Introduction
Malignant melanoma, a class of highly malignant
tumors derived from melanocytes, and is a type of skin cancer with
the highest metastasis and mortality rate (1–3). The cure
rate of early melanoma may be >90% when it is surgically
resected (1–3), while metastatic malignant melanoma often
requires chemotherapy, radiotherapy, targeted therapy,
immunotherapy or other kinds of combination therapy. Current
therapeutic interventions for metastatic melanoma are not
sufficient, and the 5-year survival rate is <20% (1–3).
Therefore, understanding the molecular mechanisms of melanoma
pathogenesis and identifying novel potential therapeutic targets
are of importance for the prevention of malignant melanoma and the
development of interventions (4–6).
Identification of the dysregulated genes in cancer
tissues is important in the study of cancer biology. Gene
expression microarrays have been applied in the high-throughput
profiling of gene expression in number of types of cancer (7,8). In
malignant melanoma tissues, a set of genes were identified to be
dysregulated compared with normal tissues, and were demonstrated to
be associated with processes involved in the carcinogenesis and
progression of melanoma, including cell growth, cell cycle
progression, apoptosis, cell migration and metastasis (9,10).
Furthermore, several of these genes were identified to be
associated with the prognosis, survival, and responses to
chemotherapy of patients with melanoma (11,12).
However, identification of dysregulated genes in melanoma tissues
and their roles in cancer development remain an ongoing process in
the study of melanoma biology.
Melanocyte-specific gene 1 (MSG1; also known
as Cbp/P30-interacting transactivator with Glu/Asp-rich
carboxy-terminal domain 1) is a transcriptional cofactor that
interacts with CREB-binding protein/p300 and modulates the
transcription of a set of downstream genes (13,14).
Previous studies have identified that MSG1 is an important
factor in the differentiation and pigmentation of melanocytes. For
example, MSG1 may promote the synthesis of melanin, thus
enhancing melanogenesis in melanocytes (15,16).
However, the roles of MSG1 in the carcinogenesis and
progression of malignant melanoma require additional investigation
and elucidation.
In the present study, in order to determine the
dysregulated gene expression present in melanoma, the gene
expression profiles of human melanoma tissues were screened using a
cDNA microarray, and compared with the expression profiles of nevus
tissues. MSG1 expression was identified to be significantly
overexpressed in melanoma tissues. The overexpression of
MSG1 in melanoma was subsequently confirmed using
immunohistochemistry (IHC) in a set of melanoma tissues. The
present study aimed to further examine the roles of MSG1 in
the carcinogenesis and progression of malignant melanoma cancer
biology, so as to elucidate novel molecular mechanisms underlying
melanoma development and potential therapeutic targets for
treatment.
Materials and methods
Clinical melanoma specimens
Human malignant melanoma tissues and melanocytic
nevus tissues (surgically resected and later histopathologically
diagnosed as benign) were obtained from patients with melanoma
during surgery, and diagnosed by pathological validation. A total
of 10 patients with nevus and melanoma tissues examined by
pathology and without other skin diseases from the Changhai
Hospital (Shanghai, China) were included in the study, including 7
male and 3 female patients with a mean age of 48 (age range,
36–65). Ten nevus and matched melanoma tissue samples were obtained
between September 2005 and September 2008. All samples were
snap-frozen in liquid nitrogen until examination. All human samples
were collected with the written informed consent of the patients,
and use of human tissues was approved by the Institutional Research
Ethics Committee of the Second Military Medical University
(Shanghai, China).
Cell culture and transfection
The human malignant melanoma A375 cell line was
obtained from the American Type Culture Collection (Manassas, VA,
USA) and cultured in DMEM (PAA; GE Healthcare, Chicago, IL, USA)
with 10% fetal bovine serum (FBS) (GE Healthcare, Chicago, IL,
USA), under 37.5°C and 5% CO2. Cells were transfected
with small interfering (si)RNAs using INTERFERin®
reagent (Polyplus-transfection SA, Illkirch, France) according to
the manufacturer's protocol, and transfected with MSG1
expressing plasmids (constructed using a pcDNA 3.1 vector)
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
using JetPEI® reagent (Polyplus-transfection SA)
following the manufacturer's protocol, and confirmed by western
blot analysis, in order to induce MSG1 overexpression. The
siRNA target sequences for human MSG1 gene were
5′-UAGCAGCACAUCAGUCGAAUA-3′ (sense) and 5′-CCCAAUAUUGUCAAUUAUUUA-3′
(antisense), and the negative control siRNA sequences were
5′-UCUCCGAACGUGUCACGUTT-3′ (sense) and 5′-ACGUGACACGUUCGGAGAATT-3′
(antisense). siRNA duplexes were transfected at a final
concentration of 10 nM.
