Introduction
Philadelphia (Ph) chromosome is named for the city
in which this special aberrant translocation of chromosome 9 and
chromosome 22 was found. Ph chromosome is a key indication to
diagnosis chronic myeloid leukemia (CML) and it is also found in
approximately 20% acute lymphoblastic leukemia (ALL) patients and
occasionally found in other hematological malignancies (1). Ph chromosome gives rise to a breakpoint
cluster region (BCR)/ABL proto-oncogene 1, non-receptor tyrosine
kinase (ABL) fusion gene that results to the constitutive
activation of proto-oncogene ABL1 and leads to oncogenesis. A
variety of BCR/ABL transcript variants have been reported such as
e13a2 (b2a2), e14a2 (b3a2) which account for more than 95% in CML
while e1a2 accounts for more than 50% in ALL (2). Rare transcript variants are occasionally
reported, including e8a2, e6a2, e14a3 etc. In terms of e14a3, most
of the cases reported are males and diagnosed with CML or atypical
CML. As to our knowledge, only 4 e14a3 (b3a3) BCR/ABL fusion
transcript in ALL cases have been reported, with two children and
two adults (3–6). Except for one lacking clinical data, all
of them received chemotherapy and two received imatinib therapy
additionally. Here we reported a case of e14a3 (b3a3) BCR/ABL
fusion transcript in an adult ALL patient who have received
chimeric antigen receptor modified (CAR-modified) T-cell therapy
and this is the first report about CAR-modified T-cell therapy
applied in a rare BCR/ABL fusion.
Materials and methods
Patient history
The present study was approved by the medical ethics
committee of Tongji Hospital (Tongi Medical College, Huazhong
University of Science and Technology, Hubei, China) and written
informed consent was obtained from the patient. A 47-year-old male
patient presented with low-grade fever and chest tightness.
Hemoglobin was 65 g/l, leucocyte count was 299×109/l,
and platelet count was 45×109/l. Bone marrow cytology
showed that the white blood cell count was significantly increased
that primitive and immature lymphocytes accounted for 87%. Clonal
immunoglobulin gene rearrangement detection showed a single clonal
peak in detection range of IGHA and IGHB respectively. Cytogenetic
analysis and fluorescent in situ hybridization (FISH)
revealed a complex karyotype and a Ph chromosome respectively. Gene
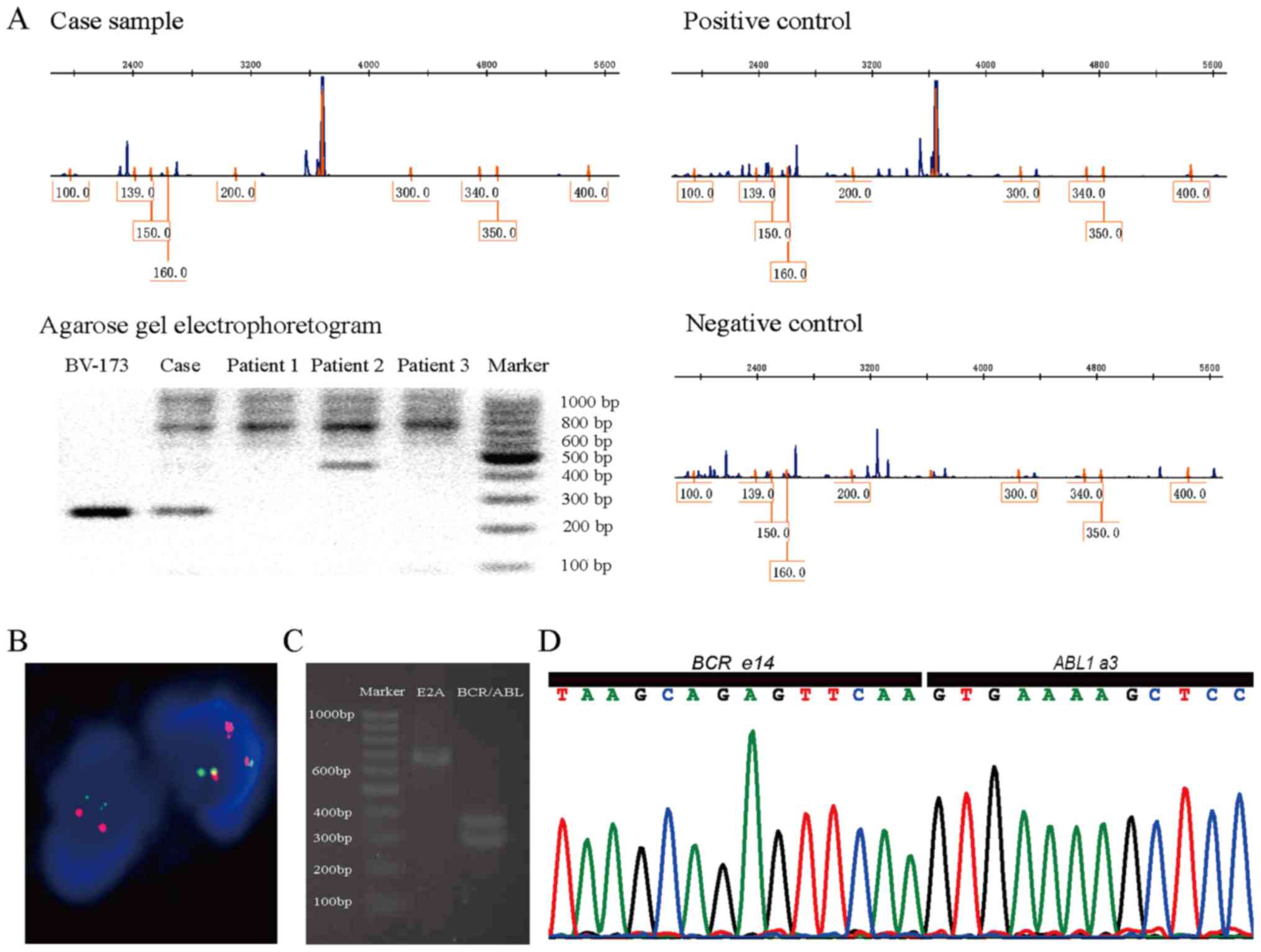
scanning and PCR found Ikaros6 transcription (Fig. 1A). After two rounds of chemotherapy he
got a cytological remission but relapsed two months later. Then he
received CAR-modified T-cell therapy that modified T cells
targeting on the lineage-specific antigens CD19 were infused into
body at the first time and followed with infusion of modified T
