Introduction
Bladder cancer is one of the most common types of
malignant tumors of the urinary and reproductive system, including
bladder urothelial cell carcinoma, squamous cell carcinoma,
adenocarcinoma and rare small cell carcinoma worldwide. Of these
subtypes, bladder urothelial cell carcinoma accounts for 90% of
cases. A series of pathogenic factors affect the occurrence of
bladder cancer, including aging, smoking and environmental
pollution (1). The mortality rate of
bladder cancer is increasing, even in childhood (1,2). The
pathogenesis of bladder cancer is complex, and involves the
activation of oncogenes and the inactivation of tumor suppressor
genes. The imbalanced activation of oncogenes and tumor suppressor
genes impairs the normal growth mechanism of cells, leading to
unrestricted cell proliferation, and increased invasiveness
(3,4).
MicroRNAs (miRNAs/miRs) are a family of endogenous
non-coding small RNA molecules with a length of 21–25 nucleotides.
miRNAs function primarily through the targeted degradation of mRNAs
and inhibition of translation into proteins. Therefore, miRNAs have
numerous biological activities (5,6). Notably,
miRNAs perform critical functions in the occurrence and development
of lung, liver, gastric, cervical, breast, and colon cancer
(7,8).
Direct evidence has also documented the abnormal expression of
miRNAs in bladder cancer tissue. miR-96 is a conserved miRNA that
may be involved in the occurrence and development of bladder cancer
(9,10). Further research also demonstrated that
miR-96 was highly expressed in bladder cancer tissue and the blood
plasma obtained from patients with bladder cancer (11–13).
Yamada et al (14) confirmed
that miR-96 expression in urine samples from bladder urothelial
carcinoma was significantly increased compared with that from
healthy subjects. Therefore, miR-96 is considered to be an
important tumor diagnostic marker for bladder cancer. However, to
the best of our knowledge, the influence of miR-96 on tumor cell
biology and its mechanisms have not been reported previously. In
the present study, miR-96 was selected as the research target; the
aim was to investigate the effects of miR-96 on bladder cancer cell
proliferation, apoptosis, invasion and metastasis and underlying
mechanisms.
Materials and methods
Cell culture
Human bladder cancer cell lines, T24 and 5637
(American Type Culture Collection, Bethesda, MD, USA) were cultured
in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal
bovine serum (FBS; Hyclone; GE Healthcare Life Sciences, Logan, UT,
USA) and 100 U/ml penicillin-streptomycin (Sigma-Aldrich; Merck
KGaA, Darmstadt, Germany) in 5% CO2 at 37°C. SV-HUC-1, a
normal uroepithelium cell line, was also obtained from the American
Type Culture Collection, and was used as the control. Cells at 60%
confluence were used in the following experiments.
Reverse transcription-polymerase chain
reaction (RT-PCR)
Total RNA was extracted according to the instruction
of the TRIzol kit (Takara Biotechnology Co., Ltd., Dalian, China)
and the purity of RNA was confirmed by optical density (OD) at
280/260 nm. Following this, RNA was amplified using a one-step
RT-PCR kit (Dalian Baosheng Biological Engineering Co., Ltd.,
Dalian, China) and PCR products were detected using 2% agarose gel
electrophoresis. The primers were added into a 25-µl PCR reaction
system following a protocol of 94°C denaturation for 45 sec, 59°C
annealing for 45 sec and 72°C extension 60 sec for 35 cycles. The
primers used are as follows: miR-96 forward,
5′-TTTGGCACTAGCACATT-3′ and reverse, 5′-TTTGGCACTAGCACATT-3′; human
ether-à-go-go-related (HERG1) potassium channel forward,
5′-TCCAGCGGCTGTACTCGGGC-3′ and reverse,
5′-TGGACCAGAAGTGGTCGGAGAACTC-3′; and GAPDH forward,
5′-AGCCACATCGCTCAGACA-3′ and reverse 5′-TGGACTCCACGACGTACT-3′.
Cell transfection
When the confluence of T24 cells reached 50–70%,
Lipofectamine™ 2000 (Thermo Fisher Scientific, Inc.) was
applied to assist the transfection of scrambled control (SC) and
miR-96 inhibitor. Then, 6 h later, the medium was changed to DMEM.
The miR-96 inhibitor (5′-AGCAAAAAUGUGCUAGUGCCAAA-3′) (1 µl) and SC
(5′-CAGUACUUUUGUGUAGUACAA-3′) were synthesized using Shanghai
GenePharma Co., Ltd. (Shanghai, China). Cells were then cultured in
DMEM containing 10% FBS for another 48 h. RT-PCR was applied to
detect miR-96 expression.
MTT assay
T24 cells were seeded in 96-well plates. When the
cell confluence reached 50–70%, Lipofectamine 2000 was applied to
transfect cells with SC and miR-96 inhibitor, as aforementioned. At
6 h later, the medium was changed into DMEM. The cells were then
cultured in DMEM containing 10% FBS for another 24, 48 and 72 h. An
MTT assay was utilized to detect cell proliferation, as previously
described (15). In total, 150 µl
DMSO (Sigma-Aldrich; Merck KGaA) was added to each well in order to
dissolve formazan. The optical density (OD) was determined using a
microplate reader at 570 nm.
Flow cytometry
Following transfection with miR-96 inhibitor and SC
for 48 h, the T24 cells were collected for Annexin V-fluorescein
isothiocyanate/propidium iodide (PI) staining using the Annexin
V-FITC Apoptosis Detection kit (cat. no. C1062; Beyotime Institute
of Biotechnology, Ningbo, China) and the proportion of apoptotic
cells was detected within 1 h using a FACSCalibur flow cytometer
(BD Biosciences, Franklin Lakes, NJ, USA) and data were analyzed
using CellQuest Pro (version 5.1; BD Biosciences, Franklin Lakes,
NJ, USA). Following transfection for 48 h, the cells were collected
for PI staining and the cell cycle distribution was assessed by
FACSCalibur (BD Biosciences) within 1 h of staining.