cDNA microarray assay
The Affymetrix GeneChip Human Genome U133 Plus 2.0
array assay was performed by Shanghai Bohao Industrial Co., Ltd.,
Shanghai, China. In brief, 5 µg total RNA samples from melanoma
tissues and paired nevus tissues were reverse transcribed into cDNA
for use in the microarray, as previously described (17). Hybridization was performed overnight
using a micro-circulation pump (Atactic Technologies, Inc.,
Houston, TX, USA), and images were collected and quantified
(17). The differentially detected
signals were gathered and presented.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
Total RNA, sourced from patient tissues, was
extracted using TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) following the manufacturer's protocol. RT-qPCR
analysis was performed using a SYBR RT-PCR kit (Takara Bio, Inc.,
Otsu, Japan) and LightCycler (Roche Diagnostics, Basel,
Switzerland). The primer sequences for MSG1 were
5′-GGCGGCACCACCATGTACCCT-3′ (sense) and 5′-AGGGGCCGGACTCGTCATACT-3′
(antisense); the primer sequences for Bcl-2 were
5′-GGTGGGGTCATGTGTGTGG-3′ (sense) and 5′-CGGTTCAGGTACTCAGTCATCC-3′
(antisense); and the internal control b-actin sequences were
5′-GGCGGCACCACCATGTACCCT-3′ (sense) and 5′-AGGGGCCGGACTCGTCATACT-3′
(antisense). The PCR cycle conditions were 95°C 15 sec, 55°C 30
sec, 72°C 30 sec for 45 cycles, and three independent experimental
repeats were performed. The relative expression level of gene mRNAs
was normalized to that of internal control β-actin by using
2−ΔΔCq cycle threshold method (18).
Cell viability analysis
The cell viability of transfected A375 cells was
examined by the MTT method. Briefly, cells (10,000) were seeded
into 96-well plates, cultured in DMEM with 10% FBS, and transfected
as described. Control cells were transfected with empty pcDNA 3.1
vectors. At the indicated time points (0, 48 and 96 h), cell
culture medium was replaced by fresh medium containing 0.5 mg/ml
MTT. Cells were then incubated at 37°C for 2 h, and the
MTT-containing medium was then replaced by 0.1 ml of dimethyl
sulfoxide to dissolve the formazan (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany). The absorbance in each well was detected at
570 nm.
Apoptosis analysis
Melanoma cells were transfected for 48 h and cell
culture medium was subsequently replaced by serum-free medium
(DMEM; PAA; GE Healthcare). At the indicated time points (0 and 48
h), cells were harvested and apoptosis was detected using a
Calbiochem® Annexin V-FITC Apoptosis Detection kit
(Merck KGaA) according to the manufacturer's protocol, and a
FACSCalibur flow cytometer (BD Biosciences, Franklin Lakes, NJ,
USA). The Annexin V-positive cells were regarded as apoptotic.
IHC
MSG1 expression in melanoma tissues was
examined by IHC. In brief, tissues were fixed with formalin and
embedded in paraffin, and then sectioned to make tissue sections (4
µm thick), which were deparaffinized in xylene for 20 min and
rehydrated in graded ethanol (95, 85, 75 and 50% for 5 min each).
Endogenous peroxidase activity was blocked by a 30 min incubation
in 3% H2O2 in PBS, and antigen retrieval was
performed in 10 mM citrate buffer (pH 6.0) by heating to boil for 5
min. The anti-MSG1 primary antibody (cat no. ab87978; Abcam,
Cambridge, UK) was diluted 1:500 and incubated at 4°C overnight.
The secondary antibody was horseradish peroxidase-conjugated goat
anti-mouse IgG secondary antibody (cat no. ab97040; Abcam,
Cambridge, UK) in 1:1,000 dilution for use and in incubation at 4°C
for 2 h. Immunostaining was visualized using a
3,3′-diaminobenzidine staining kit (Dako; Agilent Technologies,
Inc., Santa Clara, CA, USA) and analyzed using HistoFAXS system and
mean DAB staining intensity was calculated using Histoquest
software (both from TissueGnostics, Vienna, Austria), and the
images are presented in Fig. 1C.