cells targeting on CD22 for the next two times. Complete remission
was soon achieved after the first CAR-modified T-cell therapy
without obvious adverse events except for a short period of fever.
Up to date, he has maintained a complete cytogenetic remission for
5 months.
Cytogenetic analysis and FISH
Cells from bone marrow were cultured for 24–48 h and
performed with Giemsa staining and GTG-banding according to
standard cytogenetic laboratory protocols. FISH probe (Beijing GP
Medical Technologies, Beijing, China) was applied to detect BCR/ABL
rearrangement following the protocol manufacturer recommended.
Gene scanning for detection of
IKaros6
Primers were 5′-ATGGATGCTGATGAGGGTCAAGAC-3′ forward
and 5′-GATGGCTTGGTCCATCACGTGG-3′ reverse. cDNA was reverse
transcribed from total RNA which was extracted from bone marrow
based on routine protocol of our laboratory. GeneScan™ 500 LIZ™ dye
Size Standard and Hi-Di™ Formamide (both from Applied Biosystems,
America) were mixed with PCR product and loaded on AB 3500 Genetic
Analyzer (Applied Biosystems; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) after denaturalization and annealing. As the
BV-173 cell line contains the IKaros6 transcript, this cell line
was employed as the positive control for these experiments (DMSZ,
Braunschweig, Germany).
Reverse transcription polymerase chain
reaction (RT-PCR)
RT was performed by use of PrimeScript RT-PCR kit
(Takara Bio, Inc., Otsu, Japan). Fusion gene fragments were
amplified by nested PCR and the primer sets and cycling conditions
were designed as Niels Pallisgaard's publication (7) (Table I).
Fifteen microliters of PCR product was added in a 1.5% agarose gel
to electrophorese for 60 min at 100 V.
 | Table I.Primer sequences and thermocycling
conditions for nested polymerase chain reaction. |
Table I.
Primer sequences and thermocycling
conditions for nested polymerase chain reaction.
|
|
| Thermocycling
conditions |
|---|
|
|
|
|
|---|
| Primer sets | Primer sequence
(5′-3′) | First round | Second round |
|---|
| First round |
| 95°C for 5 min | 1 cycle | 95°C for 5 min | 1 cycle |
|
BCR:3060U23 |
GAGTCACTGCTGCTGCTTATGTC | 95°C for 30 sec | 25 cycles | 95°C for 30 sec | 20 cycles |
|
ABL:661L20 |
TTTTGGTTTGGGCTTCACAC | 58°C for 30 sec |
| 58°C for 30 sec |
|
| Second round |
| 72°C for 1 min |
| 72°C for 1 min |
|
|
BCR:3128U22 |
CACGTTCCTGATCTCCTCTGAC | Held at 4°C |
| 72°C for 10 min | 1 cycle |
|
ABL:642L23 |
ACACCATTCCCCATTGTGATTAT |
|
| Held at 4°C |
|
Cloning and sequencing
PCR product was purified from the agarose gel
mentioned above and inserted in pEASY-T1 Cloning Vector provided by
pEASY-T1 Cloning kit (TransGen Biotech Co., Ltd., Beijing, China).
