Introduction
Pediatric adrenocortical tumors (ACT) are a rare but
aggressive endocrine malignancy accountable for ~0.2% of childhood
cancer cases (1). ACTs comprise
benign adenomas and highly malignant carcinomas whose pathogenesis
remains incompletely understood (2).
The majority of pediatric ACTs are functional, with symptoms of
excessive androgen production, in comparison to adult ACT (1). Though histopathological demarcation
between adenoma and carcinoma is complicated, 80–90% of pediatric
ACTs are carcinomas (1). Complete
surgical resection remains the only potent treatment for pediatric
ACT (3).
The elements responsible for sporadic pediatric ACT
remain unknown, yet the resemblance of cases of ACT to cases with
inherent susceptibility indicates a common method of tumorigenesis
(4). Germline TP53 mutations
(Li-Fraumeni syndrome) or genetic and/or epigenetic modifications
affecting chromosome 11p15 (Beckwith-Wiedemann syndrome) are
commonly associated with childhood ACT (4). Insulin-like growth factor 2 (IGF2)
overexpression has been identified in pediatric adrenocortical
adenoma and carcinoma. In addition, IGF1R mRNA levels have been
demonstrated to be higher in pediatric adrenocortical carcinomas
(ACC) (5). Carcinomas have been
revealed to possess more chromosomal alterations when compared with
adenomas (6). In a prior study, it
was hypothesized that a number of genomic changes are responsible
for progression from normal tissue, to adenoma to carcinoma
(7).
A considerable number of disease-associated genes
have been identified by expression studies in previous years
(8). In the present study,
bioinformatics methodologies, such as protein-protein interaction
(PPI) network analyses (9), that are
based upon gene-encoded proteins interactions and gene module
analysis have been utilized to investigate the underlying
biological processes of progressing pediatric adrenocortical
adenoma. In the present study, differentially expressed genes
(DEGs) from pediatric adrenocortical adenoma and carcinoma were
utilized to construct PPI networks based on gene-encoded protein
interaction information. The networks were analyzed to identify
common nodes with high connectivity (hubs) among the adenoma and
carcinoma PPI networks, in order to isolate the genes responsible
for disease progression. Also, an overlapping gene module among
adenoma and carcinoma was identified to obtain prospective
molecules associated with the progressing disease. Additionally,
the association between common disease modules and pediatric ACC
was outlined.
Materials and methods
Gene expression data
The raw gene expression data was retrieved from Gene
Expression Omnibus (GEO; www.ncbi.nlm.nih.gov/geo/). The chip dataset GSE75415
(10) included 7 samples from healthy
individuals, 5 samples from pediatric adrenocortical adenoma and 18
samples from pediatric ACC. Gene expression profiling for adenoma
and carcinoma were performed using Affymetrix Human Genome U133
array (HG-U133A) chips (Affymetrix; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA).
Screening the DEGs
Pre-processing and normalization were performed to
remove noise from the expression dataset. For the normalization and
summarization of the expression dataset, Robust Multiarray
Averaging (11) in Affy package
version 1.46.1 (Bioconductor, Buffalo, NY, USA) (12) of R was utilized. Linear Models for
Microarray data (Limma) version 3.24.15 (Bioconductor) (13) package of R was employed to explore the
normalized data for differential analysis. In order to obtain the
adjusted P-values, multiple hypothesis testing correction was
performed using the Benjamini-Hochberg (14) method. Fold-change >2 and adjusted
P<0.05 were considered the be demarcating parameters for the
identification of DEGs in adenoma and carcinoma, as compared with
in normal tissues.
