Introduction
Globally, colorectal cancer (CRC) is the third most
commonly diagnosed cancer in males and the third most commonly
diagnosed cancer in females according to a previous epidemiological
survey data (1). It is also one of
the leading causes of cancer-associated mortality worldwide
(1). Each year, ~1 million new cases
are diagnosed and 30% of the 600,000 people who succumb to the
disease each year reside in China (2,3). Over the
past ten years, the incidence and mortality rates of CRC have
rapidly increased due to the prevalence of obesity, which could
partially have attributed to the development of China's economy
(4). Despite developments in
diagnostic and therapeutic strategies, which have already improved
the survival rates of patients with early stage CRC, the prognosis
of patients with late stage CRC remains poor (5,6).
Therefore, further investigations are required to gain an improved
understanding of the molecular characteristics and associated
biological mechanisms underlying the proliferation, migration and
metastasis of CRC cells. This may also enable the identification of
early screening markers and therapeutic targets.
The human serine/threonine kinase 33 (STK33) enzyme
belongs to calcium/calmodulin-dependent kinase family and is
located on chromosome 11p15.3, which is a gene-rich region
associated with several diseases, including cancer (7). A previous study investigating the
expression of STK33 mRNA and protein in normal human adult and
embryonic tissues demonstrated that STK33 is expressed in a variety
of normal tissues but at very low levels (8). However, it was observed to be highly
expressed in the testis, particularly in the spermatogenic
epithelium. It has also been demonstrated that STK33 involved in
the ‘synthetic lethality’ process in a variety of tumor cells,
which occurs when deficiency in the expression of multiple genes
results in cell death and depends on the Ras oncogene (9). This finding implies that STK33 may serve
a significant role in molecular targeted therapy for KRAS-dependent
tumors (9). By contrast, a different
study demonstrated that the activity of STK33 may be nonessential
in KRAS-dependent cell lines (10).
Therefore, the role of STK33 in tumor cells remains controversial
and the mechanisms underlying the function of STK33 in tumor
biology are complex. Previous studies have demonstrated that STK33
is overexpressed in hypopharyngeal squamous cell carcinoma (HSCC)
(11), hepatocellular carcinoma (HCC)
(12) and human large cell lung
cancer (LC) (13), and the increased
expression of STK33 may subsequently promote tumorigenesis and
disease progression. However, previous research aiming to identify
hypermethylated genes in CRC, demonstrated that the STK33 gene is
hypermethylated and its expression is subsequently downregulated
when compared with normal colorectal tissues (14). This is inconsistent with the results
of the aforementioned studies and therefore requires further
investigation.
To date, the role of STK33 in CRC tumorigenesis and
its clinical significance remains unclear. Therefore, the aim of
the current study was to investigate the methylation status of
STK33 in CRC cell lines and patients with CRC. The expression of
STK33 mRNA and protein in the collected tumor tissues and its
association with cell proliferation was also investigated. In
addition, the association between STK33 methylation status and the
clinicopathological characteristics of patients with CRC was
evaluated to determine the clinical significance of STK33 in
CRC.
Materials and methods
Patients and samples
A total of 94 patients (46–72 years old, 49 males
and 45 females) with CRC who underwent surgical treatment at the
Liaoning Cancer Hospital & Institute (Shenyang, China) between
October 2007 and May 2010, were recruited to this study. In order
to be included in the study, patients were required to have not
received any prior anticancer therapies, including chemotherapy,
radiotherapy or surgery prior to enrollment to the present study.
Follow up began the day after the surgery and lasted for 60 months.
Fresh tumor tissues and adjacent non-cancerous tissues obtained
from patients with CRC were frozen in liquid nitrogen following
resection and stored at −80°C for subsequent experiments. The
clinicopathological features of each patient are shown in Table I. Tumor grades were defined in
accordance with the criteria of the World Health Organization
(15). The pathological tumor, node,
metastasis (TNM) status of all CRC cases was defined according to
the criteria of the sixth edition of the TNM classification of the
International Union Against Cancer (2002) (16). The current study was approved by the
Ethics Committee of Liaoning Cancer Hospital & Institute, and
written informed consent was obtained from all recruited
patients.
 | Table I.Association between STK33 methylation
and the clinicopathological features of patients with colorectal
cancer. |
Table I.
Association between STK33 methylation
and the clinicopathological features of patients with colorectal
cancer.
|
|
| STK33 methy lation
status |
|
|---|
|
|
|
|
|
|---|
| Variable | No. of cases | Methylated | Unmethylated | P-value |
|---|
| Gender |
|
|
|
|
| Male | 49 | 30 | 19 | NS |
|
Female | 45 | 27 | 18 |
|
| Age (years) |
|
|
|
|
| ≥50 | 54 | 29 | 25 | NS |
|
<50 | 40 | 26 | 14 |
|
| Tumor location |
|
|
|
|
|
Colon | 59 | 38 | 21 | NS |
|
Rectum | 35 | 17 | 18 |
|
| Lymph node
metastases |
|
|
|
|
|
Absent | 58 | 39 | 19 | 0.041 |
|
Present | 36 | 16 | 20 |
|
| Tumor invasion |
|
|
|
|
|
T1/T2 | 52 | 30 | 22 | 0.032 |
|
T3/T4 | 42 | 25 | 17 |
|
| CEA level |
|
|
|
|
| No | 50 | 31 | 19 | NS |
| Yes | 44 | 24 | 20 |
|
| Distant
metastasis |
|
|
|
|
|
Absent | 63 | 43 | 20 | 0.006 |
|
Present | 31 | 12 | 19 |
|
| Tumor stage |
|
|
|
|
| I–II | 65 | 30 | 35 | 0.002 |
|
III–IV | 29 | 25 | 4 |
|
Cell lines
The normal colon cell line, NCM460, and DLD-1 and
HCT-116 CRC cell lines were obtained from the American Type Culture
Collection (Manassas, VA, USA). The NCM460 cell line was cultured
in complete Dulbecco's modified Eagle's medium (Gibco; Thermo
Fisher Scientific, Inc., Waltham, MA, USA), and the CRC cell lines
were cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific,
Inc.). All media was supplemented with 10% fetal bovine serum
(Gibco; Thermo Fisher Scientific, Inc.) and 1% streptomycin and
penicillin (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). All
cells were cultured in a humidified 37°C incubator supplemented
with 5% CO2.
