Introduction
Glioblastoma multiforme (GBM) is the most common
type of primary malignant brain tumor in adults worldwide (1). Despite advance in its treatment,
prognosis of patients with GBM remains poor: Patients have a median
survival time of 1 year and <5% of patients survive for 5 years
(2). As tumor cells can infiltrate
into adjacent normal brain tissues, allowing them to evade
therapeutic interventions and proliferate rapidly; tumor cell
proliferation and invasion are regarded as primary causes of poor
prognosis in patients with GBM. Therefore, it is essential to
identify genes that contribute to GBM cell proliferation and
invasion.
Homeobox genes are regulatory genes that encode
transcription factors during embryogenesis and normal development,
in which they regulate cell differentiation, and proliferation
(3). There are >20 subclasses of
homeobox genes; most notable among these is homeobox (HOX) gene
family that consists of 39 genes (4).
These genes are subdivided into four groups: A, B, C and D
(5,6).
HOX genes are reportedly implicated in tumor progression. HOXA7,
HOXB7 and HOXA10 were highly expressed in human lung cancer
(7). HOXD9 mRNA was present in
cervical cancer, but absent in the normal cervix (8,9).
Overexpression of HOXB6, HOXB8, HOXC8, and HOXC9 were observed in
human colon cancer (10). High
expression of Hoxa1, along with the downregulation of Hoxc9, was
observed in the neoplastic mouse mammary gland (11). Upregulation of HOXD3 and HOXD4, and
the concomitant downregulation of HOXA11 and HOXB5, were observed
in human breast cancer (9). In
addition to the from the aforementioned HOX members, HOXB3 has
recently attracted attention as its altered expression has been
observed in a variety of cancer types. For instance, HOXB3 was
upregulated in primary prostate cancer tissues compared with normal
prostate tissues. This upregulation was associated with higher
Gleason grades and with poor patient survival outcome (12). Overexpression of HOXB3 in prostate
cancer LNCaP cells promoted cellular proliferation and migration
(12). Immunocytochemical analysis of
lung carcinoma tissues exhibited high expression of HOXB3 (13). Silencing of HOXB3 in non-small cell
lung cancer A549 cells resulted in decreased growth, whereas
overexpression of HOXB3 in non-small cell lung cancer NCI-H1437
cells markedly increased tumor growth rate (14). Although HOXB3 has been investigated in
multiple cancer types, its role in GBM remains unknown. The present
study was therefore designed to investigate whether HOXB3
contributes to cell proliferation and invasion in GBM.
Materials and methods
Cell lines and cell culture
The glioma cell line U251-MG was obtained from the
Cell Bank of the Chinese Academy of Sciences (Shanghai, China), and
U87-MG from American Type Culture Collection (Manassas, VA, USA).
The two human glioma cell lines were cultured in Dulbecco's
modified Eagle's medium Dulbecco's modified Eagle's medium (DMEM;
Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) containing
2 mM glutamine, 10% fetal calf serum (FBS) (both Thermo Fisher
Scientific, Inc.), 100 U/ml penicillin and 100 µg/ml streptomycin
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). Cells were
maintained in an incubator at 37°C in a 5% CO2 atmosphere. HEB
cells were cultured at 37°C for 3 days in DMEM (Gibco; Thermo
Fisher Scientific, Inc.) containing 2 mM glutamine, 10% FBS (both
Thermo Fisher Scientific, Inc.), 100 U/ml penicillin
(Sigma-Aldrich; Merck KGaA) and 100 µg/ml streptomycin
(Sigma-Aldrich; Merck KGaA).
Tumor samples
Glioblastoma specimens were obtained from patients
(4 male and 2 female, ranging from 44–67 years old and admitted
between September 2015 and September 2016) who underwent surgical
resection at Hainan Provincial People's Hospital. Normal brain
specimens were acquired from 3 patients undergoing surgery for
epilepsy and were reviewed to verify the absence of tumor. All
samples were collected under protocols approved by the
Institutional Review Board of Hainan Provincial People's Hospital
and each patient provided written informed consent for inclusion in
the present study.
Establishment of stably transfected
HOXB3-knockdown cell lines
DNA oligos designed by the BLOCK-iT RNAi designer
(Invitrogen; Thermo Fisher Scientific, Inc.) encoding human
HOXB3-short hairpin RNAs (sequence, 5′-CGGTAAAGCCCACCAGAAT-3′)
(HOXB3-knockdown) were synthesized and cloned into the pHY-LV-KD1.1
vector (Shanghai Hanyu Biotechnology Co., Ltd., Shanghai, China) to
generate pHY-FoxO3a-KD. A vector expressing short hairpin RNA
(shRNA) targeted against an irrelevant sequence (shRNA-NC) was used
as a negative control. The Trans-Lentiviral Packaging system and
ViraPower Lentiviral Expression system (Invitrogen; Thermo Fisher
Scientific, Inc.) were used to produce shRNA. These constructs were
co-transfected with 15 ug packaging plasmids (Invitrogen; Thermo
Fisher Scientific, Inc.) into 293 cells using Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol, and viral particles were harvested 48 h
later. Lentiviruses containing shRNAs were used to silence FoxO3a
expression in U87-MG and U251-MG cells. Stably transfected cells
that consecutively silence HOXB3 were generated using one week of
puromycin selection.