Construction of PPI networks
The extracted DEGs of adenoma and carcinoma were
mapped to construct PPI networks. The Search Tool for Retrieval of
Interacting Genes/Proteins (15) v10
database (https://string-db.org/) was explored to
identify the interacting proteins and elucidate their function at a
molecular level. Interacting proteins with a confidence score
>0.4 were selected for constructing PPI networks and were
visualized in Cytoscape version 3.2.0 (National Institute of
General Medical Sciences, Bethesda, MD, USA) (16). Network topological parameters, such as
network density, network centralization and network diameter, were
evaluated with the Network Analyzer (17) plug-in of Cytoscape. Common hub genes
among the PPI networks were also identified as being those with a
connectivity degree >10. For the construction of the common
sub-network, overlapping nodes and edges were mined by comparing
the nodes and edges of the adenoma and carcinoma disease
networks.
Identification of common modules
The common sub-network among adenoma and carcinoma
was scrutinized to explore high modularity clusters in the network,
as genes in the module tend to possess similar biological function.
The Molecular Complex Detection (MCODE) (18) plug-in of Cytoscape was employed to
identify the functional modules in the disease system with degree
cutoff =2, k-score =2, maximum depth =100 and node score
cutoff=0.2. Modules with an MCODE score ≥4 and nodes ≥6 were
considered for additional analysis. Furthermore, overlapping
genes/proteins shared between common modules and common hubs of the
adenoma and carcinoma PPI networks were also identified.
Enrichment analysis
Kyoto Encyclopedia of Genes and Genomes (KEGG)
pathway and Gene Ontology (GO) enrichment analysis was performed
through WEB-based gene set analysis toolkit (WebGestalt; http://www.webgestalt.org/) (19) on genes in the common module, to
identify the biological processes and pathways underlying the
disease system. WebGestalt incorporates proteomic, genomic and
functional categories from numerous public resources. The
Benjamini-Hochberg method was utilized to obtain adjusted P-values.
The cut-off criteria included P<0.05 and number of genes
>2.
Results
Identification of DEGs
Identifying and analyzing the dysregulated genes was
indispensable for evaluation of the common underlying mechanisms of
ACT adenoma and carcinoma. A total of 550 DEGs, including 376
upregulated and 174 downregulated genes, were identified in
adrenocortical adenoma tissues compared with in healthy
individuals, with an adjusted P<0.05 and fold-change >2.
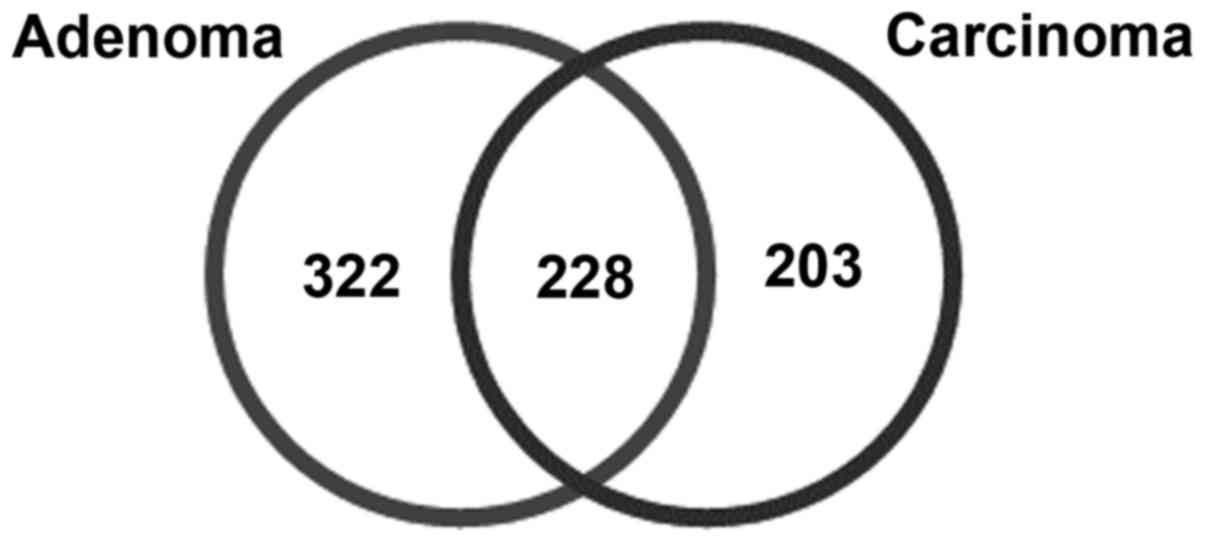
Additionally, 431 DEGs, including 228 upregulated and 203
downregulated genes, were documented for ACC, as compared with in
normal tissues, using the aforementioned predefined thresholds. A
total of 228 common DEGs were also identified among pediatric
adrenocortical adenoma and carcinoma (Fig. 1).