Transwell assay
Following transfection with miR-96 inhibitor and SC
for 48 h, T24 cells (3×103) were digested and seeded
into the upper chamber of Transwell with one-day starvation of FBS
(Hyclone; GE Healthcare Life Sciences). The lower chamber contained
DMEM with 10% FBS. the cells were seeded in the upper chamber. The
lower chamber contained only DMEM. At 48 h after seeding, the cells
in lower chamber were fixed in 4% paraformaldehyde at room
temperature for 30 min and stained with 1% crystal violet for 5 min
at room temperature. Images were captured using a light microscope
(magnification, ×200). At least five fields in each image were
counted.
Western blotting
Following transfection with miR-96 inhibitor or SC
for 48 h, the cells were collected for biochemical experiments.
Protein concentration was quantified using the BCA method. An equal
amount of protein per lane (20 µg) separated using 10% SDS-PAGE.
Following electrophoresis, proteins were transferred to a
nitrocellulose membrane. Non-specific protein binding was blocked
with 4% non-fat milk (2 h at room temperature). Membranes were
incubated with anti-HERG1 (1:1,000; cat. no. ab196301; Epitomics;
Abcam, Cambridge, MA, USA) and anti-GAPDH (1:1,000; cat. no. AG019;
Beyotime Institute of Biotechnology, Haimen, China) primary
antibodies. Subsequent to rinsing with 0.1% phosphate-buffered
saline (0.1% Tween-20), the membranes were incubated with a
horseradish peroxidase-labeled secondary antibody (1:100; cat. no.
ab131368; Abcam) at room temperature for 2 h. The signal was
detected using an Enhanced Chemiluminescence Detection kit (GE
Healthcare, Chicago, IL, USA) and scanned using
ChemiDoc™ XRS (Bio-Rad Laboratories, Inc., Hercules, CA,
USA). The densities of the blots were analyzed using Quantity One
analysis (version 1.4.6; Bio-Rad Laboratories, Inc.).
Dual-luciferase reporter assay
T24 cells (3×105) were seeded into
24-well plates and transfected with miR-96 and/or plasmid HERG1
using Lipofectamine 2000 according to the manufacturer's protocol,
as aforementioned. Plasmids (cat. no. 60847; Wuhan Miaoling Biotech
Science Co., Ltd., Wuhan, China) encoding an effective sequence to
overexpress wild-type or mutant HERG1 were constructed. Experiments
were divided into four groups: miR-96 inhibitor + wild-type HERG1
[pGL-ENNRA 3′-untranslated region (3′UTR)-Wt], SC + wild-type
HERG1, miR-96 inhibitor + mutated HERG1 (pGL-ENNRA 3′UTR-Mut) and
SC + mutated HERG1 (Shanghai Jima Industrial Co., Ltd., Shanghai,
China). A Dual-Luciferase® Reporter Assay system (cat.
no. E1960; Promega Corporation, Madison, WI, USA) was used to
detect luciferase activity. Following transfection for 24 h, the
medium was removed, cells were washed with PBS and 100 µl passive
lysis buffer (provided by the Dual-Luciferase® system)
was added and incubated for 10 min. Following centrifugation
(11,656 × g) for 10 min at 4°C and the supernatant was collected.
Next, 20 µl supernatant and 100 µl luciferase assay reagent were
mixed. Firefly luciferase activity was determined using a Glomax
20/20 fluorescence luminometer. Next, 100 µl Stop & Glo reagent
(provided by the Dual-Luciferase system) was added to detect
Renilla luciferase activity, to which the data were
normalized.
Statistical analysis
The data are presented as the mean ± standard
deviation. Statistical analyses of the data were performed using
one-way analysis of variance followed by a Bonferroni post hoc test
or unpaired Student's t-test (version 17; SPSS, Inc., Chicago, IL,
USA). P<0.05 was considered to indicate a statistically
significant difference.