Genomic DNA extraction
Genomic DNA from the NCM460, DLD-1 and HCT-116 cell
lines and fresh frozen tissues was extracted using the QIAamp DNA
Mini kit (Qiagen, Inc., Valencia, CA, USA) according to the
manufacturer's protocols, and the procedures described previously
(14). Briefly, cell pellets
harvested from cell culture medium through centrifugation for 2 min
at 6,000 × g and 4°C, and tissue samples minced using 3-mm diameter
grinders, were mixed with 700 µl lysis buffer containing 20 µg/ml
protease K (Sigma-Aldrich; Merck KGaA), 20 mM Tris HCl (pH 8.0), 5
mM EDTA (pH 8.0), 400 mM NaCl, and 1% SDS solution (Sigma-Aldrich;
Merck KGaA). The mixture was then incubated at 42°C for ~12 h.
Genomic DNA was purified using the phenol/chloroform extraction
method. Purified DNA was then eluted in 100 µl water and quantified
using a Thermo Scientific NanoDrop 2000/2000C Spectrophotometer
(Thermo Fisher Scientific, Inc., Pittsburgh, PA, USA).
Quantitative methylation-specific
polymerase chain reaction (QMSP)
The genomic DNA obtained from the cell lines and
fresh frozen tissues was bisulfite-treated using the EpiTect fast
DNA bisulfite kit (Qiagen, Inc.), in order to convert any
unmethylated cytosine residues in CpG islands into uracil residues,
while any non-methylated cytosines remained unchanged. All
procedures were performed according to the manufacturer's
protocols. The reaction mixture consisted of 85 µl bisulfite mix
solution and 35 µl DNA protection buffer, and was incubated at room
temperature for 30 min. The bisulfite-converted genomic DNA was
eluted from columns with 50 µl ddH2O and stored at −80°C
for further experiments.
The methylation status of the bisulfite-converted
genomic DNA was measured by methylation-specific PCR using the
Applied Biosystems 7500 Real-Time PCR System (Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. The
primer pairs used to detect methylation status were as follows:
STK33, forward, 5′-GTGCGTATTTGTCGGAGATTC-3′, and reverse,
5′-TACCATAACAACGACCTAACCG-3′; β-actin, forward,
5′-TGGTGATGGAGGAGGTTTAGTAAGT-3′, and reverse,
5′AACCAATAAAACCTACTCCTCCCTTAA-3′. Reactions were performed using
PCR vials containing 15 µl 2X Maxima SYBR Green/ROX qPCR Master Mix
(Thermo Fisher Scientific, Inc.), 250 nM each forward and reverse
primer, 30 ng bisulfite-converted genomic DNA template and
ddH2O to a final volume of 30 µl. Experiments were
performed in triplicate for each sample. The QMSP program was set
as follows: Denaturation at 94°C for 3 min followed by 40 cycles at
94°C for 15 sec, 60°C for 10 sec and 72°C for 10 sec. There was an
additional extension step at 72°C for 7 min prior to the completion
of PCR. Relative quantification of the amplified gene levels in the
bisulfite-converted DNA sample was achieved by measuring the
quantification cycle (Cq) values of STK33 and β-actin
and applying the 2−ΔΔCq method (17). PCR products were loaded directly onto
3% agarose gels and visualized with ethidium bromide. The gel was
visualized using a gel image analysis system (JC-300; Shanghai
Peiqing Science and Technology Co., Ltd., Shanghai, China).
STK33 RNA inhibition
The expression of STK33 was knocked down in NCM460,
DLD-1 and HCT-116 cell lines using an STK33-RNA inference
(STK33-RNAi) vector constructed by Shanghai GeneChem Co, Ltd.
(Shanghai, China). The STK33 small interfering RNA (siRNA)
oligonucleotide sequences were as follows: Forward,
5′-GATCCCAGAGAATGAGACAAGGTGTTCAAGAGACACCTTGTCTCATTCTCTGAGA-3′, and
reverse,
5′-AGCTTCTCAGAGAATGAGACAAGGTGTCTCTTGAACACCTTGTCTCATTCTCTGG-3′. The
plasmid was transfected into cells according to a previously
published protocol (12). Following
transfection with Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) for 24 h, cells were washed
with PBS for reverse transcription-quantitative polymerase chain
reaction (RT-qPCR) analysis and the cell proliferation assay. The
cells without siRNA transfection were used as controls.