PPI network construction
The PPI network for adenoma was formed with 464
nodes and 1,947 edges with a confidence score >0.4. In addition,
a PPI network with 366 nodes and 1,858 edges was constructed for
carcinoma. The PPI networks followed a power law of distribution
with R2=0.841 and 0.836 for adenoma and carcinoma,
respectively. Various network parameters for the adenoma and
carcinoma PPI networks are summarized in Table I. A total of 53 common hub genes were
identified among adenoma and carcinoma PPI networks, with
connectivity degrees >10 (Table
II). A common sub-network with 186 nodes and 544 edges was
identified by scrutinizing overlapping nodes and edges among the
two PPI networks (Fig. 2).
 | Table I.Topological parameters of adenoma and
carcinoma PPI networks. |
Table I.
Topological parameters of adenoma and
carcinoma PPI networks.
| PPI network | No. of nodes | No. of edges | R2 | Correlation | Clustering
coefficient | Network
centralization | Network density | Network diameter |
|---|
| Adenoma | 467 | 1947 | 0.841 | 0.800 | 0.254 | 0.177 | 0.018 | 9 |
| Carcinoma | 366 | 1858 | 0.836 | 0.880 | 0.304 | 0.214 | 0.028 | 9 |
 | Table II.Common hubs and common module genes
among adenoma and carcinoma protein-protein interaction
networks. |
Table II.
Common hubs and common module genes
among adenoma and carcinoma protein-protein interaction