Results
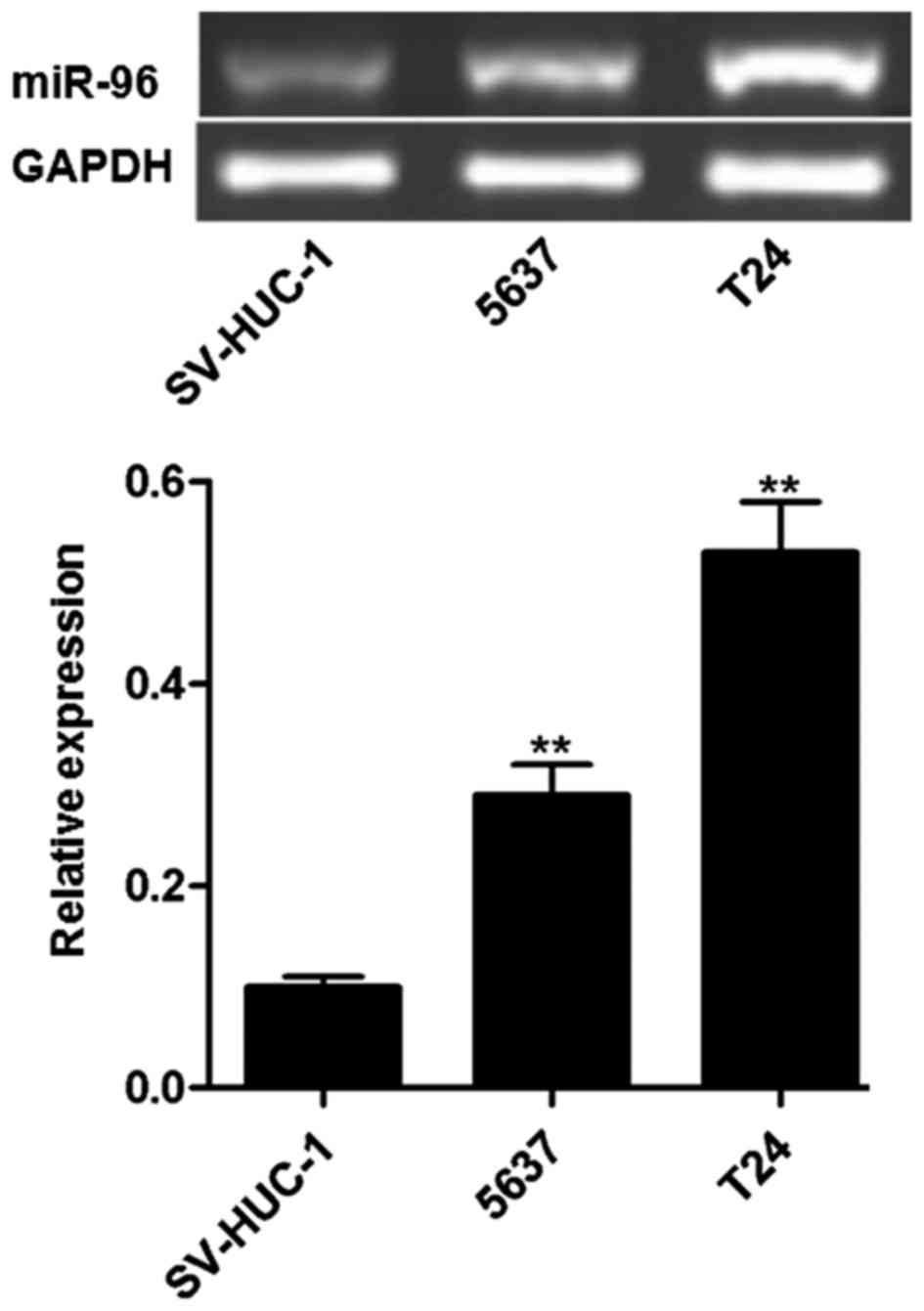
miR-96 is highly expressed in bladder
cancer cell lines
As presented in Fig.
1, miR-96 was highly expressed in T24 and 5,637 cells,
particularly in T24 cells. As a control, miR-96 was rarely
expressed in the uroepithelium SV-HUC-1 cell line. In the
subsequent experiments, T24 was selected as the cell line to be
used.
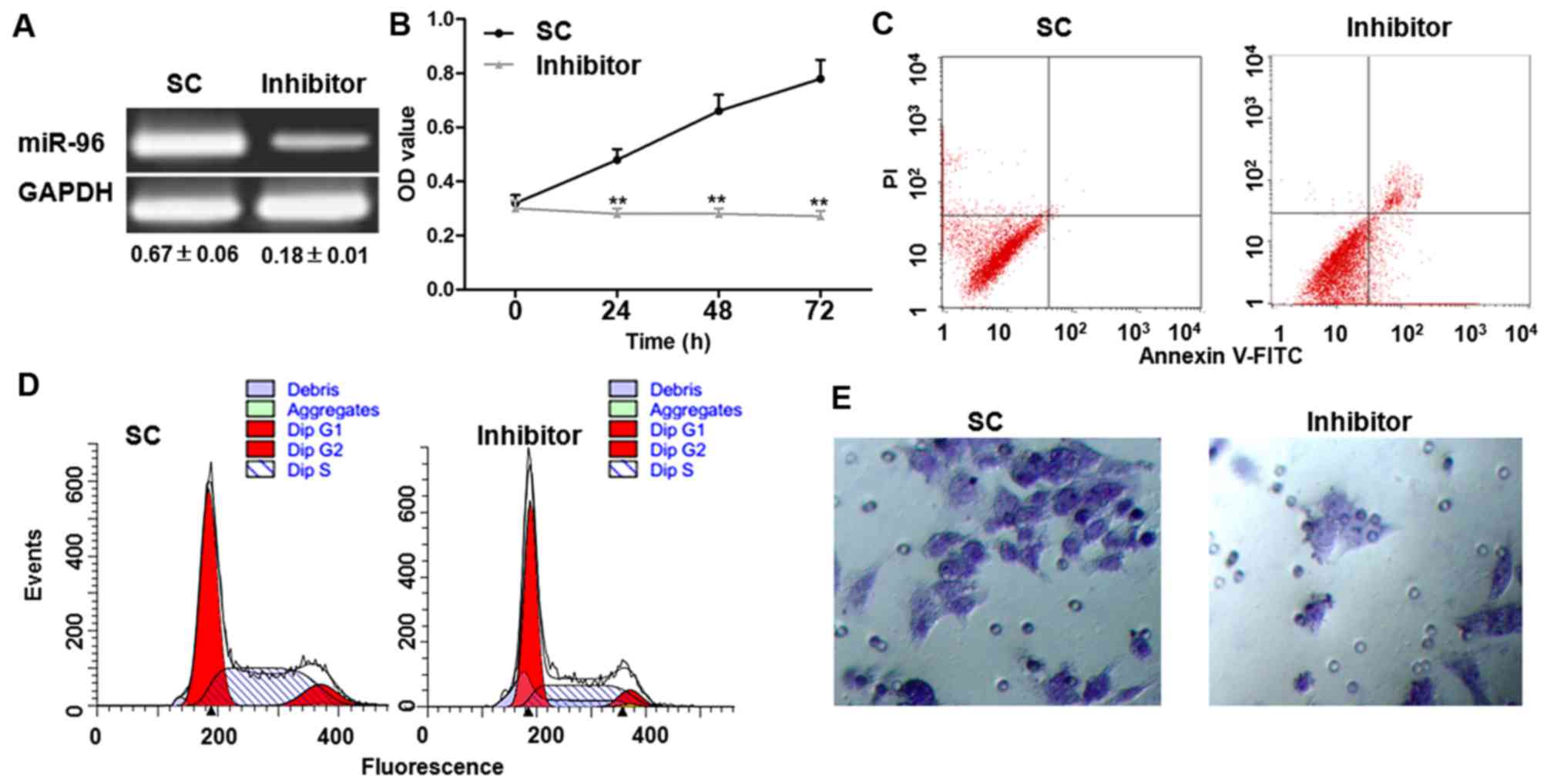
Transfection with miR-96 inhibitor
suppresses cell proliferation, induces apoptosis, arrests cell
cycle and inhibits cell invasion
Following transfection with miR-96 inhibitor for 6
h, expression miR-96 was markedly reduced (Fig. 2A). The effects of miR-96 inhibitor
transfection on cell viability were assessed. As demonstrated using
an MTT assay, cell viability was significantly reduced in cells
transfected with the miR-96 inhibitor 24, 48 and 72 h after
transfection, compared with those transfected with SC (Fig. 2B). Following transfection with miR-96
inhibitor for 48 h, the proliferation inhibition was ~50%, and
therefore, the 48-h time point was selected for the other