Cell proliferation
Cell counts were determined using Cell Counting
Kit-8 (Dojindo Molecular Technologies, Inc., Kumamoto, Japan). A
total of 1×103 GBM cells were plated onto 96-well
culture plates in triplicate, and cell growth was determined daily
for 5 days using a tetrazolium salt-based colorimetric assay
(Dojindo Molecular Technologies, Inc.) according to the
manufacturer's protocol. Absorbance was measured at 450 nm. Three
independent experiments were performed.
Transwell invasion assay
For the Transwell assay, 2×104 stably
transfected cells were plated into 24-well Boyden chambers (Corning
Incorporated, Corning, NY, USA) with an 8-µm pore polycarbonate
membrane coated with 30 µg Matrigel (BD Biosciences, San Jose, CA,
USA). Cells were plated in the upper chamber with 200 µl of
serum-free medium (DMEM; Gibco; Thermo Fisher Scientific, Inc.),
and the medium containing 20% FBS was added to the lower chamber to
serve as a chemoattractant. After 36 h, the cells were washed three
times with PBS. Non-invasive cells were removed from the upper well
using cotton swabs, and the invasive cells were then fixed with
paraformaldehyde for 15 min at room temperature, air-dried, and
stained with 0.1% crystal violet for 15 min at room temperature.
The results were observed under a light microscope in 5
predetermined fields (magnification, ×200).
Western blot analysis
Cells were lysed in radioimmunoprecipitation assay
buffer (50 mM Tris-HCl pH 8.0,1 mM EDTA pH 8.0, 5 mM DTT, 2% SDS),
and protein concentration was determined using a BCA assay
(Beyotime Institute of Biotechnology, Haimen, China). Total protein
(30 µg per lane) was resolved using a 10% SDS-PAGE and
electrotransferred to polyvinylidene fluoride membranes
(Invitrogen; Thermo Fisher Scientific, Inc.), then blocked with 5%
non-fat dry milk in Tris-buffered saline, pH 7.5 for 1 h at room
temperature. Membranes were incubated with primary antibodies
overnight at 4°C. The primary antibodies used were HOXB3 (dilution,
1:400; Santa Cruz Biotechnology, Inc., Dallas, TX, USA; cat. no.
sc28606), N-cadherin (dilution, 1:800 Cell Signaling Technology,
Inc., Danvers, MA, USA; cat. no. 4061), vimentin (dilution, 1:200;
Cell Signaling Technology, Inc.: cat. no. 3932), E-cadherin
(dilution, 1:1,000; Cell Signaling Technology, Inc.; cat. no. 3195)
and β-actin (dilution, 1:4,000; ProteinTech Group, Inc., Chicago,
IL, USA; cat. no. HRP60008). A horseradish peroxidase-conjugated
IgG secondary antibody (dilution, 1:2,000; cat. no. ab6721, Abcam,
Cambridge, MA, USA) was added and incubated at room temperature for
1 h. Bound antibodies were detected using the BeyoECL system (cat.
no. P0018; Beyotime Institute of Biotechnology), and densitometry
was performed using ImageQuant 5.2 software (GE Healthcare Life
Sciences, Little Chalfont, UK) was used for analysis.
Immunocytochemistry
Immunocytochemistry was performed to detect the
expression of HOXB3. Initially, 5-µm sections of formalin-fixed
paraffin-embedded tissues were dewaxed with xylene and rehydrated
in 100 and 95% ethanol. Antigen retrieval was performed by boiling
slides in 10 mM sodium citrate buffer pH 6.0 for 10 min.
Non-specific antibody binding in cells and slides was blocked by
incubation with blocking buffer (0.2% Triton X-100, 5% normal
donkey serum, and 5% non-fat milk in Tris-buffered saline) for 1 h
at room temperature prior to overnight incubation with
affinity-purified specific primary antibody HOXB3 (dilution, 1:200;
Santa Cruz Biotechnology, Inc.; cat. no. sc28606) in blocking
buffer at 4°C. Human GBM sections were then incubated with alkaline
phosphatase-conjugated secondary antibody (dilution, 1:500; Cayman
Chemical Company, Ann Arbor, MI, USA; cat. no. 13460-1) in the
blocking buffer for 1 h at room temperature. Immunoreactivity was
visualized by incubating the sections with 3,3-diaminobenzidine
tetrahydrochloride (Dako; Agilent Technologies, Inc., Santa Clara,
CA, USA) to produce brown precipitates. The slides were
counterstained with hematoxylin at room temperature for 2 min for
viewing negatively stained cells.
Statistical analysis
Data are presented in the graphs as the mean ±
standard deviation of three independent experiments. The
differences among groups were determined using analysis of variance
with post hoc contrasts determined using the Student-Newman-Keuls
test, and comparisons between two groups were analyzed using the
Student's t-test. P<0.05 was considered to indicate a
statistically significant difference. All statistical analyses were
performed using GraphPad Prism 6 software (GraphPad Software, Inc.,
La Jolla, CA, USA).