networks.
| Type | Gene symbol |
|---|
| Common hub genes | GAPDH, ACLY, AURKA,
FOS, HDAC1, EGR1, CAT, MMP2, SIRT1, SREBF1, H2AFZ, SKP2, CDC23,
SCD, HMGCR, BUB3, TCP1, TXN, JUNB, CREM, FDFT1, DICER1, PSMD14,
FDPS, PSMB2, SQLE, HMGCS1, DHCR7, DCN, CYP51A1, IGF2, PSMB4, PSMB7,
SPARC, NR4A1, MSMO1, HSD3B2, ACAT2, MARS, LSS, SC5DL, TARS, PGD,
RAD23B, VCAN, NR4A2, GEM, CANX, PTGIS, MAPK6, FOSB, IARS, CDH2 |
| Common module
genes | SQLE, PTGIS, SCD,
HSD3B2, DHCR7, MSMO1, C14orf1, HMGCR, FDFT1, SC5DL, LSS, CYP51A1,
HMGCS1, FDPS, ACLY, SKP2, PSMD14, GAPDH, NR4A1, EGR1, JUNB, PSMB2,
CREM, PSMB7, VCAN, PSMB4, DCN, CDC23, BUB3, SPARC, LUM, ODC1, GMNN,
AURKA |
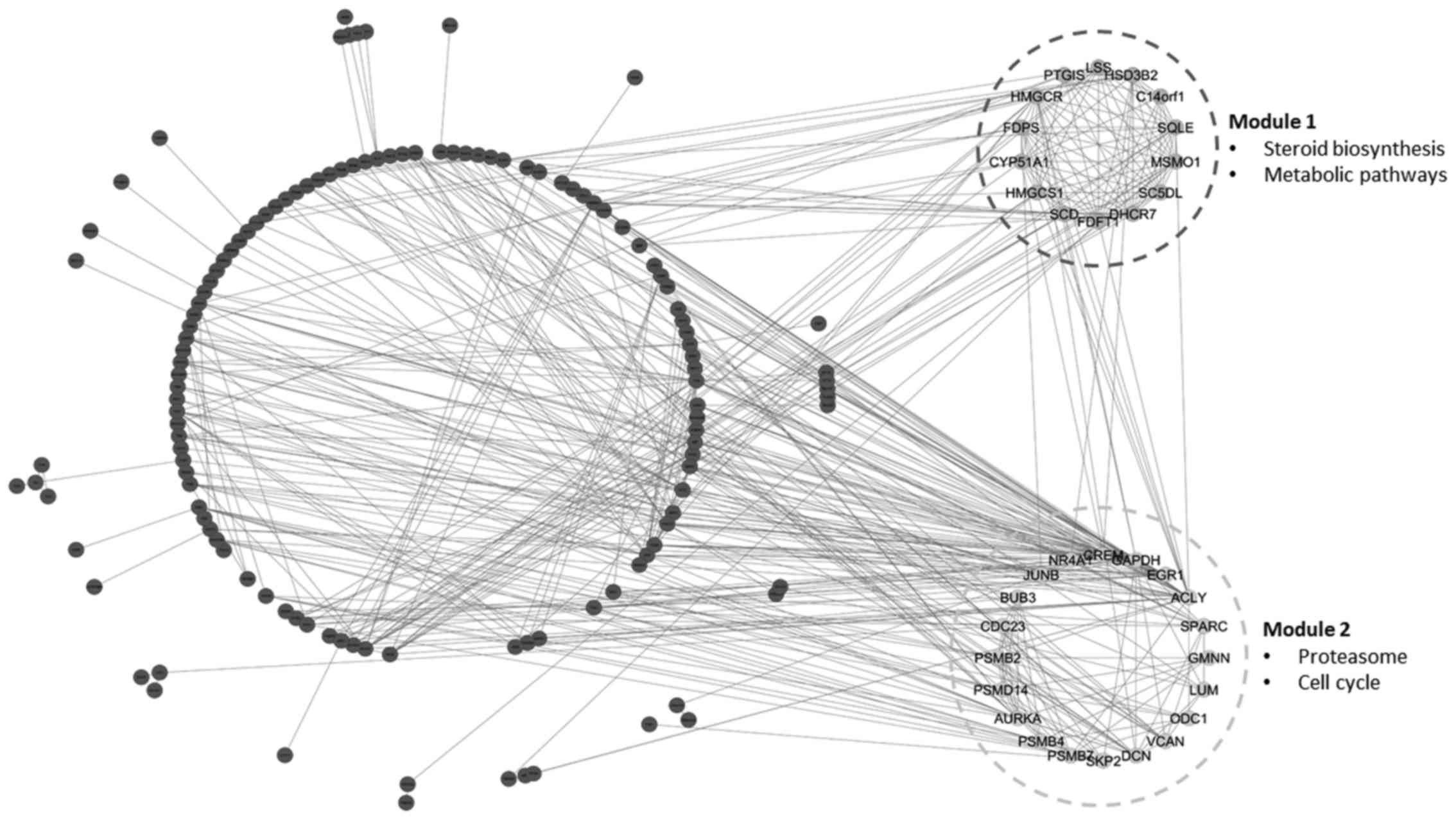
Common module identification
As an important step for identifying the common
functional module among adenoma and carcinoma disease models, the
common sub-network among adenoma and carcinoma was additionally
explored. A total of two common functional modules, module-1 (MCODE
score =12) with 14 nodes and module-2 (MCODE score =6.421) with 20
nodes, were observed among the PPI networks of adenoma and
carcinoma (Fig. 2). Notably, 12 genes
of module-1 and 13 genes of module-2 were upregulated in adenoma
and in carcinoma. Additionally, the majority of the genes of the
common modules were included in the common hub genes of PPI
networks (Table II).