experiments. The proportion of apoptosis was detected using Annexin
V-FITC/PI staining. Compared with cells transfected with SC, miR-96
inhibitor significantly increased the apoptosis rate (Fig. 2C and Table
I). PI staining revealed that transfection with the miR-96
inhibitor arrested the cell cycle at the G1 phase
(Fig. 2D and Table I). In addition, the Transwell assay
demonstrated that transfection with a miR-96 inhibitor markedly
decreased the cell invasion rate (Fig.
2E).
 | Table I.miR-96 inhibitor arrests the cell
cycle at the G1 phase and induces apoptosis in T24
cells. |
Table I.
miR-96 inhibitor arrests the cell
cycle at the G1 phase and induces apoptosis in T24
cells.
|
| Cell cycle, % | Apoptosis, % |
|---|
|
|
|
|
|---|
| Group | G1 | S | G2 | Early | Late |
|---|
| SC |
68.38±6.69 |
20.13±2.01 |
11.49±1.15 |
3.35±0.32 |
8.29±1.83 |
| miRI-96 |
87.37±8.46a |
6.13±0.56a |
6.50±0.58a |
12.28±0.28a |
19.46±1.92a |
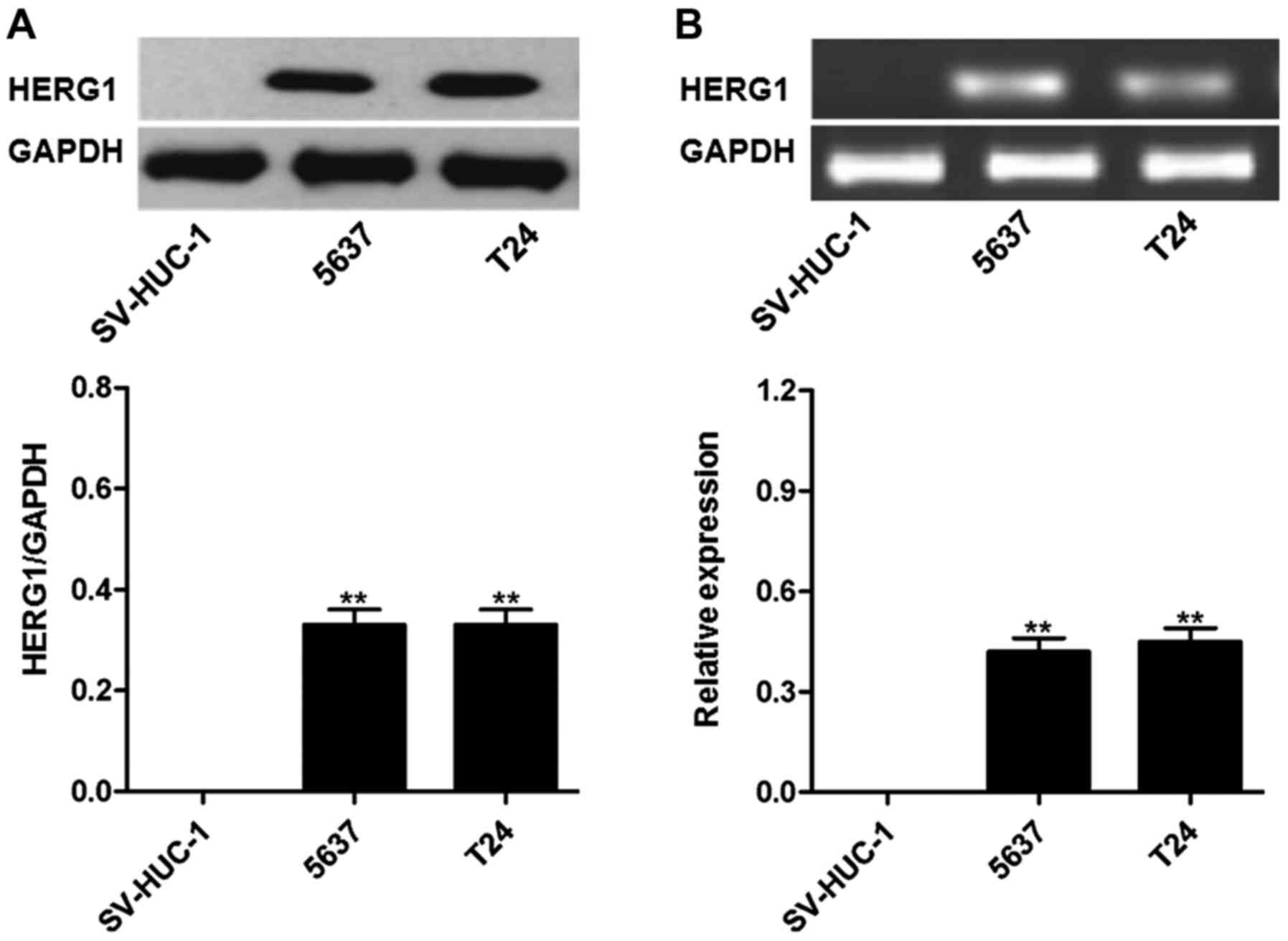
HERG1 is highly expressed in bladder
cancer cell lines
As presented in Fig.