Western blotting
Cells were lysed using Passive Lysis Buffer (Cell
Signaling Technology, Inc., Danvers, MA, USA). Protein
concentrations were measured using the BCA Protein Assay kit
(Takara Bio., Inc.) and equal amounts of extracts (30 µg) were
subjected to SDS-PAGE (10% gel), transferred onto a polyvinylidene
fluoride membrane, and then blotted. The MSG1 antibody (cat
no. ab87978) was purchased from Abcam. B-cell lymphoma 2 (Bcl-2;
cat no. CST 2872) and β-actin (cat no. CST 3700) antibodies, all at
a 1:1,000 dilution, and horseradish peroxidase-coupled secondary
antibodies were purchased from Cell Signaling Technology, Inc.
Antibody incubation was performed for 3 h at 4°C. At least 3
replicates were performed. Blocking was performed using
tris-buffered saline with Tween-20, with 5% bovine serum albumin
for 1 h at 20°C. Imaging was performed using SuperSignal West Femto
Maximum Sensitivity Substrate (Thermo Fisher Scientific, Inc.), and
densitometric analysis was performed using Labworks Image
Acquisition and Analysis Software (UVP, Upland, CA, USA).
Statistical analysis
Results are presented as the mean ± standard
deviation. Statistical analyses were performed using a Student's
t-test. P<0.05 was considered to indicate a statistically
significant difference.
Results
MSG1 expression is upregulated in
malignant melanoma
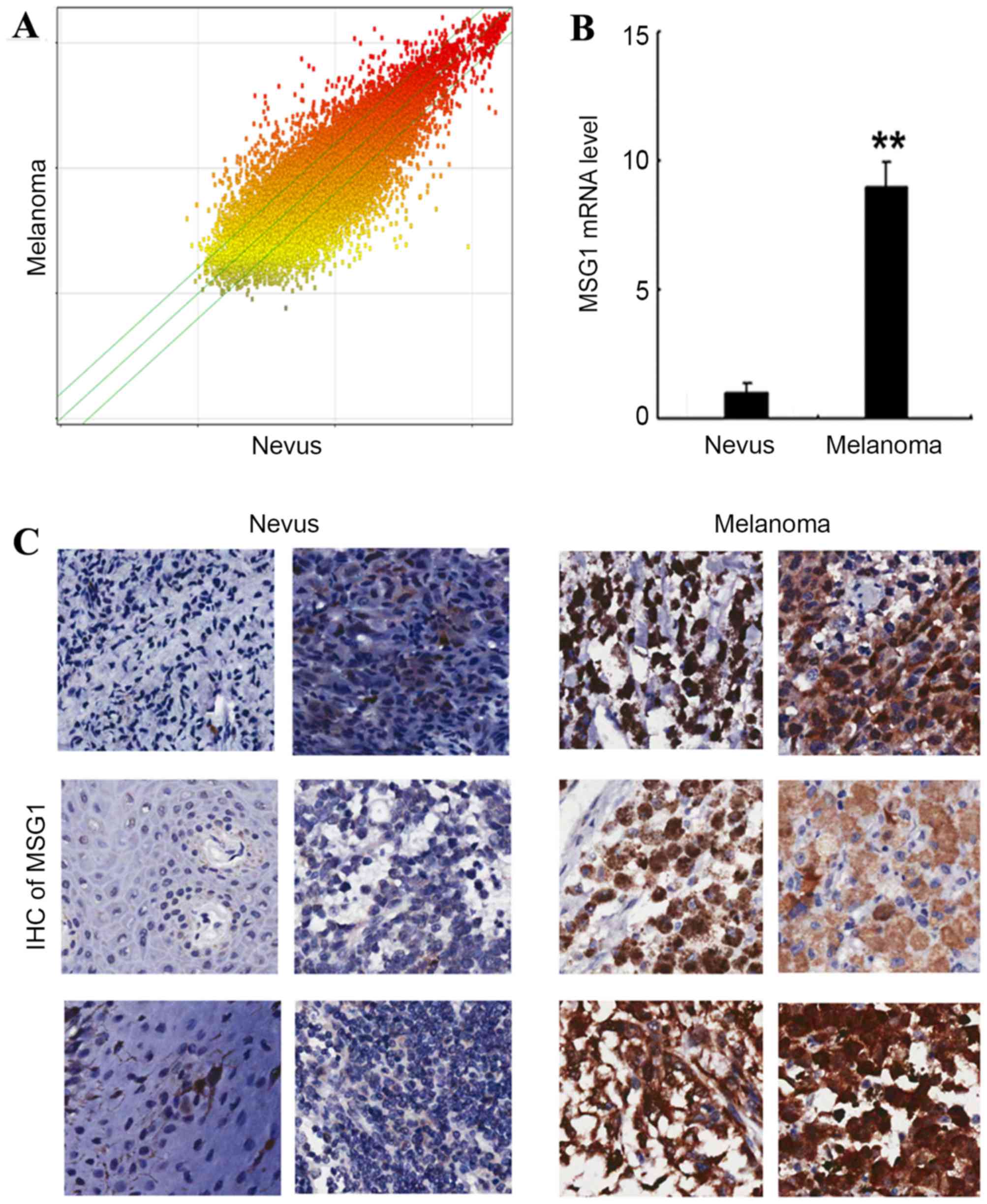
In order to determine the gene expression profile in
malignant melanoma, a cDNA microarray was performed in the melanoma
and paired nevus tissues, and the differential gene expression is
presented in Fig. 1A. Among the
differentially expressed genes, it was identified that the
melanocyte differentiation-associated gene MSG1 was
significantly upregulated in melanoma tissue compared with nevus
tissue (P<0.01; Fig. 1B).