Results
HOXB3 is highly expressed in U87-MG
and U251-MG cells
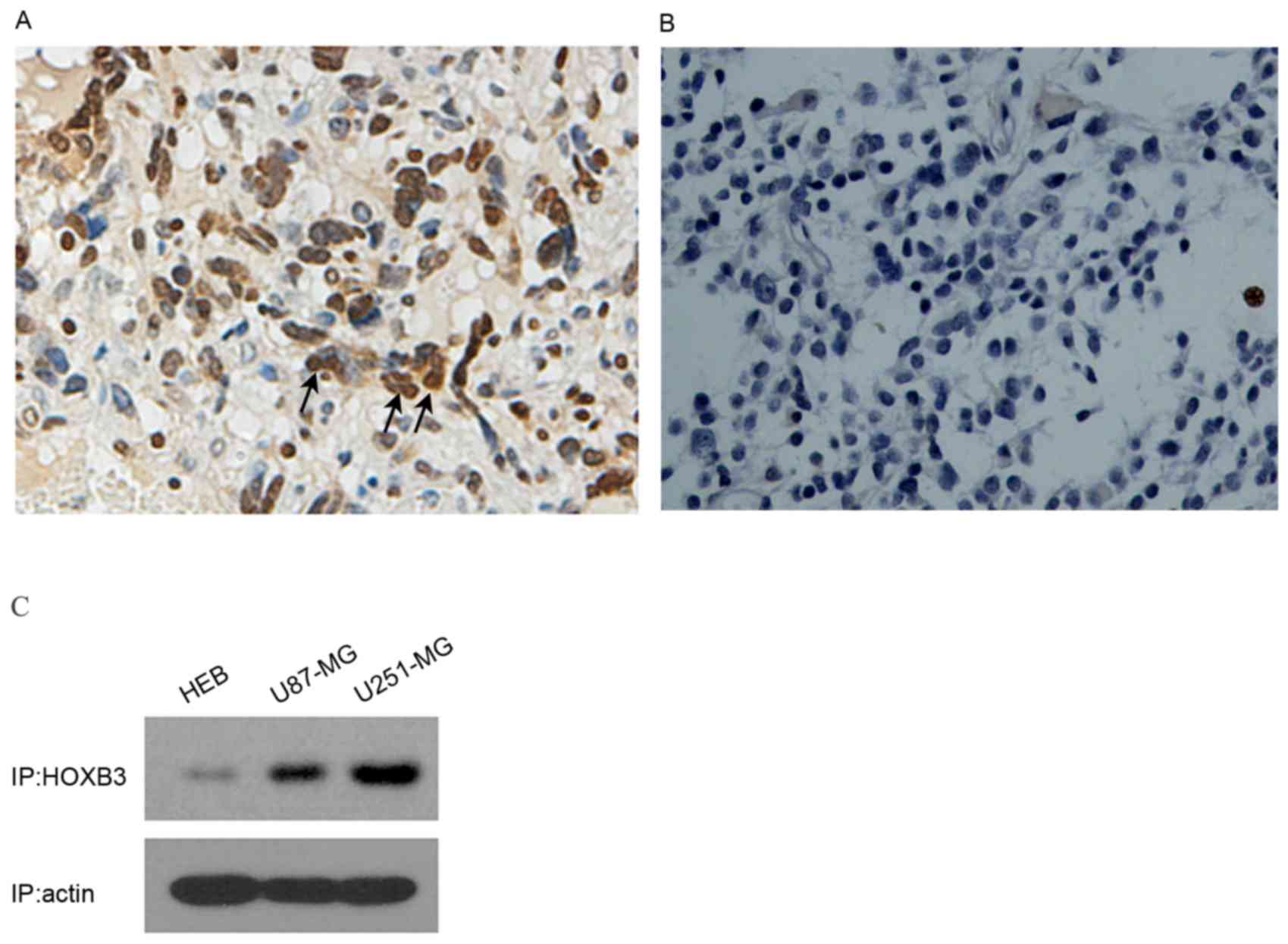
To analyze the expression level of HOXB3 in GBM, 6
tissue samples of GBM (from surgical biopsy specimens) were
initially examined using immunocytochemistry. Strongly positive
staining was observed in human GBM patient specimens (n=6); by
contrast, HOXB3 was hardly detectable in normal brain samples (n=3)
(Fig. 1A and B). These results
indicated that HOXB3 is predominantly present in cancerous tissue,
but not in normal brain tissue. As U87-MG and U251-MG are typical
in vitro models of GBM that can authentically recapitulate
GBM cancerous phenotypes, specifically cell proliferation and
invasion, these two cell lines were used to examine the expression
level of HOXB3. As expected, high expression of HOXB3 was observed
in U87-MG and U251-MG cells. By contrast, HEB cells exhibited mild
expression of HOXB3 that was barely detectable by western blot
analysis (Fig. 1C). These findings
coincide with previous reports, which demonstrated that HOXB3 was
highly expressed in cancerous cell lines (13,15).