3, HERG1 protein and mRNA was highly expressed in the bladder
cancer T24 and 5,637 cell lines, but not in uroepithelium SV-HUC-1
cells.
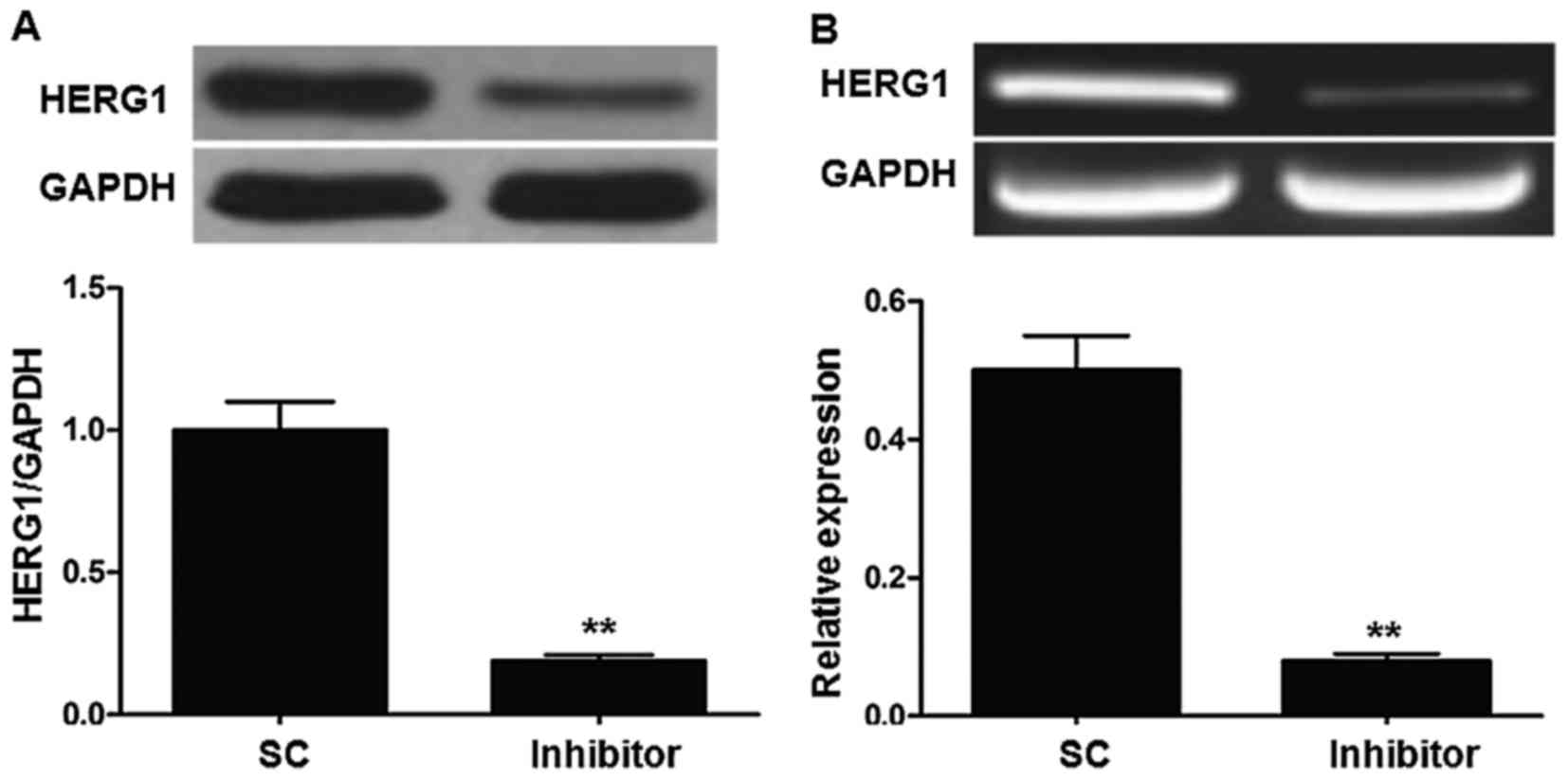
miR-96 inhibitor regulates HERG1
expression in T24 cells
As miR-96 and HERG1 were highly expressed in bladder
cancer cell lines, whether HERG1 was regulated by the expression of
miR-96 was investigated. The T24 cell line was selected for use in
these experiments. As demonstrated in Fig. 4, transfection with the miR-96
inhibitor significantly decreased HERG1 expression at the mRNA and
protein level. To assess the direct regulation of HERG1 by miR-96,
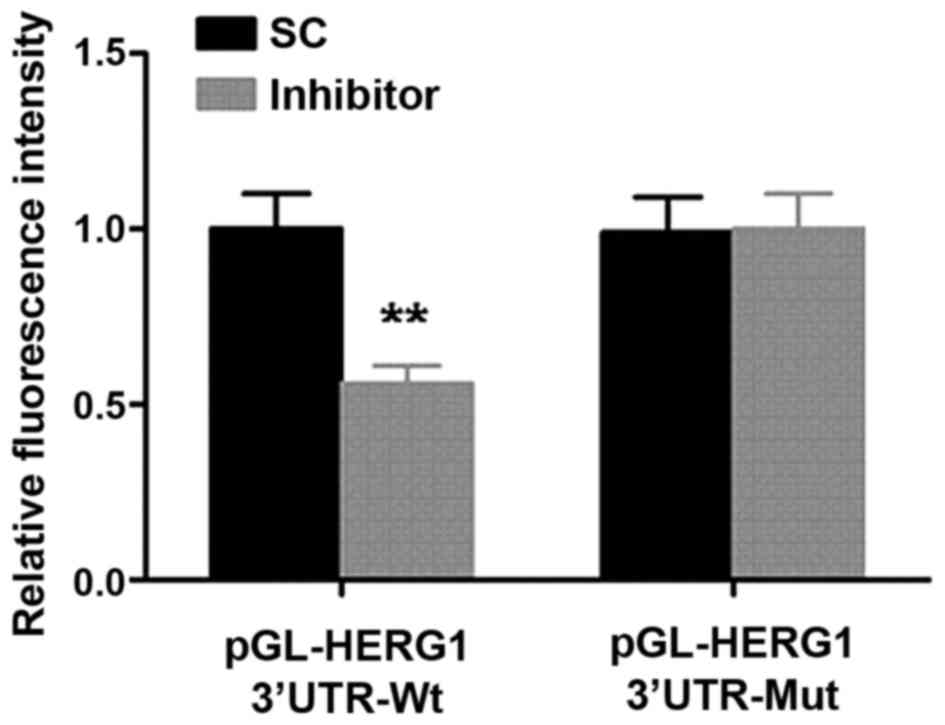
a dual-luciferase reporter assay was performed. As demonstrated in
Fig. 5, transfection with the miR-96
inhibitor significantly decreased the relative fluorescence in the
group transfected with the wild-type HERG1 3′UTR compared with the
SC group.
Discussion
The pathogenesis of cancer is a complex process;
various aspects affect its genesis and development. A commonly held
view is that the imbalanced expression of oncogenes and tumor
suppressor genes contributes to tumor cell proliferation, and
invasion. Genes promoting the tumor development are known as
oncogenes and the opposite are considered tumor suppressor genes
(16). miRNAs are a class of highly
conserved small RNAs that bind the 3′-UTR region of its target gene
and regulate the expression of target genes. The present study
demonstrated that miR-96 was a potential pro-oncogenic factor in
bladder cancer cells. Inhibition of miR-96 expression was able to
suppress cellular proliferation and invasion, promote apoptosis,
and arrest the cell cycle at the G1 phase.
Recent studies have indicated that miRNAs function
either as oncogenes or tumor suppressor genes, regulating the
proliferation, apoptosis, differentiation, migration and invasion
of tumor cells (17,18). miR-96 belongs to the miR-183 family,
the other two members of which are miR-182 and miR-183. These three
miRNAs are found at similar locations in the genome. miR-96 is
located at human chromosome locus 7q32.2, and the amplification of
human chromosome 7 is important in lung cancer (19), hepatocellular carcinoma (20), prostate cancer (21), gastric cancer (22), esophageal cancer (23), breast cancer (24) and other types of tumor (9–13). miR-96
is highly expressed in those types of cancer and has been