Functional enrichment of modules
Utilizing the recommended threshold of P<0.05 and
number of genes >2, the KEGG pathway enrichment analysis
revealed the association of the genes in module-1 with steroid
biosynthesis and metabolic pathways. Genes in module-2 were
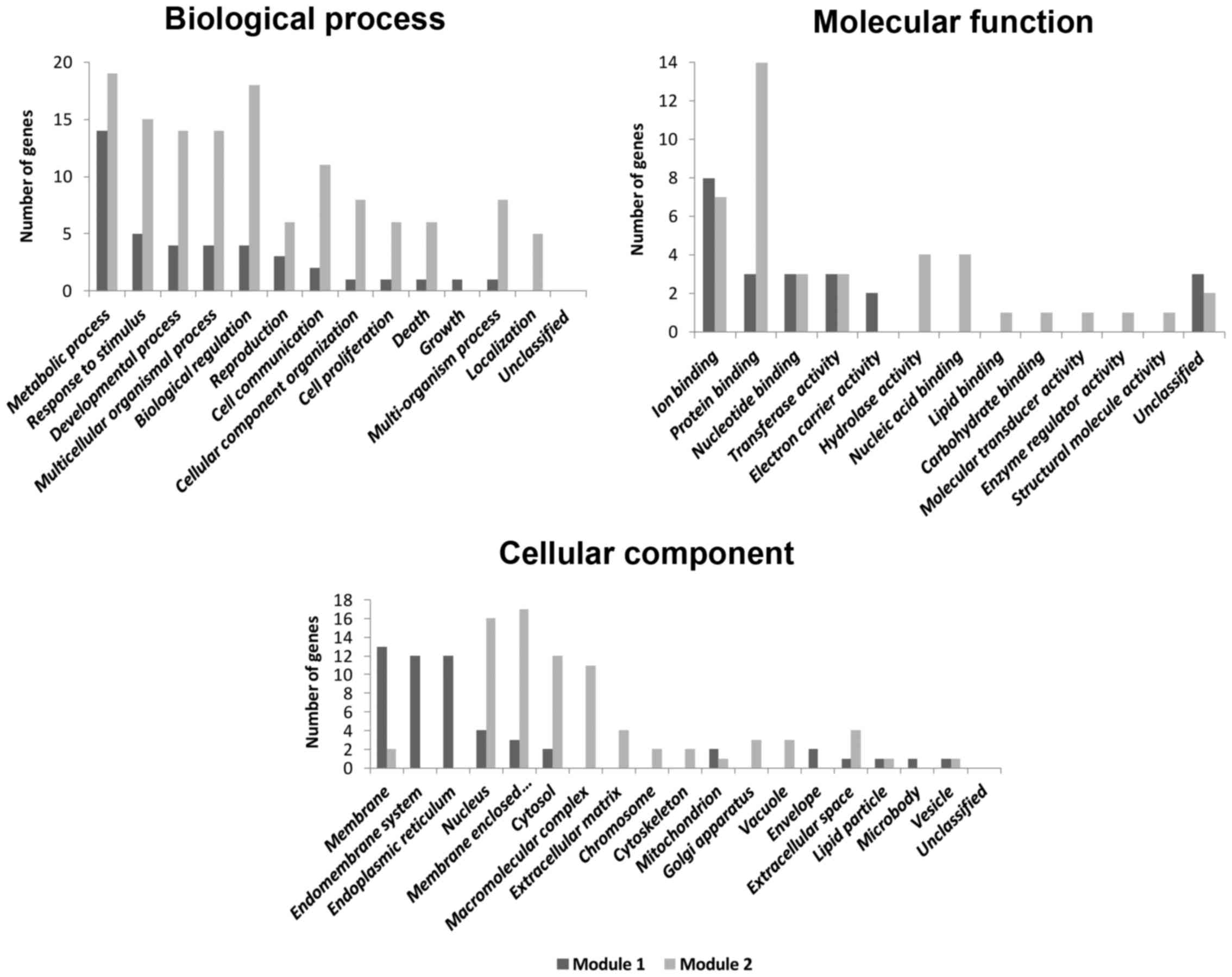
enriched in proteasome and cell cycle pathways (Table III). GO enrichment analysis
identified major biological processes, cellular components and
molecular functions associated with the genes of module-1 and
module-2 (Fig. 3).