Additionally, the upregulation of MSG1 was confirmed by IHC
in the melanoma tissues compared with nevus tissues (Fig. 1C). The results indicated that
MSG1 is upregulated in melanoma, and may potentially
participate in melanoma carcinogenesis and progression.
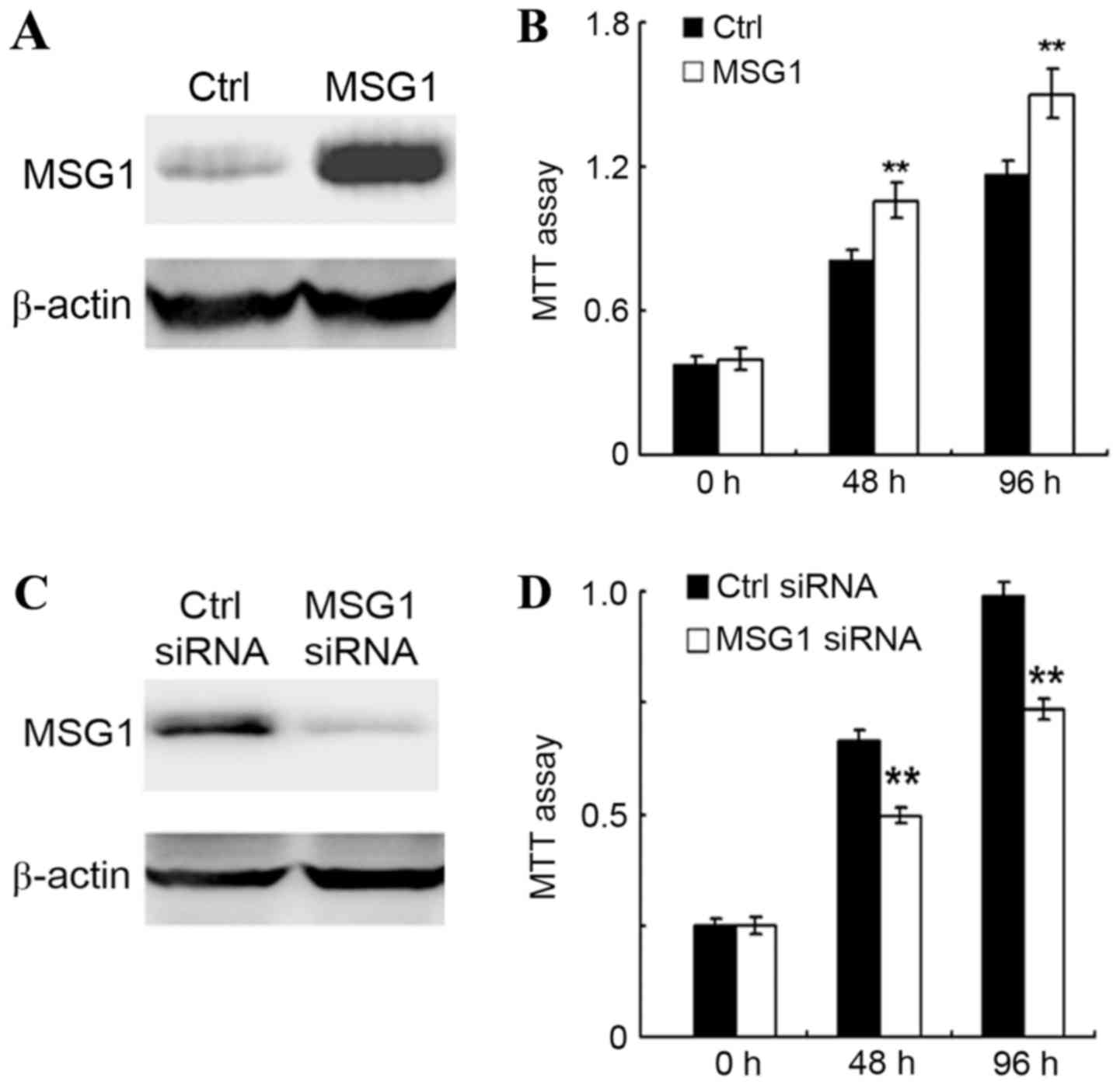
MSG1 promotes cell viability in
melanoma
As MSG1 was upregulated in melanoma, the
roles of MSG1 in melanoma development were further examined.