considered to be an oncogenic miRNA (25). Han et al (9) applied the gene expression microarray to
validate that miR-96 expression was upregulated in bladder cancer
tissue. Yoshino et al (13)
also confirmed that miR-96 was highly expressed in bladder cancer
tissue. Scheffer et al (12)
confirmed that the expression of 22 miRNAs in bladder cancer
tissues was upregulated, including miR-96. Kriebel et al
(11) reported that the expression of
plasma miR-96 in patients with bladder cancer was significantly
increased. Eissa et al (26)
demonstrated that miR-96 was highly expressed in bladder cancer
tissues, but not in benign bladder tumors. An additional report
stated that miR-96 levels in the urine samples of patients with
bladder cancer were increased (14).
Therefore, it appears that miR-96 may serve important roles in the
development and progression of bladder cancer. The present study
demonstrated that miR-96 expression was upregulated in bladder
cancer cell lines. These cell lines served as in vitro
models, allowing for the study of the biological activity of miR-96
in bladder cancer.
In the present study, RT-PCR was used to detect
miR-96 expression, which demonstrated high expression of miR-96 in
normal bladder cells, particularly in bladder cancer cells. In the
subsequent experiments, transfection with miR-96 inhibitor was
found to suppress the cell proliferation and invasion, promote
apoptosis and arrest the cell cycle at the G1 phase.
Tumor cells must complete the cell cycle to replicate. Thus, cells
that do not pass the G1 checkpoint ‘loop out’ of the
cell cycle and into a resting state termed the G0 phase.
Data from the present study were consistent with a previously
published study, in which it was verified that transfection with
miR-96 inhibitor arrested the Panc-1 cells at the G1
phase and inhibited cellular proliferation (27). Apoptosis is a process of programmed
cell death that occurs in multicellular organisms. In tumor cells,
apoptosis is impaired, leading to unrestricted cell proliferation
(28). Apoptosis is a method of
eliciting cell death of cancer cells during chemotherapy or
radiotherapy (29). The present study
also found that miR-96 inhibitor promoted apoptosis of bladder
cancer cells. In addition, transfection with the miR-96 inhibitor
also prohibited the migration of bladder cancer cells.
Consistently, Yu et al (30)
also verified that transfection with the miR-96 inhibitor
suppressed the invasion of renal cancer cell lines Caki-1 and
786-O.