Transformation and positive clone detection were performed by
recommended protocol for the kit. Plasmid DNA of positive clone was
extracted by GenElute Plasmid Miniprep kit (Sigma-Aldrich; Merck
KGaA, Darmstadt, Germany) and sequenced on AB 3500 Genetic Analyzer
(Applied Biosystems; Thermo Fisher Scientific, Inc.). PCR primers
for positive clone detection and sequencing primer were provided in
pEASY-T1 Cloning kit.
Probe detection
To monitor the load of BCR/ABL fusion gene, a TaqMan
probe was designed on BCR exon 14
(5′-CGTCCACTCAGCCACTGGATTTAAGCA-3′). PCR primer sequences were
5′-GGGCTCTATGGGTTTCTGAATGT-3′ forward and
5′-AGACCCGGAGCTTTTCACTTG−3′ reverse. RCR ran in StepOnePlus
Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific,
Inc.) and condition was default settings.
Results
The karyotype was 43–47, XY, +6, t(9,22)(q34;q11),
del(9)(p22), +mar, inc [cp5]/46, XY[5]. FISH confirmed the presence
of the BCR-ABL translocation with 92% of cells reporting positive
for the translocation with the pattern of 1O1G2F (Fig. 1B). RT-PCR followed with agarose gel
electrophoresis showed two abnormal bands approximately 300 bp
(Fig. 1C). Cloning and Sanger
sequencing revealed a fusion of BCR e14 and ABL a3 without any
extra insertion or deletion (Fig.
1D). During the course of disease, BCR/ABL fusion gene probe
detection have been done for 5 times which depicted the rise and
fall of the Ph positive cell clones in profile (Table II).
 | Table II.Breakpoint cluster region/ABL
proto-oncogene 1, non-receptor tyrosine kinase fusion transcript
detected by reverse transcription-quantitative polymerase chain
reaction during the clinical course. |
Table II.
Breakpoint cluster region/ABL
proto-oncogene 1, non-receptor tyrosine kinase fusion transcript
detected by reverse transcription-quantitative polymerase chain
reaction during the clinical course.
| Clinical course | Material | Transcript
detected | Quantitative
result |
|---|
| Initial
diagnosis | Bone marrow | e14a3 | 108.30% |
| One month after
chemotherapy completed | Bone marrow | e14a3 |
0.19% |
| Two months after
chemotherapy completed | Bone marrow | e14a3 |
2.44% |
| Three months after
chemotherapy completed | Bone marrow | e14a3 | 110.48% |
| Two months after the
1st CAR-T therapy | Bone marrow | e14a3 |
0.01% |
| One month after the
2nd CAR-T therapy | Bone marrow | – | 0 |
| One month after the
3rd CAR-T therapy | Bone marrow | – | 0 |
Discussion
The reason why BCR/ABL e14a3 is so rare has not been
discussed too much. Considering the intron between a1 and a2 has a
large size of approximately 140 kb and the intron between a2 and a3
has only approximately 0.5 kb, the answer seems to be clear. The
breakpoint may randomly locate in any part of the region, and based
on the principle of probability, there is a little chance to
generate a e14a3 fusion.
Partially due to the limited number of the peculiar
cases, the influence of lacking exon a2 in BCR/ABL fusion to
patients' clinical outcome is still controversial (8). More reliable results and conclusions
hang on the accumulation of a large number of related cases from
laboratories all over the world, and which calls for the extensive
use of primer sets that can efficiently detect those rare fusion
transcripts. As for ALL, condition gets more complicated
considering the heterogeneity of patients and complicated
pathogenesis. Kurita et al (5)
compared the outcome of ALL patients with BCR/ABL fusions lacking
exon a2 that received imatinib treatment and those who did not, and
he found that lacking exon a2 proved to be an adverse factor to
prognosis but imatinib therapy brought a good prognosis remarkably.
But it's not that every ALL patient with BCR/ABL fusion lacking
exon a2 achieved a complete remission by the additional usage of
imatinib, including the one Kurita had reported and the present
case.
Most reported ALL cases with e14a3 BCR/ABL fusion
transcript had a progressive progression and unfavorable outcome
(Table III). Except for BCR/ABL
fusion gene, patient of the present case additionally had an IKZF1
deletion which is conformed as a significant adverse prognosis
factor (9). After receiving a common
chemotherapy, he achieved a cytological remission but soon
relapsed. Other than the cases previously published dead without
further therapy could be applied, he got a chance and consented to
the CAR-T clinical trial, which brought him a complete remission.