The cell viability of melanoma A375 cells was examined in
control-transfected or MSG1-overexpressing cells, revealing
that MSG1 overexpression could significantly increase cell
viability in A375 cells (P<0.01; Fig.
2A and B). In addition, this result was confirmed by the
knockdown of MSG1 expression. Transfection of the A375 cells
with an MSG1-specific siRNA significantly decreased the cell
viability compared with the control-siRNA-transfected cells
(P<0.01; Fig. 2C and D). Thus, it
was concluded that overexpression of MSG1 in melanoma may
increase cell viability.
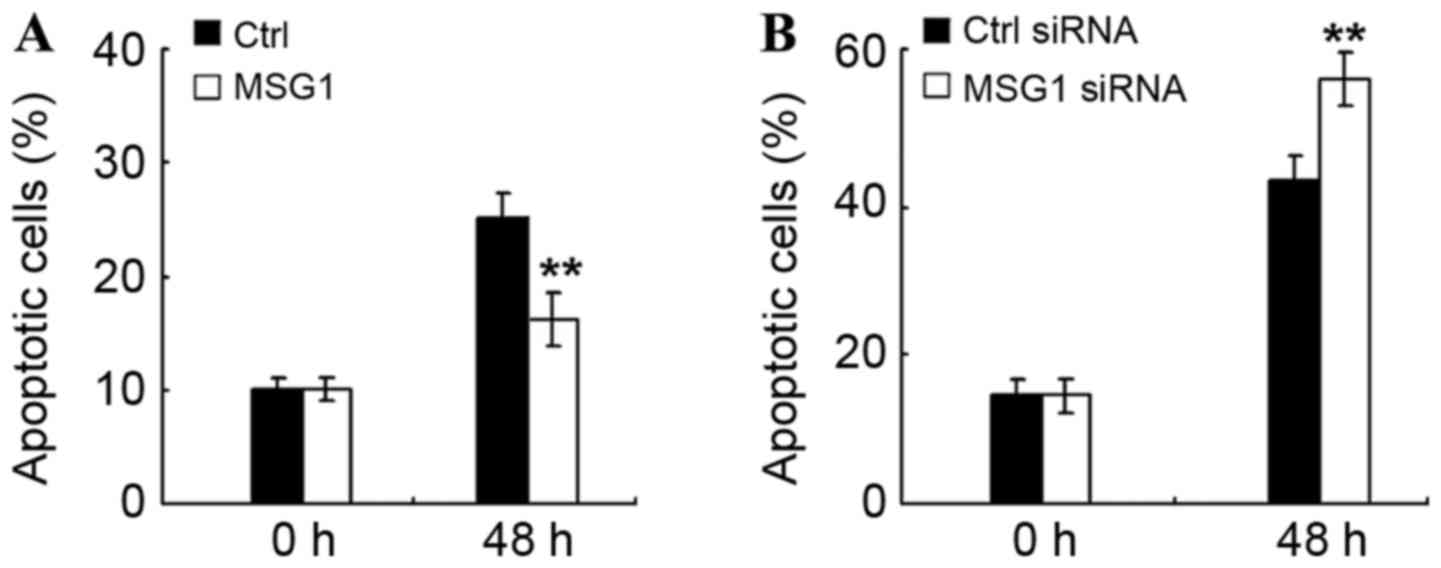
MSG1 inhibits cell apoptosis in
melanoma
In order to determine the mechanism of
MSG1-promoted cell viability, the effect of MSG1 on
cell apoptosis were additionally examined in melanoma cells. As
demonstrated in Fig. 3A, MSG1
overexpression significantly inhibited cell apoptosis induced by
serum deprivation (P<0.01), whereas knockdown of MSG1
expression significantly promoted the serum deprivation-induced
cell apoptosis (P<0.01; Fig. 3B).
Therefore, MSG1 may inhibit cell apoptosis of melanoma
cells, thus enhancing cell viability and promoting melanoma
progression.