RT-qPCR
Total RNA from cell lines and fresh frozen tissues
was isolated using TRIzol reagent (Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocols. Total RNA was eluted in
20 µl DEPC-treated water and the concentration was determined using
the Thermo Fisher NanoDrop 2000/2000C Spectrophotometer (Thermo
Fisher Scientific, Inc.). cDNA was synthesized from 1 µg RNA
obtained from each sample using the PrimeScript RT Reagent kit
(Takara Biotechnology Co., Ltd., Dalian, China). The reaction
mixture consisted of 1 µg RNA, 4 µl 5X PrimeScript buffer, 1 µl 50
µM oligo-dT primer, 1 µl PrimeScript RT Enzyme mix and 11.5 µl
ddH2O, and the reaction was conducted at 37°C for 1 h.
cDNA was eluted with ddH2O and stored at −80°C.
qPCR was performed to determine the expression of
STK33 in cell lines and fresh frozen tissues using SYBR Green qPCR
SuperMix UDG with ROX kit (Invitrogen; Thermo Fisher Scientific,
Inc.). The Applied Biosystems 7500 Real-Time PCR System (Thermo
Fisher Scientific, Inc.) was used and the following thermal cycling
parameters were applied: 95°C for 10 min followed by 35 cycles of
95°C for 10 sec and 60°C for 1 min. The primers used were as
follows: STK33, forward, 5′-CTTCGGTGAGACCAACCAAT-3′ and reverse,
5′-TGTAATTGGCATCAGGGACA-3′; and β-actin, forward,
5′-AGAGCTACGAGCTGCCTGAC-3′ and reverse, 5′-AGCACTGTGTTGGCGTACAG-3′.
The expression of β-actin was used as an endogenous standard to
normalize the expression of STK33 using the 2−ΔΔCq
method (17). Each sample was tested
at least 3 times simultaneously.
Western blot analysis
Total proteins were extracted from cell lines and
fresh tissues using radioimmunoprecipitation assay lysis buffer
(Santa Cruz Biotechnology, Inc., Dallas, TX, USA) and centrifuged
at 14,000 × g for 10 min at 4°C to obtain crude protein extracts.
The concentration of the protein extracts was measured by BCA
Protein Assay Kit (Thermo Fisher Scientific Inc.). Protein samples
(50 µg) were separated by 10% sodium dodecyl sulfate-polyacrylamide
gel electrophoresis and then transferred onto a PVDF membrane. The
membrane was blocked with 5% skimmed milk at room temperature for 1
h and then incubated with the following primary antibodies: STK33
mouse anti-human monoclonal antibody (dilution, 1:1,000; cat no.
ab57693) and β-actin mouse anti-human monoclonal antibody
(dilution, 1:1,000; cat no. ab8226; both Abcam, Cambridge, MA, USA)
at 4°C for ~12 h. The membrane was then incubated with horseradish
peroxidase-conjugated goat anti-mouse secondary antibody (1:1,000;
cat no. ab97040; Abcam) at room temperature for 2 h. The bands were
visualized using enhanced chemiluminescence reagent (EMD Millipore,
Billerica, MA, USA) and quantified by densitometry analysis using
ImageJ software (version 1.48; National Institutes of Health,
Bethesda, MD, USA).
Cell proliferation assay
An MTT assay was performed to measure the
proliferation of the normal colon and CRC cell lines, as well as
STK33-RNAi transfected cells in the current study. Briefly, cells
were seeded in a 96-well plate at a cell density of
3×103 cells/ml for 24 h. A total of 20 µl MTT solution
(5 mg/ml; Sigma-Aldrich; Merck KGaA) was then added to each well at
indicated time points (12, 24, 48 and 72 h), and cells were
incubated for 4 h at 37°C. Following incubation, the cell culture
medium was removed and 150 µl DMSO was added to each well to
dissolve the formazan crystals. The optical density was measured at
a wavelength of 570 nm using a Thermo Multiskan Spectrum
spectrophotometer (Thermo Fisher Scientific, Inc.). Each
measurement was performed in triplicate under the same
conditions.
Immunohistochemistry analysis
(IHC)
IHC analysis was performed on the frozen tumor and
adjacent normal tissues obtained from patients with CRC. Tissues
were fixed in 4% formalin at room temperature for 48 h and embedded
in paraffin. The paraffin-embedded tissues (5 µm) were then
deparaffinized in 100% xylene (Sigma-Aldrich; Merck KGaA) and
rehydrated using a graded ethanol series. Tissues were subsequently
washed using citrate buffer (pH 6.0) 3 times (1 min each time) and
incubated with 5% goat serum (Beyotime, Shanghai, China) for 30 min
at 37°C. Following washing with PBS, sections were incubated with
the primary mouse anti-human STK33 monoclonal antibody for 24 h at
4°C (dilution, 1:1,000; cat no. ab57693), followed by the
horseradish peroxidase conjugated goat anti-mouse secondary
antibody (dilution, 1:1,000; cat no. ab97040; both Abcam) for 2 h
at 4°C. All sections were counterstained with 0.2% hematoxylin for
5 min at room temperature, dehydrated with ethanol and washed with
100% xylene for 10 sec at room temperature. The Olympus CX31-LV320
microscope (Olympus Corporation, Tokyo, Japan) was used to observe
the stained tissues.
Statistical analysis
The results were analyzed using GraphPad Prism 6.0
statistical software (GraphPad Software, Inc. La Jolla, CA, USA).
Data are presented as the mean ± standard deviation. A Student's
t-test or one-way analysis of variance followed by Tukey's test
were conducted to determine the significance between or among
different groups. A χ2 test and Fisher's exact test were
used to examine the association of STK33 methylation and various
clinicopathological parameters. Overall survival was analyzed using
the Kaplan-Meier method and the statistical significance between
survival curves was assessed by log-rank test. Univariate Cox
proportional hazard regressions were applied to estimate the
individual hazard ratios for overall survival. The variables that
were significant in the univariate analysis were included in the
multivariate analysis. P<0.05 was determined to indicate a
statistically significant difference.