As a model of new therapies emergent in recent years, CAR-modified
T-cell therapy has achieved a series of splendid success. Despite
severe associated toxicities occurring occasionally, remarkable
therapy effect remains shiny and promises a hopeful future of
hematological neoplasm treatment. In theory, CAR-modified T cells
archer and destroy target cells accurately by the identification of
cell surface antigens and therefore it's independent of whether the
BCR/ABL fusion transcript or drug resistance exists. Patients who
do not have a good response with imatinib may have more clinical
benefit from CAR-modified T-cell therapy. There is much likelihood
that CAR-modified T-cell therapy would be the first-line treatment
instead of being the last hope for ALL in the near future and
brought an altered choose with TKI for those who carried Ph
chromosome.
 | Table III.Clinical characteristics of all
published cases of ALL with breakpoint cluster region/ABL
proto-oncogene 1, non-receptor tyrosine kinase e14a3 fusion
transcript. |
Table III.
Clinical characteristics of all
published cases of ALL with breakpoint cluster region/ABL
proto-oncogene 1, non-receptor tyrosine kinase e14a3 fusion
transcript.
| Author, year | Patient no. | Gender | Age (years) | Type of ALL | WBC count
(×109/l) | Treatment | Clinical outcome | Duration of follow-up
(months) | (Refs.) |
|---|
| Inukai et al,
1993 | 1 | Female | 3 | cALL | 11.4 | Chemotherapy | Succumbed | 41 | (3) |
| Picard et al,
2006 | 2 | Unknown | Unknown | Unknown | Unknown | Unknown | Unknown | Unknown | (6) |
| Kim et al,
2009 | 3 | Male | 12 | pre-B ALL | 190 | Chemotherapy followed
with imatinib | CR | 5 | (4) |
| Kurita et al,
2016 | 4 | Male | 43 | cALL | 203 | Chemotherapy followed
with HSCT | Succumbed | 11 | (5) |
| Present case | 5 | Male | 47 | cALL | 299 | Chemotherapy and
imatinib followed with CAR-T | CR | 5 mo | – |
References
|
1
|
Faderl S, Talpaz M, Estrov Z, O'Brien S,
Kurzrock R and Kantarjian HM: The biology of chronic myeloid
leukemia. N Engl J Med. 341:164–172. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kurzrock R, Shtalrid M, Romero P, Kloetzer
WS, Talpas M, Trujillo JM, Blick M, Beran M and Gutterman JU: A
novel c-abl protein product in Philadelphia-positive acute
lymphoblastic leukaemia. Nature. 325:631–635. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Inukai T, Sugita K, Suzuki T, Ijima K, Goi
K, Tezuka T, Kojika S, Hatakeyama K, Kagami K, Mori T, et al: A
novel 203 kD aberrant BCR-ABL product in a girl with Philadelphia
chromosome positive acute lymphoblastic leukaemia. Br J Haematol.
85:823–825. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kim J, Park TS, Lyu CJ, Song J, Lee KA,
Kim SJ, Lee HJ and Choi JR: BCR/ABL rearrangement with b3a3 fusion
transcript in a case of childhood acute lymphoblastic leukemia.
Cancer Genet Cytogenet. 189:132–137. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kurita D, Hatta Y, Hojo A, Kura Y, Sawada
U, Kanda Y and Takei M: Adult acute lymphoblastic leukemia with a
rare b3a3 type BCR/ABL1 fusion transcript. Cancer Genet.
209:161–165. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Picard C, Hayette S, Bilhou-Nabera C,
Cayuela JM, Delabesse E, Frenoy N, Preudhomme C, Dupont M, Bastard
C, Bories D, et al: Prospective multicentric molecular study for
poor prognosis fusion transcripts at diagnosis in adult B-lineage
ALL patients: the LALA 94 experience. Leukemia. 20:2178–2181. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Pallisgaard N, Hokland P, Riishøj DC,
Pedersen B and Jørgensen P: Multiplex reverse
transcription-polymerase chain reaction for simultaneous screening
of 29 translocations and chromosomal aberrations in acute leukemia.
Blood. 92:574–588. 1998.PubMed/NCBI
|
|
8
|
Snyder DS, McMahon R, Cohen SR and Slovak
ML: Chronic myeloid leukemia with an e13a3 BCR-ABL fusion: Benign
course responsive to imatinib with an RT-PCR advisory. Am J
Hematol. 75:92–95. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Martinelli G, Iacobucci I, Storlazzi CT,
Vignetti M, Paoloni F, Cilloni D, Soverini S, Vitale A, Chiaretti
S, Cimino G, et al: IKZF1 (Ikaros) deletions in BCR-ABL1-positive
acute lymphoblastic leukemia are associated with short disease-free
survival and high rate of cumulative incidence of relapse: A GIMEMA
AL WP report. J Clin Oncol. 27:5202–5207. 2009. View Article : Google Scholar : PubMed/NCBI
|