 | Table III.Top five enriched KEGG pathways of the
genes of the common modules. |
Table III.
Top five enriched KEGG pathways of the
genes of the common modules.
| Term | Category | Description | Gene count | Adjusted P-value |
|---|
| Module 1 |
|
|
|
|
| KEGG | hsa00100 | Steroid
biosynthesis | 7 |
9.42×10−21 |
| KEGG | hsa01100 | Metabolic
pathways | 12 |
1.29×10−17 |
| KEGG | hsa00900 | Terpenoid backbone
biosynthesis | 3 |
1.24×10−8 |
| Module 2 |
|
|
|
|
| KEGG | hsa03050 | Proteasome | 4 |
2.26×10−8 |
| KEGG | hsa04110 | Cell cycle | 3 |
6.38×10−5 |
| KEGG | hsa04114 | Oocyte meiosis | 2 |
2.00×10−3 |
| KEGG | hsa04120 | Ubiquitin mediated
proteolysis | 2 |
2.30×10−3 |
| KEGG | hsa01100 | Metabolic
pathways | 3 |
1.47×10−2 |
Discussion
The present study aimed to recognize the genes that
serve an important role in the advancement of disease from normal,
to adenoma, to carcinoma. This required an integrative approach
combining differential expression and interaction network analyses.
In the present study, two networks were constructed based on the
differential expression of gene-encoded protein interaction
information. The degree (number of connected nodes to the disease
specific nodes) of disease-associated genes is appreciably higher
compared with the degree of the overall human interactome,
proposing their significant role when compared with other genes.
The data also support the hypothesis that disease-associated genes
possess higher numbers of connections (20,21). A
total of 53 genes were identified to be common hubs of the disease
system, and may perform important biological functions.
A total of 228 common DEGs were identified among
pediatric adrenocortical adenoma and carcinoma. There were two
common functional modules identified within the adenoma and
carcinoma disease models. Pathway enrichment of the genes in the
common module-1 revealed that the genes were enriched in steroid
biosynthesis and metabolic pathways. Genes in common module-2 were
associated with the proteasome and cell cycle-associated pathways.
The result indicates that there are two common gene modules that
support steroid biosynthesis, proteasome, metabolic pathways and
cell cycle, which are associated with adenoma and carcinoma. These
common modules may provide prospective molecular bases of disease
progression. Nodes of the common modules were identified to possess
greater average degrees compared with other nodes of the disease
models, indicating that the genes in the common modules may be
markedly more associated with disease processes compared with other
genes in the disease system. Overlap between common module genes
and common hubs of networks suggest that the deregulation of
steroid biosynthesis, metabolic pathways, proteasome and cell cycle
may assist in the development and progression of this disease.
These pathways also present prospective targets for future
research.
In the present study, steroid biosynthesis was one
of the affected pathways. Virilization and androgen hypersecretion,
primarily of cortisol, due to hormone excesses may be observed in
~90% of pediatric patients, providing an improved prognosis
compared with adult patients (22).