Previous studies indicated that potassium ion
channels serve important roles in the process of cellular
proliferation (31,32). In addition, calcium and chloride
channels also have roles in the occurrence and development of
tumors (33). The HERG1 channel is a
potassium ion channel and a member of the fast delayed rectifier
voltage-gated potassium ion family. HERG1 is located on chromosome
7q35-36 and was identified by Warmke and Ganetzky in 1994 (34) in Drosophila and in mammals.
HERG1 consists of six transmembrane proteins, and is
inward-rectifying and ion-selective. HERG1 regulates the entry of
calcium into the cell, promotes calcium second messenger signaling
transduction, regulates cytoskeletal changes, and alters cellular
proliferation and migration (31,32). HERG1
was initially reported to serve important roles in the process of
repolarizing cardiac membrane potential (32). HERG1 was identified to be highly
expressed in various tumor tissues, and was closely associated with
tumor cell proliferation, apoptosis, differentiation, migration and
invasion (35–37).
Downregulation of HERG1 in tumor cells can inhibit
the proliferation, migration and invasion of tumor cells, all of
which are malignant biological activities (38). The present study screened the
expression of HERG1 in bladder cancer T24 and 5,637 cell lines, and
the normal uroepithelium SV-HUC-1 cell line. The results of the
current study revealed that HERG1 expression was significantly
increased in bladder cancer cell lines compared with that in
SV-HUC-1 cells. Bioinformatic analysis has demonstrated that HERG1
may be a target gene of miR-96 (39).
The present study also utilized a dual luciferase reporter assay to
verify that HERG1 was a target gene of miR-96. In addition, results
of western blot analysis and RT-PCR also revealed that miR-96
inhibitor significantly reduced HERG1 expression.
The present study demonstrated that miR-96 was
highly expressed in bladder cancer cell lines. miR-96 inhibition
resulted in a number of anticancer effects on bladder cancer cells,
including inhibition of proliferation and invasion, and promotion
of apoptosis. Notably, evidence revealed that HERG1 was a target of
miR-96. These results provided experimental evidence that support
the role of miR-96 as a therapeutic target for bladder cancer.
References
|
1
|
Gan X, Lin X, He R, Lin X, Wang H, Yan L,
Zhou H, Qin H and Chen G: Prognostic and clinicopathological
significance of downregulated p16 expression in patients with
bladder cancer: A systematic review and meta-analysis. Dis Markers.
2016:52596022016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yin M, Joshi M, Meijer RP, Glantz M,
Holder S, Harvey HA, Kaag M, Fransen van de Putte EE, Horenblas S
and Drabick JJ: Neoadjuvant chemotherapy for muscle-invasive
bladder cancer: A systematic review and two-step meta-analysis.
Oncologist. 21:708–715. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cauberg Evelyne CC, de la Rosette JJ and
de Reijke TM: Emerging optical techniques in advanced cystoscopy
for bladder cancer diagnosis: A review of the current literature.
Indian J Urol. 27:245–251. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fahmy N, Lazo-Langner A, Iansavichene AE
and Pautler SE: Effect of anticoagulants and antiplatelet agents on
the efficacy of intravesical BCG treatment of bladder cancer: A
systematic review. Can Urol Assoc J. 7:E740–E749. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mizuguchi Y, Takizawa T, Yoshida H and
Uchida E: Dysregulated miRNA in progression of hepatocellular
carcinoma: A systematic review. Hepatol Res. 46:391–406. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang QX, Zhu YQ, Zhang H and Xiao J:
Altered MiRNA expression in gastric cancer: A systematic review and
meta-analysis. Cell Physiol Biochem. 35:933–944. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gambari R, Brognara E, Spandidos DA and
Fabbri E: Targeting oncomiRNAs and mimicking tumor suppressor
miRNAs: Nuew trends in the development of miRNA therapeutic
strategies in oncology (Review). Int J Oncol. 49:5–32. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Srivastava K and Srivastava A:
Comprehensive review of genetic association studies and
meta-analyses on miRNA polymorphisms and cancer risk. PLoS One.
7:e509662012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Han Y, Chen J, Zhao X, Liang C, Wang Y,
Sun L, Jiang Z, Zhang Z, Yang R, Chen J, et al: MicroRNA expression
signatures of bladder cancer revealed by deep sequencing. PLoS One.
6:e182862011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wu Z, Liu K, Wang Y, Xu Z, Meng J and Gu
S: Upregulation of microRNA-96 and its oncogenic functions by
targeting CDKN1A in bladder cancer. Cancer Cell Int. 15:1072015.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kriebel S, Schmidt D, Holdenrieder S,
Goltz D, Kristiansen G, Moritz R, Fisang C, Müller SC and Ellinger
J: Analysis of tissue and serum microRNA expression in patients
with upper urinary tract urothelial cancer. PLoS One.
10:e01172842015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Scheffer AR, Holdenrieder S, Kristiansen
G, von Ruecker A, Müller SC and Ellinger J: Circulating microRNAs
in serum: Novel biomarkers for patients with bladder cancer? World
J Urol. 32:353–358. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yoshino H, Seki N, Itesako T, Chiyomaru T,
Nakagawa M and Enokida H: Aberrant expression of microRNAs in
bladder cancer. Nat Rev Urol. 10:396–404. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yamada Y, Enokida H, Kojima S, Kawakami K,
Chiyomaru T, Tatarano S, Yoshino H, Kawahara K, Nishiyama K, Seki N
and Nakagawa M: MiR-96 and miR-183 detection in urine serve as
potential tumor markers of urothelial carcinoma: Correlation with
stage and grade, and comparison with urinary cytology. Cancer Sci.