Silencing of HOXB3 inhibits cell
proliferation in U87-MG and U251-MG cells
HOXB3 has been shown to serve a role in cell
proliferation in multiple types of cancer, as demonstrated by its
ability to induce cancer cell growth and tumor formation (16). To investigate the ability of HOXB3 to
modulate cell proliferation in GBM, two typical GBM cell lines,
U251-MG and U87-MG, were engineered using lentiviral gene transfer
to express a specific shRNA against HOXB3. Upon transfection, GBM
cells stably silencing HOXB3 were obtained through puromycin
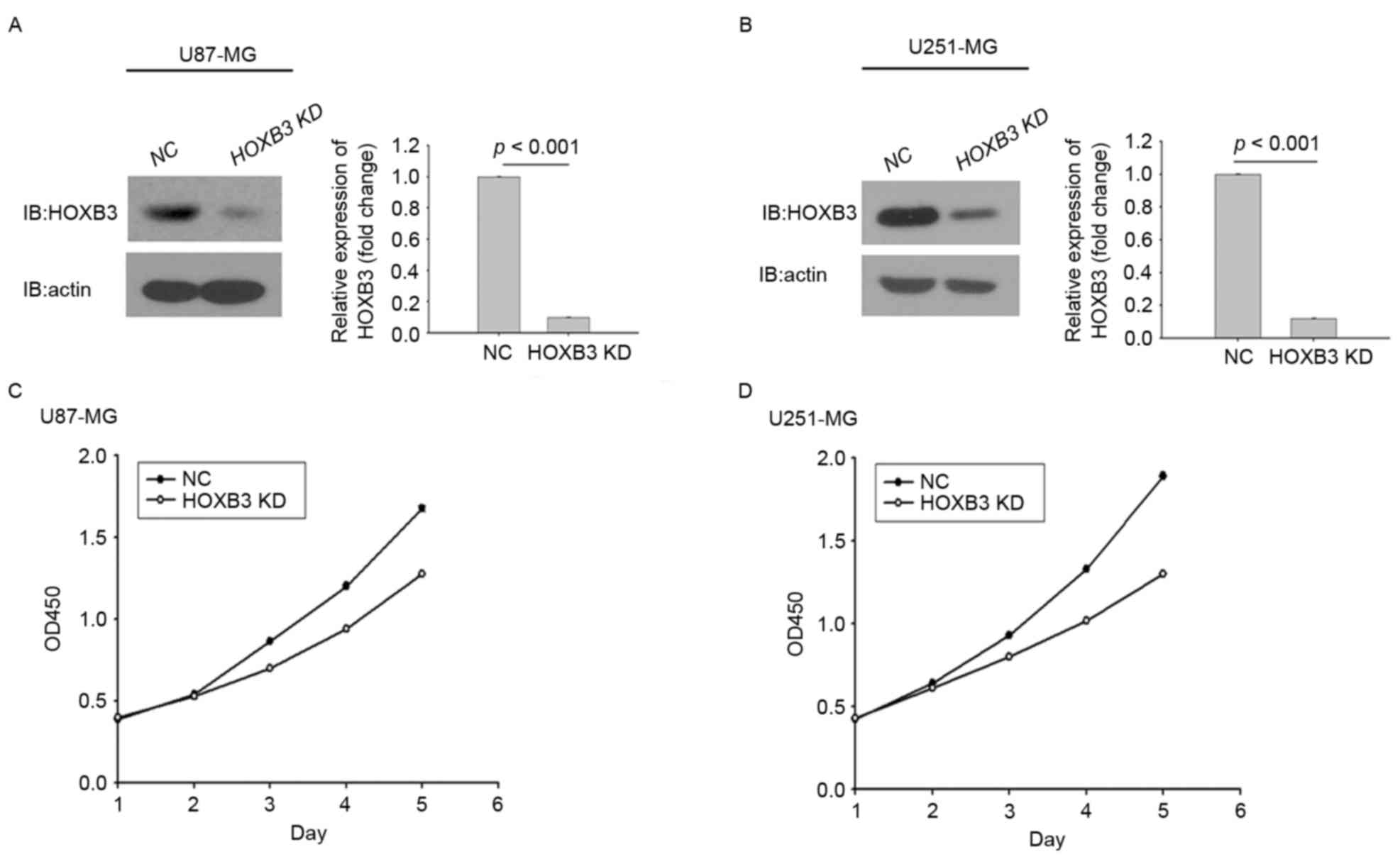
selection and assessed for their proliferation capacities. Fig. 2A depicts stably transfected U87-MG
cells expressing significantly lower protein level of HOXB3
compared with the parental U87-MG cells, in which HOXB3 protein
level were analyzed by western blot analysis. Similarly, this
result was observed in stably transfected U251-MG cells (Fig. 2B).
These results indicated that the two stably
transfected GBM cells had been successfully generated. Whether
altered expression of HOXB3 had an effect on GBM cell proliferation
was next investigated. Stably transfected U87-MG cells that
consistently silenced endogenous HOXB3 exhibited lower cell
proliferation capacity compared with parental U87-MG cells. This
trend did not appear until day 3, and this difference was further
enhanced over time (Fig. 2C).