Results
STK33 is hypermethylated in CRC cell
lines
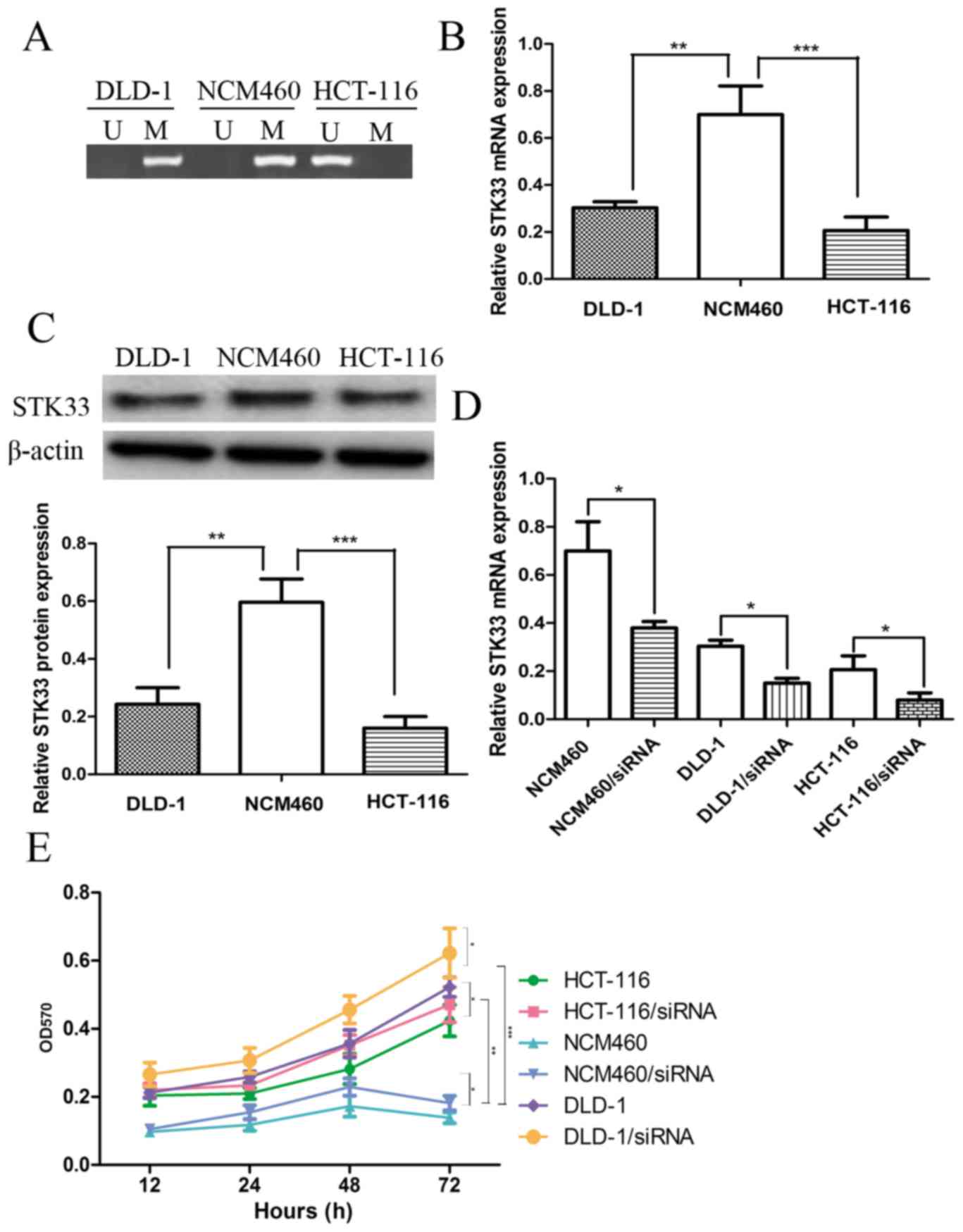
The methylation status of STK33 in CRC cell lines
was investigated by performing QMSP in normal colon cells, NCM460,
and CRC cell lines, DLD-1 and HCT-116. The STK33 in CRC cell lines
was methylated while STK33 in normal colon cell was unmethylated
(Fig. 1A). The expression of STK33
mRNA and protein in NCM460, DLD-1 and HCT-116 cells was then
determined to investigate the effect of STK33 hypermethylation on
STK33 expression. The expression of STK33 mRNA and protein was
significantly decreased in the DLD-1 (both P<0.01) and HCT-116
(P<0.001) cell lines compared with NCM460 cells (Fig. 1B and C). These results suggest that
STK33 may serve a crucial role in the development of CRC, and may
also present a useful biomarker.
STK33 hypermethylation may promote
cell proliferation
The hypermethylation status of STK33 in CRC cell
lines DLD-1 and HCT-116 was established through QMSP analysis. The
results indicated that STK33 hypermethylation may lead to the
downregulation of STK33 at the mRNA and protein expression levels.
siRNA was used to knock down the expression of STK33 in NCM460,
DLD-1 and HCT-116 cell lines. The results of RT-qPCR demonstrated
that STK33 siRNA efficiently downregulated the expression of STK33
in these cell lines (Fig. 1D). The
role of STK33 hypermethylation on the progression and development
of CRC was investigated by analyzing the cell proliferation rate of
CRC and normal colon cells, as well as the STK33-downregulated cell
lines, using an MTT assay. The results demonstrated that the cell
proliferation rate of DLD-1 and HCT-116 CRC cells was increased
compared with normal NCM460 cells (Fig.