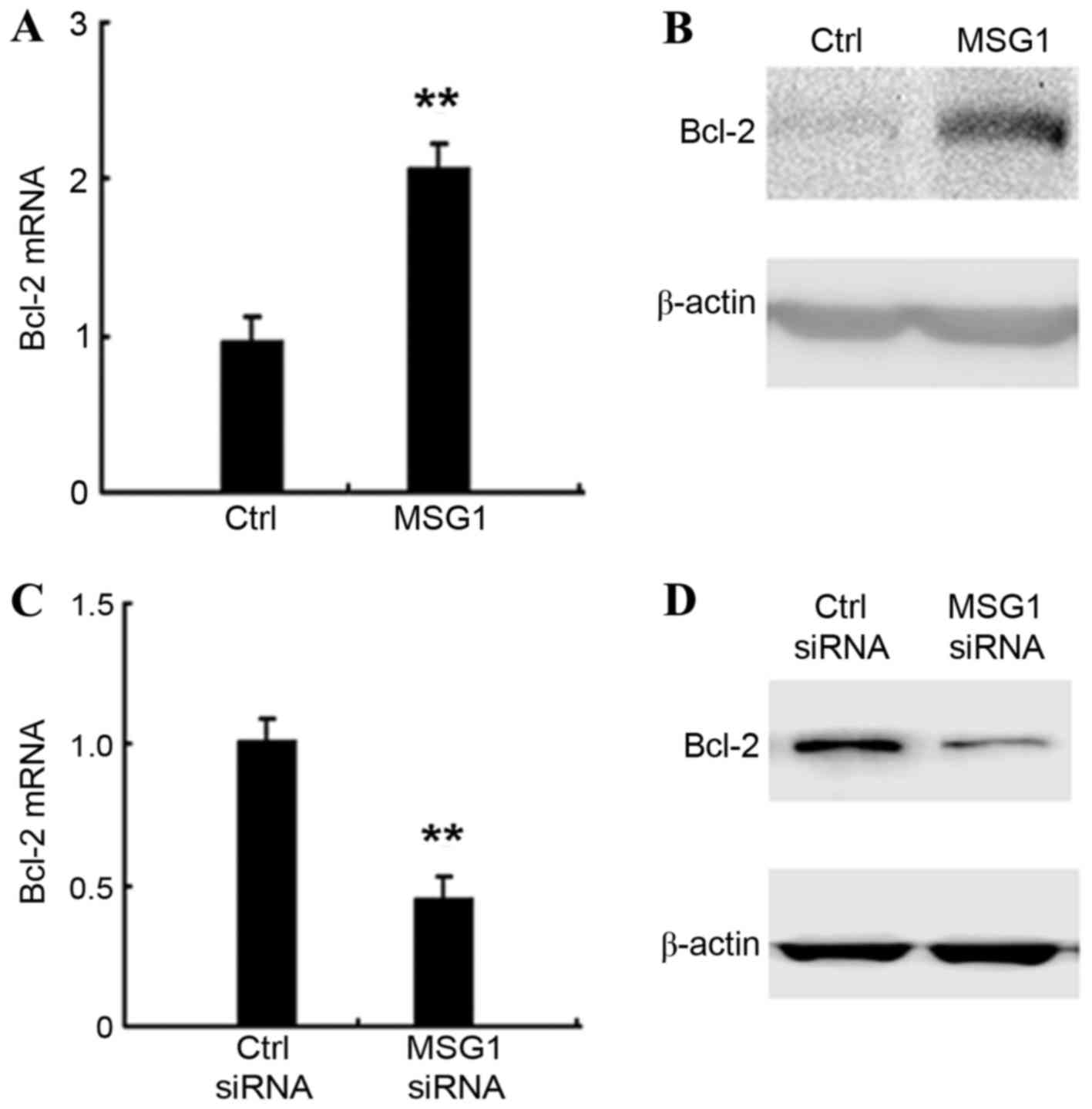
MSG1 enhances anti-apoptotic Bcl-2
expression
The molecular mechanisms responsible for the
inhibition of apoptosis mediated by MSG1 overexpression were
explored. The expression of apoptosis-associated intracellular
proteins in MSG1-overexpressing A375 cells were screened,
and it was identified that the expression of the anti-apoptotic
protein Bcl-2 was significantly increased by MSG1
overexpression at the mRNA (P<0.01; Fig. 4A) and protein levels (Fig. 4B). Furthermore, the knockdown of
MSG1 expression significantly inhibited the expression of
Bcl-2 in A375 cells (P<0.01; Fig. 4C
and D). Taken together, the results indicate that MSG1
may enhance the expression of anti-apoptotic Bcl-2, thus inhibiting
cell apoptosis and promoting melanoma progression.