Similarly, in the cohort of the U251 cells, the proliferative
capacity of U251 cells with HOXB3-knockdown was severely diminished
when compared with that determined for its parental cells. Notably,
the turning point at which the growth rate of the parental cells
exceeded that of the stably transfected cells engineered to silence
HOXB3 also emerged on day 3 (Fig.
2D). It was evident that silencing of HOXB3 alone was able to
confer to GBM cells, at least in the in vitro models in the
present study, a reduced capacity for cell proliferation,
indicating that HOXB3 serves an important role in GBM cell
proliferation.
Silencing of HOXB3 inhibits cell
invasion in U87-MG and U251-MG cells
In addition to promoting cell proliferation, HOXB3
has also been reported to be involved in cell invasion (16,17). To
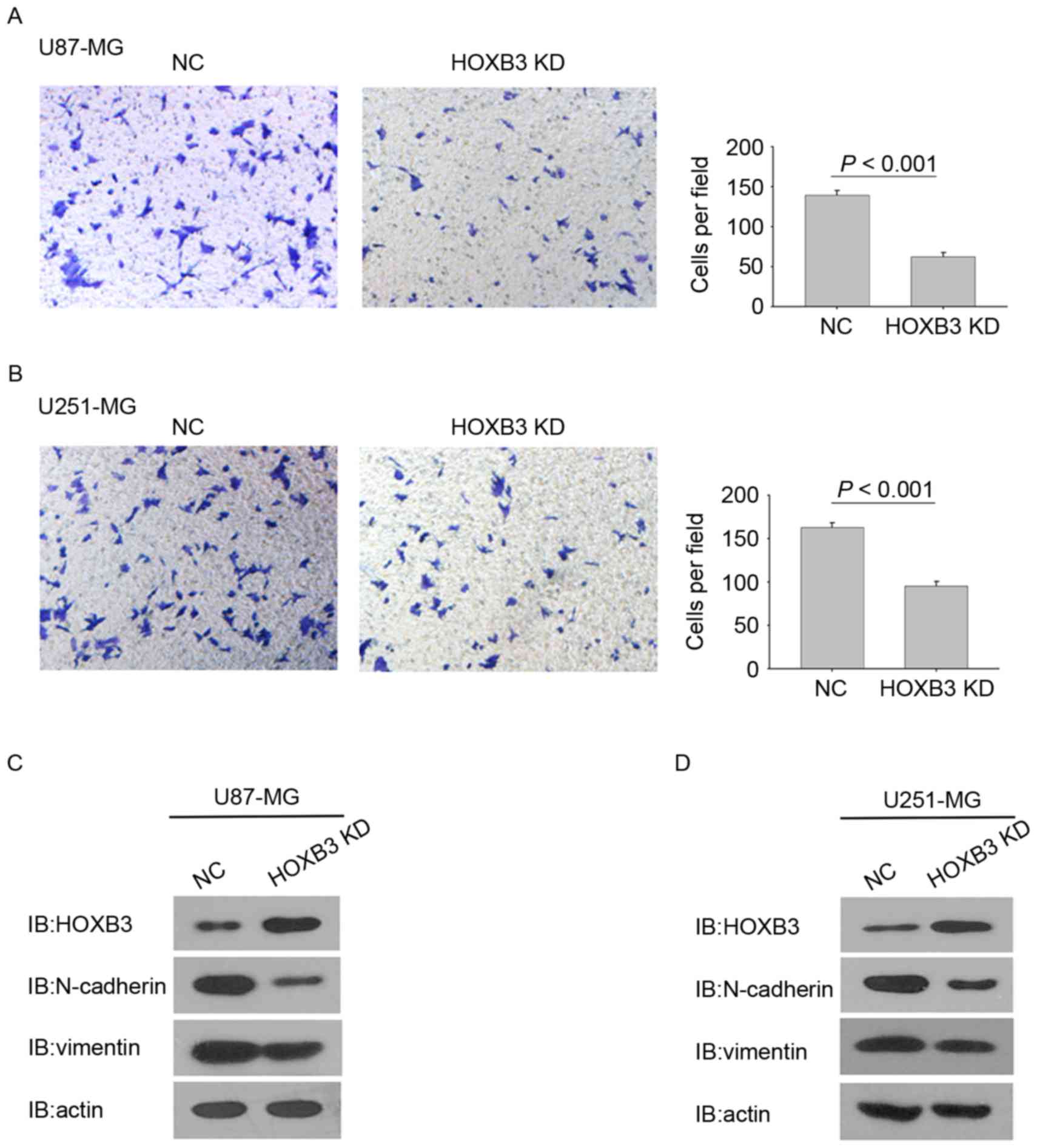
determine the effect of HOXB3 on GBM cell invasion, a Matrigel
invasion assay was performed to analyze the invasive capacities of
the GBM cells stably expressing HOXB3 shRNA and their corresponding
parental cells. Identical numbers of cells from the stably
transfected and parental cohorts were plated into the upper
chambers and incubated for 36 h. Fig.