It has been proposed that targeting steroidogenic factor-1 may
decrease cortisol production and provide an antitumor effect
(23). Measurements of
dehydroepiandrosterone sulfate, hydroxyprogesterone, aldosterone,
urinary 17-ketosteroids and 17-hydroxycorticosteroid, testosterone,
plasma cortisol and free cortisol are routinely evaluated for
suspected patients with ACT (24). In
the present study, proteasome-associated pathways were also
demonstrated to be affected by this disease model. The
Wnt/β-catenin signaling pathway is crucial for cellular growth and
regulation of the adrenal gland (25). Abnormal accumulation of β-catenin
(CTNNB1) causes activation of the Wnt signaling pathway and
prevents its degradation by the ubiquitin-proteasome system
(25). CTNNB1-activating mutations
have been commonly identified in pediatric ACTs (4). Additionally, it has been proposed that
Wnt signaling inhibitors may assist in the treatment of childhood
ACC (26). Cell cycle-associated
pathways were revealed to be commonly affected by the disease
system in the present study. TP53 is a tumor suppressor gene
involved in cell cycle regulation, thereby causing cell cycle
arrest or cell death by DNA damage. Germline TP53 mutation is
markedly associated with pediatric ACT by promoting chromosomal
instability (4). It has been proposed
that loss of the normal inhibitory role of TP53 protein in the cell
cycle is associated with the development of ACC (27).
The majority of the genes of common module-1 and
module-2 are located in the membrane and membrane-enclosed lumen,
respectively, suggesting prospective targets for disease treatment.
Genes of module-1 and module-2 are enriched in biological processes
associated with metabolism, which provides mechanisms for
additional studies. The enriched molecular functions, such as ion
binding and protein binding, represent prospective drug
targets.
In conclusion, pediatric adrenocortical adenoma and
carcinoma disease models were explored to identify common hub genes
among adenoma and carcinoma, which serve roles in
disease-associated pathological processes. Additionally, two common
gene modules were identified among the adenoma and carcinoma
networks. The genes of these modules were also the common hubs of
the disease model. The present study exhibited certain limitations,
as the entire study was based on a bioinformatics approache, and
the results must be verified by further experiments.
References
|
1
|
Gulack BC, Rialon KL, Englum BR, Kim J,
Talbot LJ, Adibe OO, Rice HE and Tracy ET: Factors associated with
survival in pediatric adrenocortical carcinoma: An analysis of the
National Cancer Data Base (NCDB). J Pediatr Surg. 51:172–177. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ronchi CL, Sbiera S, Leich E, Henzel K,
Rosenwald A, Allolio B and Fassnacht M: Single nucleotide
polymorphism array profiling of adrenocortical tumors-evidence for
an adenoma carcinoma sequence? PLoS One. 8:e739592013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pinto EM, Morton C, Rodriguez-Galindo C,
McGregor L, Davidoff AM, Mercer K, Debelenko LV, Billups C, Ribeiro
RC and Zambetti GP: Establishment and characterization of the first
pediatric adrenocortical carcinoma xenograft model identifies
topotecan as a potential chemotherapeutic agent. Clin Cancer Res.
19:1740–1747. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pinto EM, Chen X, Easton J, Finkelstein D,
Liu Z, Pounds S, Rodriguez-Galindo C, Lund TC, Mardis ER, Wilson
RK, et al: Genomic landscape of paediatric adrenocortical tumours.