Discussion
In the present study, the gene expression profile in
human melanoma tissues was screened using a cDNA microarray, and it
was identified that the melanocyte differentiation-associated gene
MSG1 was significantly overexpressed in melanoma tissues. It
was also identified that MSG1 may promote malignant melanoma
progression by the inhibition of cell apoptosis, which is mediated
by enhanced expression of anti-apoptotic Bcl-2. Therefore, these
data suggest a novel molecular mechanism for malignant melanoma
carcinogenesis and progression, which may suggest potential
therapeutic strategies for the treatment of patients with
melanoma.
The dysregulation of a set of genes has previously
been identified in the carcinogenesis and progression of malignant
melanoma, and several of these genes have been demonstrated to be
correlated with the survival of melanoma patients (19–22). For
example, caveolin-1 has been demonstrated to be upregulated in
melanoma tissues, and correlated with melanoma metastasis and
prognosis (23). In the present
study, the upregulation of MSG1 was revealed to be
associated with melanoma progression (23), but it remains unknown whether high
MSG1 expression in melanoma tissues predicts a poor survival
time in patients with melanoma. Our future studies will further
examine this issue in a larger cohort of patients with
melanoma.
The promotion of cell viability and the inhibition
of cell apoptosis are important aspects of cancer biology, and
malignant melanoma has demonstrated a set of mechanisms for
survival, including the inhibition of apoptosis and immune evasion
(24–26). The present study indicated that
expression of the important anti-apoptotic protein Bcl-2 is
significantly induced by MSG1 expression, and may contribute
to the MSG1-mediated inhibition of apoptosis. However, the
detailed mechanism responsible for the MSG1-induced Bcl-2
expression remains unknown. We hypothesized that MSG1 may
enhance or participate in the initiation of Bcl-2 gene
transcription, which requires additional investigation.
In conclusion, the upregulation of MSG1
expression in melanoma tissues has been identified in the present
study, suggesting that MSG1 may function as an oncogene in
the carcinogenesis and progression of malignant melanoma. However,
the detailed mechanism responsible for the upregulated expression
of MSG1 in melanoma cells remains unknown. At present,
genetic and epigenetic mechanisms have been identified to underlie
the dysregulation of genes in cancer biology (19,27–29). We
intend to investigate the detailed mechanism of the regulation of
MSG1 expression in melanoma cells, with the aim of
elucidating the potential MSG1-mediated regulatory loop in
melanoma development.
Acknowledgements
The authors would like to thank Dr Hao Zhang and Dr
Hao Tang from Changhai Hospital (Shanghai, China) for their
extensive assistance in the present project. The present study was
supported by the Project of Shanghai Science and Technology
Commission (grant no. 13JC1401403) and the SMMU Start-up Foundation
for Youths (grant no. 2013QN08).
References
|
1
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar
|
|
3
|
Balch CM, Gershenwald JE, Soong SJ,
Thompson JF, Atkins MB, Byrd DR, Buzaid AC, Cochran AJ, Coit DG,
Ding S, et al: Final version of 2009 AJCC melanoma staging and
classification. J Clin Oncol. 27:6199–6206. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ortega E, Marti RM, Yeramian A, Sorolla A,
Dolcet X, Llobet D, Abal L, Santacana M, Pallares J,
Llombart-Cussac A and Matias-Guiu X: Targeted therapies in
gynecologic cancers and melanoma. Semin Diagn Pathol. 25:262–273.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Terheyden P, Tilgen W and Hauschild A:
Recent aspects of medical care of malignant melanoma. J Dtsch
Dermatol Ges. 6:868–878. 2008.(In English, German). View Article : Google Scholar : PubMed/NCBI
|
|
6
|
di Pietro A, Tosti G, Ferrucci PF and
Testori A: Oncophage: Step to the future for vaccine therapy in
melanoma. Expert Opin Biol Ther. 8:1973–1984. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hou J, Zhou Y, Zheng Y, Fan J, Zhou W, Ng
IO, Sun H, Qin L, Qiu S, Lee JM, et al: Hepatic RIG-I predicts
survival and interferon-α therapeutic response in hepatocellular
carcinoma. Cancer Cell. 25:49–63. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hou J, Lin L, Zhou W, Wang Z, Ding G, Dong
Q, Qin L, Wu X, Zheng Y, Yang Y, et al: Identification of miRNomes
in human liver and hepatocellular carcinoma reveals miR-199a/b-3p
as therapeutic target for hepatocellular carcinoma. Cancer Cell.