3A shows that HOXB3-silencing U87-MG cells exhibited a
statistically significant reduction in cell invasion. Consistently,
the stable silencing of HOXB3 in U251-MG cells also led to a
significant reduction in cell invasion when compared with the
parental U251-MG cells (Fig. 3B).
These findings indicate that HOXB3 alone is capable
of modulating GBM cell invasion. Notably, silencing of HOXB3 in
U87-MG and U251-MG cells changed some cells' elongated,
fibroblastic morphology to become relatively rounded and cobbled
(Fig. 3A and B), reminiscent of
epithelial cells undergoing an epithelial-mesenchymal transition
(EMT) (18,19). Several prior studies have verified
that upregulation of mesenchymal genes promotes cell invasion in
GBM (20–24). It was therefore examined whether
silencing of HOXB3 in GBM cells led to evident changes in the
expression of EMT-associated genes. As expected, U87-MG cells that
are stably silencing HOXB3 exhibited a marked decrease in the level
of N-cadherin and vimentin proteins, which are typical mesenchymal
markers. By contrast, E-cadherin, which is a classical epithelial
marker, was upregulated in the stably transfected U87-MG cells
compared with its parental cells (Fig.
3C). A similar result was observed in the cohort of U251-MG
cells (Fig. 3D). These findings
demonstrate that HOXB3 is a critical driver of GBM invasion, and
its effect on cell invasion may result from triggering the
increased expression of N-cadherin and vimentin.
Discussion
High expression of HOXB3 has been reported in a
variety of cancer types, including lung and colon cancer, and its
relevance to poor prognosis of cancer has also been established
(13,15,17).
However, the role for HOXB3 in GBM remains unknown. To the best of
our knowledge, this is the first report to reveal that HOXB3
promotes cell proliferation and invasion in GBM.
GBM cells often invade into adjacent 1–2 cm of the
normal brain tissue, which allows tumor cells to evade standard
treatment interventions (2). Thus,
there is a pressing requirement to define the molecular mechanisms
responsible for invasion. The present study focused on HOXB3, and
demonstrated that silencing of HOXB3 was able to inhibit tumor cell
invasion in glioblastoma. These findings indicate that HOXB3 serves
a role in GBM cell invasion. In this regard, it is unexpected in
that HOXB3 has been reported to suppress or has no effect on cell
invasion and migration. Weiss et al (25) observed that the specific silencing of
HOXB3 was sufficient to reduce the migration of pancreatic cancer
cells in vitro, and invasion and metastasis of pancreatic
cancer in vivo, indicating that HOXB3 functions as a
suppressor of cell invasion. In colorectal cancer, high expression
of HOXB3 had no effect on cell invasion, but instead participates
in regulation of cell proliferation (18). Therefore, the present findings
increase the body of understanding regarding the role of HOXB3 in
cancer.
EMT is an important regulatory program by which
transformed epithelial cells acquire enhanced invasiveness
(26). Carcinoma cells that arise
from normal epithelial cells acquire multiple attributes of EMT,
including alterations to their shape, and abrogation of cell-cell
interactions (14) (such as altered
expression of cadherins), that enable their invasion. Whereas GBMs
are non-epithelial tumors, highly invasive GBM cells necessarily
undergo a switch to a more mesenchymal and aggressive state,
referred to as an EMT-like program, similar to the classical EMT
characterized in epithelial cells (27). The present study initially observed
that silencing of HOXB3 led to a resulting change in cell shape, a
typical sign of EMT, and concomitant reduction in invasiveness.
Therefore, we hypothesized that reduced cell invasion may result
from molecular events associated with the EMT-like program. The
results of the present study indicated that expression of molecules
associated with EMT, including N-cadherin and vimentin, were
downregulated in the cancer cells that silence HOXB3 expression. By
contrast, the epithelial marker E-cadherin was upregulated in the
HOXB3-silencing cells. These findings are consistent with those
obtained from previous studies (28,29), in
which GBM cell invasion is associated with the altered expression
of key EMT-associated genes.
Although multiple examples of aberrantly high HOXB3
expression, combined with the data presented in the present report,
demonstrate that HOXB3 has been implicated in tumor development and
progression, its effect on cancerous phenotypes, particularly
cancer cell proliferation, remains poorly characterized. Several
findings otherwise provide evidence against the positive role of
HOXB3 in cancer cell proliferation, as exemplified by: i) 19
malignant mammary cell lines, including MDA-MB-231 and MCF-7 cells,
exhibited downregulated level of HOXB3 compared with the
non-malignant SVCT cell line and human breast tissue; ii) malignant
melanoma cell lines also exhibited downregulation of HOXB3
expression (30). Additionally, the
biological effects of Hox genes vary and exhibit tissue specificity
(31). It is therefore possible that
the HOXB3 functions as a pro-proliferative factor depending on the
cell type. In certain types of cancer, including leukemia, a set of
Hox genes acting cooperatively promote clonal expansion (32). By contrast, Zhang et al
(33) observed that forced expression
of a single Hox gene can affect tumorigenesis in neuroblastoma.