1E). The cell proliferation rate of cells with RNAi-induced
knockdown of STK33 expression was significantly increased when
compared with the untransfected cells (P<0.01, Fig. 1E). Taken together these results
suggest that the hypermethylation of STK33 in CRC cell lines may
promote CRC cell proliferation.
STK33 hypermethylation in patients
with CRC
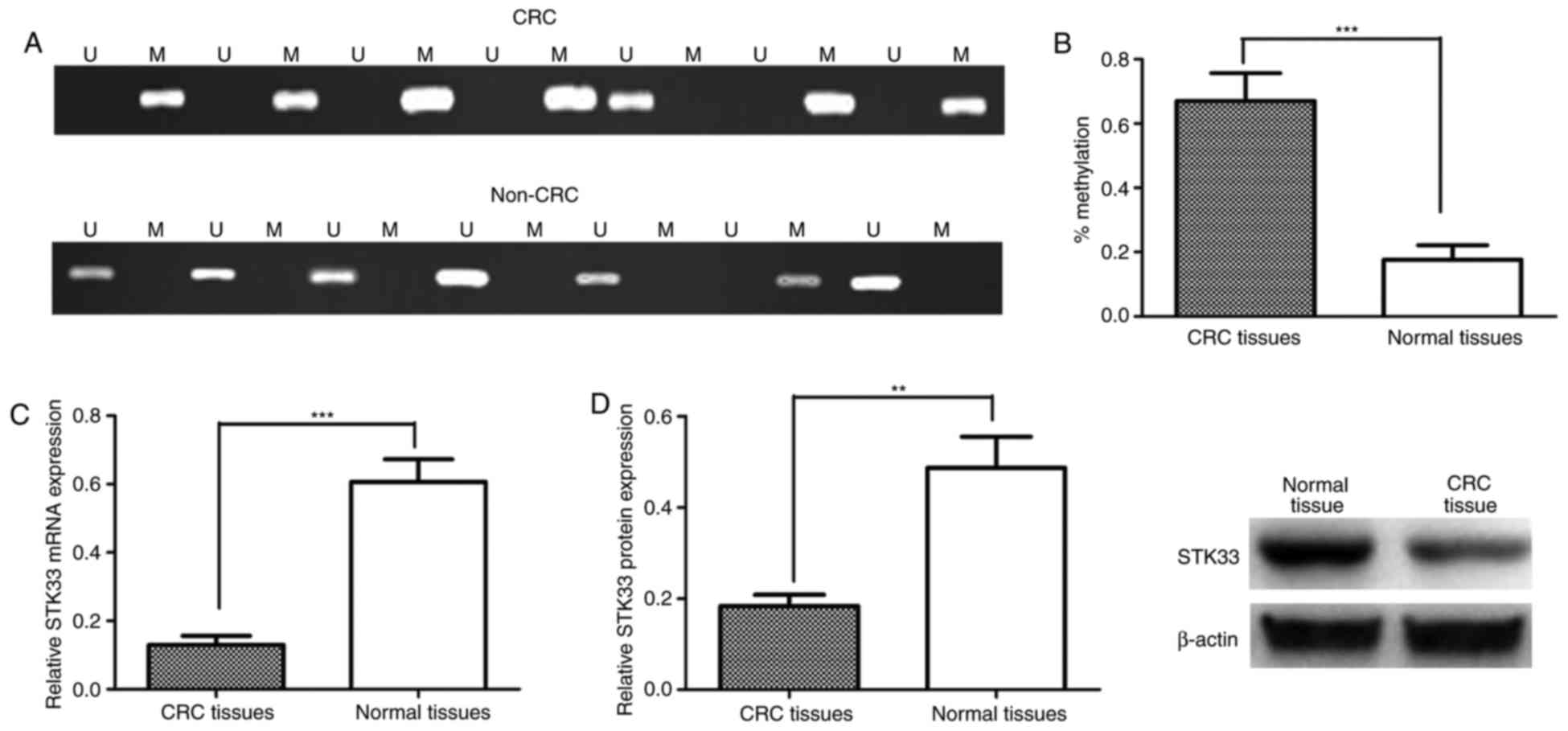
The potential role of STK33 in the development and
progression of CRC was investigated further by determining the
methylation status of STK33 in 94 pairs of CRC tissues and adjacent
noncancerous tissues obtained from patients with CRC enrolled in
the current study. STK33 was observed to be hypermethylated in
55/94 (58.51%) CRC tissue samples and 18/94 adjacent noncancerous
tissue samples (19.15%; Fig. 2A). The
methylation status of STK33 in CRC and adjacent noncancerous tissue
samples was then compared. The results demonstrated that
methylation of STK33 was significantly increased in CRC tissues
when compared with adjacent noncancerous tissues (P<0.001;
Fig. 2B). The expression of STK33
mRNA and protein in CRC and adjacent noncancerous tissues was then
measured using RT-qPCR and western blot analysis, respectively, to
examine the effect of hypermethylation on the expression of STK33.
The expression of STK33 mRNA and protein was significantly
decreased in CRC tissues compared with adjacent noncancerous
tissues (P<0.001 and P<0.01, respectively; Fig. 2C and D). The association between STK33
hypermethylation and mRNA expression was also investigated
(Table II). The results indicated
that downregulation of STK33 mRNA was associated with STK33
hypermethylation (P<0.001).
 | Table II.Univariate analysis of the association
between STK33 methylation and mRNA expression levels. |
Table II.