19:232–243. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sumantran VN, Mishra P and Sudhakar N:
Microarray analysis of differentially expressed genes regulating
lipid metabolism during melanoma progression. Indian J Biochem
Biophys. 52:125–131. 2015.PubMed/NCBI
|
|
10
|
Dadras SS, Lin RJ, Razavi G, Kawakami A,
Du J, Feige E, Milner DA, Loda MF, Granter SR, Detmar M, et al: A
novel role for microphthalmia-associated transcription
factor-regulated pigment epithelium-derived factor during melanoma
progression. Am J Pathol. 185:252–265. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Minca EC, Tubbs RR, Portier BP, Wang Z,
Lanigan C, Aronow ME, Triozzi PL, Singh A, Cook JR, Saunthararajah
Y, et al: Genomic microarray analysis on formalin-fixed
paraffin-embedded material for uveal melanoma prognostication.
Cancer Genet. 207:306–315. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chiu CG, Nakamura Y, Chong KK, Huang SK,
Kawas NP, Triche T, Elashoff D, Kiyohara E, Irie RF, Morton DL and
Hoon DS: Genome-wide characterization of circulating tumor cells
identifies novel prognostic genomic alterations in systemic
melanoma metastasis. Clin Chem. 60:873–885. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Han B, Liu N, Yang X, Sun HB and Yang YC:
MRG1 expression in fibroblasts is regulated by Sp1/Sp3 and an Ets
transcription factor. J Biol Chem. 276:7937–7942. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yahata T, de Caestecker MP, Lechleider RJ,
Andriole S, Roberts AB, Isselbacher KJ and Shioda T: The
MSG1 non-DNA-binding transactivator binds to the p300/CBP
coactivators, enhancing their functional link to the Smad
transcription factors. J Biol Chem. 275:8825–8834. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nair SS, Chaubal VA, Shioda T, Coser KR
and Mojamdar M: Over-expression of MSG1 transcriptional
co-activator increases melanin in B16 melanoma cells: A possible
role for MSG1 in melanogenesis. Pigment Cell Res.
14:206–209. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ahmed NU, Shioda T, Coser KR, Ichihashi M
and Ueda M: Aberrant expression of MSG1 transcriptional
activator in human malignant melanoma in vivo. Pigment Cell Res.
14:140–143. 2001.PubMed/NCBI
|
|
17
|
Hou J, Wang P, Lin L, Liu X, Ma F, An H,
Wang Z and Cao X: MicroRNA-146a feedback inhibits RIG-I-dependent
Type I IFN production in macrophages by targeting TRAF6, IRAK1, and
IRAK2. J Immunol. 183:2150–2158. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Martinez-Cardús A, Vizoso M, Moran S and
Manzano JL: Epigenetic mechanisms involved in melanoma pathogenesis
and chemoresistance. Ann Transl Med. 3:2092015.PubMed/NCBI
|
|
20
|
Elder DE: Pathology of melanoma. Surg
Oncol Clin N Am. 24:229–237. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bartlett EK and Karakousis GC: Current
staging and prognostic factors in melanoma. Surg Oncol Clin N Am.
24:215–227. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Higgins HW II, Lee KC, Galan A and Leffell
DJ: Melanoma in situ: Part II. Histopathology, treatment, and
clinical management. J Am Acad Dermatol. 73:193–203. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Stenzel M, Tura A, Nassar K, Rohrbach JM,
Grisanti S, Lüke M and Lüke J: Analysis of caveolin-1 and
phosphoinositol-3 kinase expression in primary uveal melanomas.
Clin Experiment Ophthalmol. 44:400–409. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Talaiezadeh A, Jalali F, Galehdari H and
Khodadadi A: Time depended Bcl-2 inhibition might be useful for a
targeted drug therapy. Cancer Cell Int. 15:1052015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mukherjee N, Schwan JV, Fujita M, Norris
DA and Shellman YG: Alternative treatments for melanoma: Targeting
BCL-2 family members to De-Bulk and kill cancer stem cells. J
Invest Dermatol. 135:2155–2161. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hartman ML and Czyz M: Pro-survival role
of MITF in melanoma. J Invest Dermatol. 135:352–358. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Brazel AJ and Vernimmen D: The complexity
of epigenetic diseases. J Pathol. 238:333–344. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ambrosone CB, Hong CC and Goodwin PJ: Host
factors and risk of breast cancer recurrence: Genetic, epigenetic
and biologic factors and breast cancer outcomes. Adv Exp Med Biol.
862:143–153. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Farooqi AA, Tang JY, Li RN, Ismail M,
Chang YT, Shu CW, Yuan SS, Liu JR, Mansoor Q, Huang CJ and Chang
HW: Epigenetic mechanisms in cancer: Push and pull between kneaded
erasers and fate writers. Int J Nanomedicine. 10:3183–3191.
2015.PubMed/NCBI
|