Coinciding with the latter, the results of the proliferation assay
in the present study revealed that HOXB3 had a noticeable effect on
cell growth. To further investigate the effect of HOXB3 signaling
on cell proliferation, future studies should specifically examine
which of the cell cycle genes may be regulated by the action of
HOXB3.
In conclusion, the findings presented in the present
study reveal that HOXB3 mediates cell proliferation and invasion in
U87-MG and U251-MG cells, providing an insight into the active role
of HOXB3 in GBM. The current study raises the possibility that
HOXB3 acts as a potential therapeutic target, information that
potentially contributes to GBM therapy in the future.
Acknowledgements
The authors are grateful to the National Natural
Science Foundation of China (grant no. 81660502) for funding this
study.
References
|
1
|
Bonavia R, Inda MM, Cavenee WK and Furnari
FB: Heterogeneity maintenance in glioblastoma: A social network.
Cancer Res. 71:4055–4060. 2011. View Article : Google Scholar
|
|
2
|
Hou LC, Veeravagu A, Hsu AR and Tse VC:
Recurrent glioblastoma multiforme: A review of natural history and
management options. Neurosurgical Focus. 20:E52006. View Article : Google Scholar
|
|
3
|
Gray S, Pandha HS, Michael A, Middleton G
and Morgan R: HOX genes in pancreatic development and cancer. JOP.
12:216–219. 2011.
|
|
4
|
Scott MP: A rational nomenclature for
vertebrate homeobox (HOX) genes. Nucleic Acids Res. 21:1687–1688.
1993. View Article : Google Scholar
|
|
5
|
Scott MP: Vertebrate homeobox gene
nomenclature. Cell. 71:551–553. 1992. View Article : Google Scholar
|
|
6
|
Levine M and Hoey T: Homeobox proteins as
sequence-specific transcription factors. Cell. 55:537–540. 1988.
View Article : Google Scholar
|
|
7
|
Calvo R, West J, Franklin W, Erickson P,
Bemis L, Li E, Helfrich B, Bunn P, Roche J, Brambilla E, et al:
Altered HOX and WNT7A expression in human lung cancer. Proc Natl
Acad Sci USA. 97:pp. 12776–12781. 2000; View Article : Google Scholar
|
|
8
|
Hung YC, Ueda M, Terai Y, Kumagai K, Ueki
K, Kanda K, Yamaguchi H, Akise D and Ueki M: Homeobox gene
expression and mutation in cervical carcinoma cells. Cancer Sci.
94:4372003. View Article : Google Scholar
|
|
9
|
Cantile M, Pettinato G, Procino A,
Feliciello I, Cindolo L and Cillo C: In vivo expression of the
whole HOX gene network in human breast cancer. Eur J Cancer.
39:257–264. 2003. View Article : Google Scholar
|
|
10
|
Vider BZ, Zimber A, Chastre E, Gespach C,
Halperin M, Mashiah P, Yaniv A and Gazit A: Deregulated expression
of homeobox-containing genes, HOXB6, B8, C8, C9, and Cdx-1, in
human colon cancer cell lines. Biochem Biophys Res Commun.
272:513–518. 2000. View Article : Google Scholar
|
|
11
|
Friedmann Y, Daniel CA, Strickland P and
Daniel CW: Hox genes in normal and neoplastic mouse mammary gland.
Cancer Res. 54:5981–5985. 1994.
|
|
12
|
Chen J, Zhu S, Jiang N, Shang Z, Quan C
and Niu Y: HoxB3 promotes prostate cancer cell progression by
transactivating CDCA3. Cancer Lett. 330:217–224. 2013. View Article : Google Scholar
|
|
13
|
Bodey B, Bodey B Jr, Gröger AM, Siegel SE
and Kaiser HE: Immunocytochemical detection of homeobox B3, B4, and
C6 gene product expression in lung carcinomas. Anticancer Res.
20:2711–2716. 2000.
|
|
14
|
Blaschuk OW and Devemy E: Cadherins as
novel targets for anti-cancer therapy. Eur J Pharmacol.
625:195–198. 2009. View Article : Google Scholar
|
|
15
|
Palakurthy RK, Wajapeyee N, Santra MK,
Gazin C, Lin L, Gobeil S and Green MR: Epigenetic silencing of the
RASSF1A tumor suppressor gene through HOXB3-mediated induction of
DNMT3B expression. Mol Cell. 36:219–230. 2009. View Article : Google Scholar
|
|
16
|
Chen H, Fan Y, Xu W, Chen J, Xu C, Wei X,
Fang D and Feng Y: miR-10b inhibits apoptosis and promotes
proliferation and invasion of endometrial cancer cells via
targeting HOXB3. Cancer Biother Radiopharm. 31:225–231. 2016.
View Article : Google Scholar
|
|
17
|
Almeida MI, Nicoloso MS, Zeng L, Ivan C,
Spizzo R, Gafà R, Xiao L, Zhang X, Vannini I, Fanini F, et al:
Strand-specific miR-28-5p and miR-28-3p have distinct effects in
colorectal cancer cells. Gastroenterology. 142:886–896.e9. 2012.