Nat Commun. 6:63022015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lima Lde O, Lerario AM, Alencar GA, Brito
LP, Almeida MQ, Domenice S, Latronico AC, Mendonca BB and Fragoso
MC: Clinical and molecular aspects of a pediatric metachronous
adrenocortical tumor. Arq Bras Endocrinol Metabol. 55:72–77. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lalli E and Figueiredo BC: Pediatric
adrenocortical tumors: What they can tell us on adrenal development
and comparison with adult adrenal tumors. Front Endocrinol
(Lausanne). 6:232015.PubMed/NCBI
|
|
7
|
Gara SK, Wang Y, Patel D, Liu-Chittenden
Y, Jain M, Boufraqech M, Zhang L, Meltzer PS and Kebebew E:
Integrated genome-wide analysis of genomic changes and gene
regulation in human adrenocortical tissue samples. Nucleic Acids
Res. 43:9327–9339. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Guo L, Du Y and Wang J: Network analysis
reveals a stress-affected common gene module among seven
stress-related diseases/systems which provides potential targets
for mechanism research. Sci Rep. 5:129392015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nair J, Ghatge M, Kakkar VV and Shanker J:
Network analysis of inflammatory genes and their transcriptional
regulators in coronary artery disease. PLoS One. 9:e943282014.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
West AN, Neale GA, Pounds S, Figueredo BC,
Rodriguez Galindo C, Pianovski MA, Oliveira Filho AG, Malkin D,
Lalli E, Ribeiro R and Zambetti GP: Gene expression profiling of
childhood adrenocortical tumors. Cancer Res. 67:600–608. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Irizarry RA, Hobbs B, Collin F,
Beazer-Barclay YD, Antonellis KJ, Scherf U and Speed TP:
Exploration, normalization and summaries of high density
oligonucleotide array probe level data. Biostatistics. 4:249–264.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gautier L, Cope L, Bolstad BM and Irizarry
RA: affy-analysis of Affymetrix GeneChip data at the probe level.
Bioinformatics. 20:307–315. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Smyth GK: Limma: Linear models for
microarray dataBioinformatics and computational biology solutions
using R and bioconductor. Springer; New York: pp. 397–420. 2005,
View Article : Google Scholar
|
|
14
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: A practical and powerful approach to
multiple testing. J Roy Stat Soc B Met. 57:289–300. 1995.
|
|
15
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43(Database Issue): D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Smoot ME, Ono K, Ruscheinski J, Wang PL
and Ideker T: Cytoscape 2.8: New features for data integration and
network visualization. Bioinformatics. 27:431–432. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Assenov Y, Ramírez F, Schelhorn SE,
Lengauer T and Albrecht M: Computing topological parameters of
biological networks. Bioinformatics. 24:282–284. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bader GD and Hogue CW: An automated method
for finding molecular complexes in large protein interaction
networks. BMC Bioinformatics. 4:22003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang J, Duncan D, Shi Z and Zhang B:
WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): Update 2013.
Nucleic Acids Res. 41(Web Server Issue): W77–W83. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jonsson PF and Bates PA: Global
topological features of cancer proteins in the human interactome.
Bioinformatics. 22:2291–2297. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sun J, Jia P, Fanous AH, van den Oord E,
Chen X, Riley BP, Amdur RL, Kendler KS and Zhao Z: Schizophrenia
gene networks and pathways and their applications for novel
candidate gene selection. PLoS One. 5:e113512010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Maluf DF, de Oliveira BH and Lalli E:
Therapy of adrenocortical cancer: Present and future. Am J Cancer
Res. 1:222–232. 2011.PubMed/NCBI
|
|
23
|
Aufforth RD and Nilubol N: Emerging
therapy for adrenocortical carcinoma. Int J Endocr Oncol.
1:173–182. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ribeiro RC, Michalkiewicz EL, Figueiredo
BC, DeLacerda L, Sandrini F, Pianovsky MD, Sampaio G and Sandrini
R: Adrenocortical tumors in children. Braz J Med Biol Res.
33:1225–1234. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Berthon A, Martinez A, Bertherat J and Val
P: Wnt/β-catenin signalling in adrenal physiology and tumour
development. Mol Cell Endocrinol. 351:87–95. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gundgurthi A, Kharb S, Dutta MK, Garg MK,
Khare A, Jacob MJ and Bhardwaj R: Childhood adrenocortical
carcinoma: Case report and review. Indian J Endocrinol Metab.
16:431–435. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pinto EM, Ribeiro RC, Figueiredo BC and
Zambetti GP: TP53-associated pediatric malignancies. Genes Cancer.
2:485–490. 2011. View Article : Google Scholar : PubMed/NCBI
|