102:522–529. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhu G, Wang X, Wu S and Li Q: Involvement
of activation of PI3K/Akt pathway in the protective effects of
puerarin against MPP+-induced human neuroblastoma
SH-SY5Y cell death. Neurochem Int. 60:400–408. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lv Q, Xue Y, Li G, Zou L, Zhang X, Ying M,
Wang S, Guo L, Gao Y, Li G, et al: Beneficial effects of evodiamine
on P2X(4)-mediated inflammatory injury of human umbilical vein
endothelial cells due to high glucose. Int Immunopharmacol.
28:1044–1049. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dijkstra S, Mulders PF and Schalken JA:
Clinical use of novel urine and blood based prostate cancer
biomarkers: A review. Clin Biochem. 47:889–896. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kaboli PJ, Rahmat A, Ismail P and Ling KH:
MicroRNA-based therapy and breast cancer: A comprehensive review of
novel therapeutic strategies from diagnosis to treatment. Pharmacol
Res. 97:104–121. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Guo H, Li Q, Li W, Zheng T, Zhao S and Liu
Z: MiR-96 downregulates RECK to promote growth and motility of
non-small cell lung cancer cells. Mol Cell Biochem. 390:155–160.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li Z and Wang Y: MiR-96 targets SOX6 and
promotes proliferation, migration and invasion of hepatocellular
carcinoma. Biochem Cell Biol. Sep 11–2017.(Epub ahead of print).
View Article : Google Scholar
|
|
21
|
Xu L, Zhong J, Guo B, Zhu Q, Liang H, Wen
N, Yun W and Zhang L: miR-96 promotes the growth of prostate
carcinoma cells by suppressing MTSS1. Tumour Biol. 37:12023–12032.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cao LL, Xie JW, Lin Y, Zheng CH, Li P,
Wang JB, Lin JX, Lu J, Chen QY and Huang CM: miR-183 inhibits
invasion of gastric cancer by targeting Ezrin. Int J Clin Exp
Pathol. 7:5582–5594. 2014.PubMed/NCBI
|
|
23
|
Xia H, Chen S, Chen K, Huang H and Ma H:
MiR-96 promotes proliferation and chemo- or radioresistance by
down-regulating RECK in esophageal cancer. Biomed Pharmacother.
68:951–958. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hong Y, Liang H, Uzair-Ur-Rehman, Wang Y,
Zhang W, Zhou Y, Chen S, Yu M, Cui S, Liu M, et al: miR-96 promotes
cell proliferation, migration and invasion by targeting PTPN9 in
breast cancer. Sci Rep. 6:374212016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ma Y, Liang AJ, Fan YP, Huang YR, Zhao XM,
Sun Y and Chen XF: Dysregulation and functional roles of
miR-183-96-182 cluster in cancer cell proliferation, invasion and
metastasis. Oncotarget. 7:42805–42825. 2016.PubMed/NCBI
|
|
26
|
Eissa S, Matboli M, Essawy NO and Kotb YM:
Integrative functional genetic-epigenetic approach for selecting
genes as urine biomarkers for bladder cancer diagnosis. Tumour
Biol. 36:9545–9552. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li C, Du X, Tai S, Zhong X, Wang Z, Hu Z,
Zhang L, Kang P, Ji D, Jiang X, et al: GPC1 regulated by miR-96-5p,
rather than miR-182-5p, in inhibition of pancreatic carcinoma cell
proliferation. Int J Mol Sci. 15:6314–6327. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lowe SW and Lin AW: Apoptosis in cancer.
Carcinogenesis. 21:485–495. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Croce CM and Reed JC: Finally, an
apoptosis-targeting therapeutic for cancer. Cancer Res.
76:5914–5920. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yu N, Fu S, Liu Y, Xu Z, Liu Y, Hao J,
Wang B and Zhang A: miR-96 suppresses renal cell carcinoma invasion
via downregulation of Ezrin expression. J Exp Clin Cancer Res.
34:1072015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
He FZ, McLeod HL and Zhang W: Current
pharmacogenomic studies on hERG potassium channels. Trends Mol Med.
19:227–238. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sanguinetti MC: HERG1 channel agonists and
cardiac arrhythmia. Curr Opin Pharmacol. 15:22–27. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Litan A and Langhans SA: Cancer as a
channelopathy: Ion channels and pumps in tumor development and
progression. Front Cell Neurosci. 9:862015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Warmke JW and Ganetzky B: A family of
potassium channel genes related to eag in Drosophila and mammals.
Proc Natl Acad Sci USA. 91:pp. 3438–3442. 1994; View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lastraioli E, Lottini T, Bencini L,
Bernini M and Arcangeli A: hERG1 potassium channels: Novel
biomarkers in human solid cancers. Biomed Res Int. 2015:8964322015.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lastraioli E, Perrone G, Sette A, Fiore A,
Crociani O, Manoli S, D'Amico M, Masselli M, Iorio J, Callea M, et
al: hERG1 channels drive tumour malignancy and may serve as
prognostic factor in pancreatic ductal adenocarcinoma. Br J Cancer.
112:1076–1087. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Pillozzi S and Arcangeli A: Physical and
functional interaction between integrins and hERG1 channels in
cancer cells. Adv Exp Med Biol. 674:55–67. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zeng W, Liu Q, Chen Z, Wu X, Zhong Y and
Wu J: Silencing of hERG1 gene inhibits proliferation and invasion,
and induces apoptosis in human osteosarcoma cells by targeting the
NF-κB pathway. J Cancer. 7:746–757. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Feng J, Yu J, Pan X, Li Z, Chen Z, Zhang
W, Wang B, Yang L, Xu H, Zhang G and Xu Z: HERG1 functions as an
oncogene in pancreatic cancer and is downregulated by miR-96.
Oncotarget. 5:5832–5844. 2014. View Article : Google Scholar : PubMed/NCBI
|