Univariate analysis of the association
between STK33 methylation and mRNA expression levels.
|
| STK33 expression |
|
|---|
|
|
|
|
|---|
| Methylation
status | Downregulated
(n=51) | No change (n=43) | P-value |
|---|
| Methylated
(n=55) | 38 | 17 | <0.001 |
| Unmethylated
(n=39) | 13 | 26 |
|
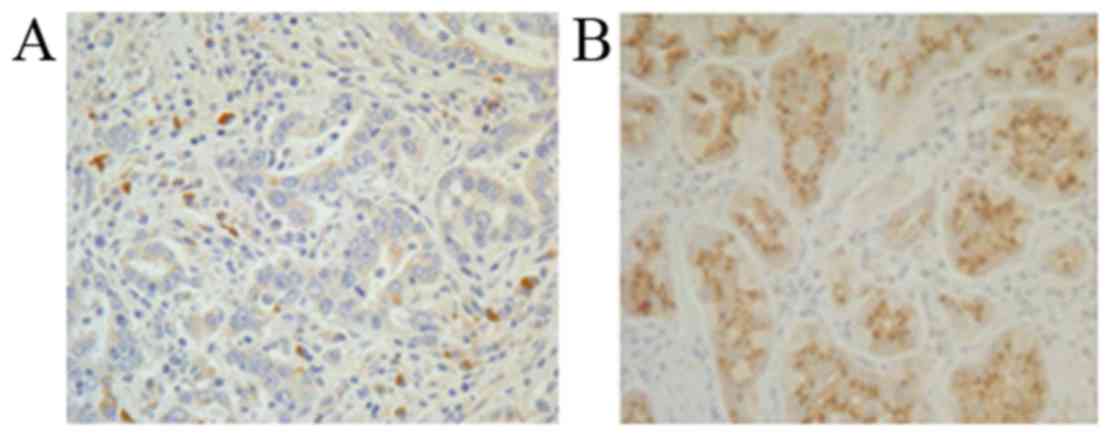
To investigate the expression and localization of
STK33 in CRC tissues, IHC analysis of the CRC and adjacent
noncancerous tissues was performed. Consistent with the conclusions
drawn from the RT-qPCR and western blot analyses, STK33 was
downregulated in CRC tissues compared with adjacent noncancerous
tissues (Fig. 3). The results suggest
that hypermethylation of STK33 in patients with CRC may be
responsible for the reduced expression of STK33 at the mRNA and
protein levels.
Clinical significance of STK33
hypermethylation in CRC
The clinical significance of STK33 hypermethylation
in CRC was determined by investigating the association between
STK33 hypermethylation and the clinicopathological parameters
recorded for patients with CRC. The results indicated the STK33
hypermethylation in CRC samples was associated with lymph node
metastasis (P<0.05), tumor invasion (P<0.05), distant
metastasis (P<0.01) and TNM stage (P<0.01; Table I). There was no association observed
between STK33 hypermethylation and the remaining
clinicopathological features, including age, gender, tumor location
and carcinoembryonic antigen (CEA) level (Table I).
STK33 hypermethylation is associated
with poor prognosis in CRC
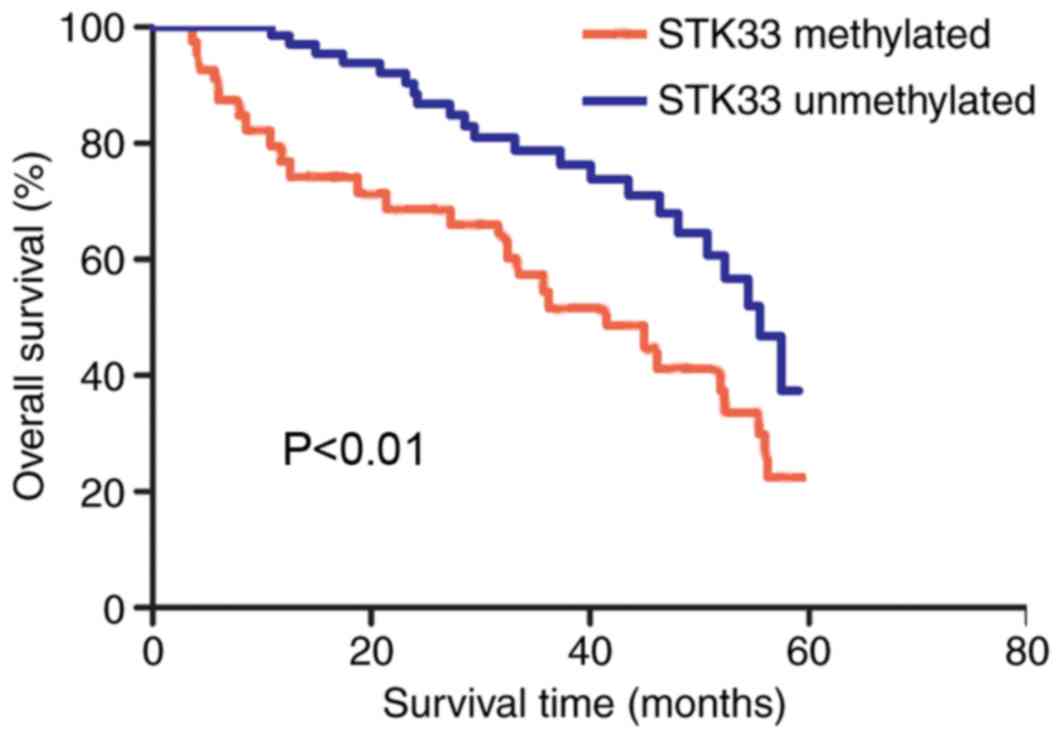
The overall survival rate of patients with
hypermethylated and unmethylated STK33 was compared using the
Kaplan-Meier method. The survival curves demonstrated that patients
with hypermethylated STK33 exhibited a significantly shorter
survival rate compared with those with unmethylated STK33
(P<0.01; Fig. 4). The univariate
analysis demonstrated that STK33 hypermethylation, lymph node
metastasis, tumor invasion, distant metastasis and TNM stage were
all associated with the overall survival of patients with CRC (all
P<0.05; Table III). However, no
association between overall survival and age, gender, tumor
location and CEA level were observed. Multivariate Cox regression
analysis was then used to analyze significant associations from the
univariate analysis. The results demonstrated that STK33
hypermethylation, lymph node metastasis, tumor invasion, distant
metastasis and TNM stage were independent predictive indicators of
a poor outcome in CRC (all P<0.05; Table III). The results therefore indicate
that STK33 hypermethylation may present a useful prognostic marker
for CRC.