View Article : Google Scholar
|
|
18
|
Mikheeva SA, Mikheev AM, Petit A, Beyer R,
Oxford RG, Khorasani L, Maxwell JP, Glackin CA, Wakimoto H,
González-Herrero I, et al: TWIST1 promotes invasion through
mesenchymal change in human glioblastoma. Mol Cancer. 9:1942010.
View Article : Google Scholar
|
|
19
|
Cheng WY, Kandel JJ, Yamashiro DJ, Canoll
P and Anastassiou D: A multi-cancer mesenchymal transition gene
expression signature is associated with prolonged time to
recurrence in glioblastoma. PLoS One. 7:e347052012. View Article : Google Scholar
|
|
20
|
Kotelevets L, Van-Hengel J, Bruyneel E,
Mareel M, Van-Roy F and Chastre E: The lipid phosphatase activity
of PTEN is critical for stabilizing intercellular junctions and
reverting invasiveness. J Cell Biol. 155:1129–1135. 2001.
View Article : Google Scholar
|
|
21
|
Xia M, Hu M, Wang J, Xu Y, Chen X, Ma Y
and Su L: Identification of the role of Smad interacting protein 1
(SIP1) in glioma. J Neurooncol. 97:225–232. 2010. View Article : Google Scholar
|
|
22
|
Asano K, Duntsch CD, Zhou Q, Weimar JD,
Bordelon D, Robertson JH and Pourmotabbed T: Correlation of
N-cadherin expression in high grade gliomas with tissue invasion. J
Neurooncol. 70:3–15. 2004. View Article : Google Scholar
|
|
23
|
Motta FJ, Valera ET, Lucio-Eterovic AK,
Queiroz RG, Neder L, Scrideli CA, Machado HR, Carlotti-Junior CG,
Marie SK and Tone LG: Differential expression of E-cadherin gene in
human neuroepithelial tumors. Genet Mol Res. 7:295–304. 2008.
View Article : Google Scholar
|
|
24
|
Lu KV, Chang JP, Parachoniak CA, Pandika
MM, Aghi MK, Meyronet D, Isachenko N, Fouse SD, Phillips JJ,
Cheresh DA, et al: VEGF inhibits tumor cell invasion and
mesenchymal transition through a MET/VEGFR2 complex. Cancer Cell.
22:21–35. 2012. View Article : Google Scholar
|
|
25
|
Weiss FU, Marques IJ, Woltering JM,
Vlecken DH, Aghdassi A, Partecke LI, Heidecke CD, Lerch MM and
Bagowski CP: Retinoic acid receptor antagonists inhibit mir-10a
expression and block metastatic behavior of pancreatic cancer.
Gastroenterology. 137:2136–2145.e1-7. 2009. View Article : Google Scholar
|
|
26
|
Polyak K and Weinberg RA: Transitions
between epithelial and mesenchymal states: Acquisition of malignant
and stem cell traits. Nat Rev Cancer. 9:265–273. 2009. View Article : Google Scholar
|
|
27
|
Hanahan D and Weinberg R: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar
|
|
28
|
Kahlert UD, Nikkhah G and Maciaczyk J:
Epithelial-to-mesenchymal(-like) transition as a relevant molecular
event in malignant gliomas. Cancer Lett. 331:131–138. 2013.
View Article : Google Scholar
|
|
29
|
Kahlert UD, Maciaczyk D, Doostkam S, Orr
BA, Simons B, Bogiel T, Reithmeier T, Prinz M, Schubert J,
Niedermann G, et al: Activation of canonical WNT/β-catenin
signaling enhances in vitro motility of glioblastoma cells by
activation of ZEB1 and other activators of
epithelial-to-mesenchymal transition. Cancer Lett. 325:42–53. 2012.
View Article : Google Scholar
|
|
30
|
Svingen T and Tonissen KF: Altered HOX
gene expression in human skin and breast cancer cells. Cancer Biol
Ther. 2:518–523. 2003. View Article : Google Scholar
|
|
31
|
Shah N and Sukumar S: The Hox genes and
their roles in oncogenesis. Nat Rev Cancer. 10:361–371. 2010.
View Article : Google Scholar
|
|
32
|
Calvo KR, Sykes DB, Pasillas MP and Kamps
MP: Nup98-HoxA9 immortalizes myeloid progenitors, enforces
expression of Hoxa9, Hoxa7 and Meis1, and alters cytokine-specific
responses in a manner similar to that induced by retroviral
co-expression of Hoxa9 and Meis1. Oncogene. 21:4247–4256. 2002.
View Article : Google Scholar
|
|
33
|
Zhang X, Hamada J, Nishimoto A, Takahashi
Y, Murai T, Tada M and Moriuchi T: HOXC6 and HOXC11 increase
transcription of S100beta gene in BrdU-induced in vitro
differentiation of GOTO neuroblastoma cells into Schwannian cells.
J Cell Mol Med. 11:299–306. 2007. View Article : Google Scholar
|