamous cell carcinoma via targeting MMP9 and
MMP14. PLoS One. 9:e1084352014. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Li G, Ren S, Su Z, Liu C, Deng T, Huang D,
Tian Y, Qiu Y and Liu Y: Increased expression of miR-93 is
associated with poor prognosis in head and neck squamous cell
carcinoma. Tumour Biol. 36:3949–3956. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Liu CJ, Shen WG, Peng SY, Cheng HW, Kao
SY, Lin SC and Chang KW: miR-134 induces oncogenicity and
metastasis in head and neck carcinoma through targeting WWOX gene.
Int J Cancer. 134:811–821. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Xu Q, Sun Q, Zhang J, Yu J, Chen W and
Zhang Z: Downregulation of miR-153 contributes to
epithelial-mesenchymal transition and tumor metastasis in human
epithelial cancer. Carcinogenesis. 34:539–549. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Baba O, Hasegawa S, Nagai H, Uchida F,
Yamatoji M, Kanno NI, Yamagata K, Sakai S, Yanagawa T and Bukawa H:
MicroRNA-155-5p is associated with oral squamous cell carcinoma
metastasis and poor prognosis. J Oral Pathol Med. 45:248–255. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Yang CC, Hung PS, Wang PW, Liu CJ, Chu TH,
Cheng HW and Lin SC: miR-181 as a putative biomarker for lymph-node
metastasis of oral squamous cell carcinoma. J Oral Pathol Med.
40:397–404. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Obayashi M, Yoshida M, Tsunematsu T, Ogawa
I, Sasahira T, Kuniyasu H, Imoto I, Abiko Y, Xu D, Fukunaga S, et
al: microRNA-203 suppresses invasion and epithelial-mesenchymal
transition induction via targeting NUAK1 in head and neck cancer.
Oncotarget. 7:8223–8239. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Chang KW, Liu CJ, Chu TH, Cheng HW, Hung
PS, Hu WY and Lin SC: Association between high miR-211 microRNA
expression and the poor prognosis of oral carcinoma. J Dent Res.
87:1063–1068. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Liu X, Yu J, Jiang L, Wang A, Shi F, Ye H
and Zhou X: MicroRNA-222 regulates cell invasion by targeting
matrix metalloproteinase 1 (MMP1) and manganese superoxide
dismutase 2 (SOD2) in tongue squamous cell carcinoma cell lines.
Cancer Genomics Proteomics. 6:131–139. 2009.PubMed/NCBI
|
|
102
|
Sun Q, Zhang J, Cao W, Wang X, Xu Q, Yan
M, Wu X and Chen W: Dysregulated miR-363 affects head and neck
cancer invasion and metastasis by targeting podoplanin. Int J
Biochem Cell Biol. 45:513–520. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Huang WC, Chan SH, Jang TH, Chang JW, Ko
YC, Yen TC, Chiang SL, Chiang WF, Shieh TY, Liao CT, et al:
miRNA-491-5p and GIT1 serve as modulators and biomarkers for oral
squamous cell carcinoma invasion and metastasis. Cancer Res.
74:751–764. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Liao L, Wang J, Ouyang S, Zhang P, Wang J
and Zhang M: Expression and clinical significance of microRNA-1246
in human oral squamous cell carcinoma. Med Sci Monit. 21:776–781.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Manikandan M, Magendhra Deva Rao AK,
Arunkumar G, Manickavasagam M, Rajkumar KS, Rajaraman R and
Munirajan AK: Oral squamous cell carcinoma: microRNA expression
profiling and integrative analyses for elucidation of
tumourigenesis mechanism. Mol Cancer. 15:282016. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Bhatlekar S, Fields JZ and Boman BM: HOX
genes and their role in the development of human cancers. J Mol Med
(Berl). 92:811–823. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Marcinkiewicz KM and Gudas LJ: Altered
epigenetic regulation of homeobox genes in human oral squamous cell
carcinoma cells. Exp Cell Res. 320:128–143. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Chen C, Zhang Y, Zhang L, Weakley SM and
Yao Q: MicroRNA-196: Critical roles and clinical applications in
development and cancer. J Cell Mol Med. 15:14–23. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Liu Y, Zhang Y, Wu H, Li Y, Zhang Y, Liu
M, Li X and Tang H: miR-10a suppresses colorectal cancer metastasis
by modulating the epithelial-to-mesenchymal transition and anoikis.
Cell Death Dis. 8:e27392017. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Chen W, Tang Z, Sun Y, Zhang Y, Wang X,
Shen Z, Liu F and Qin X: miRNA expression profile in primary
gastric cancers and paired lymph node metastases indicates that
miR-10a plays a role in metastasis from primary gastric cancer to
lymph nodes. Exp Ther Med. 3:351–356. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Harris T, Jimenez L, Kawachi N, Fan JB,
Chen J, Belbin T, Ramnauth A, Loudig O, Keller CE, Smith R, et al:
Low-level expression of miR-375 correlates with poor outcome and
metastasis while altering the invasive properties of head and neck
squamous cell carcinomas. Am J Pathol. 180:917–928. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Veit JA, Scheckenbach K, Schuler PJ, Laban
S, Wiggenhauser PS, Thierauf J, Klussmann JP and Hoffmann TK:
MicroRNA expression in differentially metastasizing tumors of the
head and neck: Adenoid cystic versus squamous cell carcinoma.