 | Table III.Univariate and multivariate analysis
of the association between clinical features and overall
survival. |
Table III.
Univariate and multivariate analysis
of the association between clinical features and overall
survival.
| A, Univariate
analysis |
|---|
|
|---|
| Variable | HR | 95% CI | P-value |
|---|
| STK33 | 2.055 | 1.145–3.687 | 0.016 |
| Age | 1.555 | 0.867–2.787 | 0.138 |
| Gender | 1.490 | 0.826–2.688 | 0.185 |
| Tumor location | 1.174 | 0.967–3.039 | 0.065 |
| Lymph node
metastases | 2.059 | 1.147–3.695 | 0.015 |
| Tumor invasion | 1.878 | 1.059–3.331 | 0.031 |
| CEA level | 1.631 | 0.915–2.908 | 0.097 |
| Distant
metastasis | 1.961 | 1.100–3.498 | 0.022 |
| Tumor stage | 2.163 | 1.198–3.905 | 0.011 |
|
| B, Multivariate
analysis |
|
|
Variable | HR | 95% CI | P-value |
|
| STK33 | 2.147 | 1.189–3.874 | 0.011 |
| Age | – | – | – |
| Gender | – | – | – |
| Tumor location | – | – | – |
| Lymph node
metastases | 1.963 | 1.101–3.502 | 0.022 |
| Tumor invasion | 1.796 | 1.018–3.166 | 0.043 |
| CEA level | – | – | – |
| Distant
metastasis | 2.057 | 1.146–3.690 | 0.016 |
| Tumor stage | 2.061 | 1.149–3.699 | 0.015 |
Discussion
STK33
encodes a human kinase enzyme that was discovered by
comparative genomic analysis of human chromosome 11p15.3 and its
orthologous region on the distal region of mouse chromosome 7
(7). The STK33 gene consists of 12
exons and a 1,545 bp open reading frame, which was identified from
the full-length transcript amplified from human uterus RNA
(7). STK33 is differentially
expressed in a variety of normal and malignant tissues (18). Protein kinases serve a major role in
the regulation of a number of fundamental cellular processes
(19). The location of STK33 in the
human genome is strongly associated with many diseases (20); therefore, studies aiming to interpret
the role of STK33 expression in cancer have been conducted
(9). It has been demonstrated that
STK33 is overexpressed in HSCC, HCC and LC (10–13).
However, the STK33 gene has been demonstrated to be hypermethylated
and in CRC cell lines, and its mRNA expression levels have been
observed to be downregulated in tumor tissues compared with normal
tissues (14). These results are not
consistent with the results demonstrated in HSCC, HCC and LC. The
role of STK33 in CRC is currently unknown; therefore, the aim of
the present study was to investigate the methylation status of
STK33 in patients with CRC, and examine its potential as a novel
diagnostic and therapeutic target.
The present study demonstrated that the STK33 gene
was hypermethylated in CRC cell lines, which is inconsistent with
previous research (14). RT-qPCR and
western blotting analyses demonstrated that the expression of STK33
mRNA and protein in CRC cell lines was significantly reduced
compared with that in the normal colon cell line. The expression of
STK33 in the cell lines was then knocked down using siRNA, which
was subsequently verified by RT-qPCR. The effect of STK33
hypermethylation on CRC progression was investigated by measuring
the cell proliferation rate of the CRC and normal colon cell lines.
The cell proliferation rate was increased in the CRC cell lines
when compared with normal colon cells, which suggests that STK33
hypermethylation may promote cell proliferation. However, the
mechanisms underlying these effects require further study. It
should be noted that the limitation of the in vitro
experiment of the present study was the lack of scramble or
negative control siRNA transfected groups. The cells transfected
with siRNA targeting STK33 were used to investigate the effect of
STK33 on cell proliferation and those without siRNA transfection
were used as control.
The methylation of STK33 and its clinical
significance in 94 patients with CRC was then investigated. The
STK33 gene was hypermethylated in the majority of tissues from
patients with CRC, and further statistical analysis indicated that
the expression of STK33 mRNA and protein was significantly
decreased in CRC tissues with hypermethylated STK33 when compared
with those with unmethylated STK33. The results also indicated that
reduced STK33 expression was associated with STK33
hypermethylation. Therefore, gene methylation may be a major
mechanism for the silencing of STK33 expression in CRC. IHC
analysis also confirmed the results obtained from the RT-qPCR and
western blotting analysis demonstrating that STK33 expression was
downregulated in CRC tissue samples with STK33
hypermethylation.