Anticancer Res. 35:1271–1277. 2015.PubMed/NCBI
|
|
113
|
Min A, Zhu C, Peng S, Rajthala S, Costea
DE and Sapkota D: MicroRNAs as important players and biomarkers in
oral carcinogenesis. Biomed Res Int. 2015:1869042015. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Zahran F, Ghalwash D, Shaker O, Al-Johani
K and Scully C: Salivary microRNAs in oral cancer. Oral Dis.
21:739–747. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Johnson JJ, Miller DL, Jiang R, et al:
Protease-activated Receptor-2 (PAR-2)-mediated Nf-κB activation
suppresses inflammation-associated tumor Suppressor MicroRNAs in
oral squamous cell carcinoma. J Biol Chem. 291:6936–6945. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Mascolo M, Siano M, Ilardi G, Russo D,
Merolla F, De Rosa G and Staibano S: Epigenetic disregulation in
oral cancer. Int J Mol Sci. 13:2331–2353. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Hayashi M, Wu G, Roh JL, et al:
Correlation of gene methylation in surgical margin imprints with
locoregional recurrence in head and neck squamous cell carcinoma.
Cancer. 121:1957–1965. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Asli NS and Kessel M: Spatiotemporally
restricted regulation of generic motor neuron programs by
miR-196-mediated repression of Hoxb8. Dev Biol. 344:857–868. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Dasen JS: Long noncoding RNAs in
development: solidifying the Lncs to Hox gene regulation. Cell Rep.
5:1–2. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Ma L: Role of miR-10b in breast cancer
metastasis. Breast Cancer Res. 12:2102010. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Wu Y, Zhang L, Zhang L, Wang Y, Li H, Ren
X, Wei F, Yu W, Liu T, Wang X, et al: Long non-coding RNA HOTAIR
promotes tumor cell invasion and metastasis by recruiting EZH2 and
repressing E-cadherin in oral squamous cell carcinoma. Int J Oncol.
46:2586–2594. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Adami GR, Tang JL and Markiewicz MR:
Improving accuracy of RNA-based diagnosis and prognosis of oral
cancer by using noninvasive methods. Oral Oncol. 69:62–67. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
123
|
He Q, Chen Z, Cabay RJ, et al: microRNA-21
and microRNA-375 from oral cytology as biomarkers for oral tongue
cancer detection. Oral Oncol. 57:15–20. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Koch FP, Kämmerer PW, Kämmerer P, Al-Nawas
B and Brieger J: Influence of class M1 glutathione S-transferase
(GST Mu) polymorphism on GST M1 gene expression level and tumor
size in oral squamous cell carcinoma. Oral Oncol. 46:128–133. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Serrao NR, Reid SM and Wilson CC:
Establishing detection thresholds for environmental DNA using
receiver operator characteristic (ROC) curves. Conservation Genet
Resour. 1–8. 2017.
|
|
126
|
Mordhorst LB, Sorbe B and Ahlin C: A study
of serum biomarkers associated with relapse of cervical cancer.
Anticancer Res. 32:4913–4922. 2012.PubMed/NCBI
|
|
127
|
Hosmer DW Jr, Lemeshow S and Sturdivant
RX: Applied Logistic Regression. 3rd edition. John Wiley &
Sons; Hoboken, NJ: pp. 173–182. 2013
|
|
128
|
Braoudaki M, Lambrou GI, Giannikou K, et
al: miR-15a and miR-24-1 as putative prognostic microRNA signatures
for pediatric pilocytic astrocytomas and ependymomas. Tumour Biol.
37:9887–9897. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Braoudaki M, Lambrou GI, Papadodima SA,
Stefanaki K, Prodromou N and Kanavakis E: MicroRNA expression
profiles in pediatric dysembryoplastic neuroepithelial tumors. Med
Oncol. 33:52016. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Hunt EA, Broyles D, Head T and Deo SK:
MicroRNA detection: Current technology and research strategies.
Annu Rev Anal Chem (Palo Alto Calif). 8:217–237. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Doleshal M, Magotra AA, Choudhury B,
Cannon BD, Labourier E and Szafranska AE: Evaluation and validation
of total RNA extraction methods for microRNA expression analyses in
formalin-fixed, paraffin-embedded tissues. J Mol Diagn. 10:203–211.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Dai L, Wang Y, Chen L, Zheng J, Li J and
Wu X: MiR-221, a potential prognostic biomarker for recurrence in
papillary thyroid cancer. World J Surg Oncol. 15:112017. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Li J, Smyth P, Flavin R, Cahill S, Denning
K, Aherne S, Guenther SM, O'Leary JJ and Sheils O: Comparison of
miRNA expression patterns using total RNA extracted from matched
samples of formalin-fixed paraffin-embedded (FFPE) cells and snap
frozen cells. BMC Biotechnol. 7:362007. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Manikandan M, Magendhra Deva Rao AK, et
al: Down regulation of miR-34a and miR-143 may indirectly inhibit
p53 in oral squamous cell carcinoma: A pilot study. Asian Pac J
Cancer Prev. 16:7619–7625. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Moratin J, Hartmann S, Brands R, et al:
Evaluation of miRNA-expression and clinical tumour parameters in
oral squamous cell carcinoma (OSCC). J Craniomaxillofac Surg.
44:876–881. 2016. View Article : Google Scholar : PubMed/NCBI
|