The analysis of the association between STK33
hypermethylation and clinicopathological features demonstrated that
lymph node metastasis, tumor invasion, distant metastasis and TNM
stage were significantly associated with STK33 hypermethylation.
During the patient follow-up period, it was observed that the
overall survival of patients with hypermethylated STK33 was
decreased compared with those with unmethylated STK33. This
suggests that STK33 hypermethylation may be associated with a poor
outcome for patients with CRC. Furthermore, the univariate and
multivariate analyses indicated that STK33 hypermethylation, lymph
node metastasis, tumor invasion, distant metastasis and TNM stage
were independent predictive factors for the poor prognosis of
CRC.
In conclusion, the results of the present study
demonstrated that the STK33 gene was hypermethylated in CRC cell
lines and tissue samples when compared with normal controls, which
may be responsible for the observed reduction in STK33 mRNA and
protein expression levels. In addition, the results suggest that
STK33 hypermethylation may present a useful biomarker for the
prognosis of patients with CRC. To the best of the author's
knowledge, this is the first study to investigate the
hypermethylation status of STK33 in CRC, and explore the
association between STK33 hypermethylation and clinicopathological
features. Limitations of the present study include the small number
of enrolled patients. Therefore, future studies should include more
patients to verify the findings we obtained. Furthermore, more
research is required to further elucidate the mechanism of how
STK33 promotes CRC progression.
References
|
1
|
Siegel R, Desantis C and Jemal A:
Colorectal cancer statistics, 2014. CA Cancer J Clin. 64:104–117.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Li S, Wang J, Lu Y and Fan D: Screening
and early diagnosis of colorectal cancer in China: A 12 year
retrospect (1994–2006). J Cancer Res Clin Oncol. 133:679–686. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhao P, Dai M, Chen W and Li N: Cancer
trends in China. Jpn J Clin Oncol. 40:281–285. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen WQ, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li L and Ma BB: Colorectal cancer in
Chinese patients: Current and emerging treatment options. Onco
Targets Ther. 7:1817–1828. 2014.PubMed/NCBI
|
|
6
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mujica AO, Hankeln T and Schmidt ER: A
novel serine/threonine kinase gene, STK33, on human chromosome
11p15.3. Gene. 280:175–181. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mujica AO, Brauksiepe B, Saaler-Reinhardt
S, Reuss S and Schmidt ER: Differential expression pattern of the
novel serine/threonine kinase, STK33, in mice and men. FEBS J.
272:4884–4898. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Scholl C, Fröhling S, Dunn IF, Schinzel
AC, Barbie DA, Kim SY, Silver SJ, Tamayo P, Wadlow RC, Ramaswamy S,
et al: Synthetic lethal interaction between oncogenic KRAS
dependency and STK33 suppression in human cancer cells. Cell.
137:821–834. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Babij C, Zhang Y, Kurzeja RJ, Munzli A,
Shehabeldin A, Fernando M, Quon K, Kassner PD, Ruefli-Brasse AA,
Watson VJ, et al: STK33 kinase activity is nonessential in
KRAS-dependent cance. Cancer Res. 71:5818–5826. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Huang Lingyan LY, Chen C, Zhang GD, Ju YR,
Zhang JZ, Wang HB and Li JF: STK33 overexpression in hypopharyngeal
squamous cell carcinoma: Possible role in tumorigenesis. BMC
Cancer. 15:132015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang P, Chen H, Wu J, Yan A and Zhang L:
STK33 plays an important positive role in the development of human
large cell lung cancers with variable metastatic potential. Acta
Biochim Biophys Sin (Shanghai). 47:214–223. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yang T, Song B, Zhang J, Yang GS, Zhang H,
Yu WF, Wu MC, Lu JH and Shen F: STK33 promotes hepatocellular
carcinoma through binding to c-Myc. Gut. 65:124–133. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Moon JW, Lee SK, Lee JO, Kim N, Lee YW,
Kim SJ, Kang HJ, Kim J, Kim HS and Park SH: Identification of novel
hypermethylated genes and demethylating effect of vincristine in
colorectal cancer. J Exp Clin Cancer Res. 33:42014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Rindi G, Arnold R, Bosman FT, Bosman T,
Carneiro F, Hruban R and Theise N: Nomenclature and classification
of neuroendocrine neoplasms of the digestive system WHO
classification of tumours of the digestive system 4th edition. Int
Agen Res Cancer (IARC). 13–14. 2010.
|
|
16
|
Greene FL, Page DL, Fleming ID, Fritz A,
Balch CM and Haller DG: AJCC cancer staging handbook from the AJCC
cancer staging manual. 6th edition. Springer; New York: 2002,
View Article : Google Scholar
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bastienne B, Mujica AO, Herrmann H and
Schmidt ER: The Serine/threonine kinase Stk33 exhibits
autophosphorylation and phosphorylates the intermediate filament
protein Vimentin. BMC Biochem. 9:252008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wilmanns M, Gautel M and Mayans O:
Activation of calcium/calmodulin regulated kinases. Cell Mol Biol
(Noisy-le-grand). 46:883–894. 2000.PubMed/NCBI
|
|
20
|
Nowak NJ and Shows TB: Genetics of
chromosome 11: Loci for pediatric and adult malignancies,
developmental disorders, and other diseases. Cancer Invest.
13:646–659. 1995. View Article : Google Scholar : PubMed/NCBI
|