Introduction
Gliomas are the most common form of central nervous
system (CNS) primary tumors, accounting for >50% of all primary
CNS tumors (1). Gliomas are
characterized by increased proliferation, invasion and malignancy.
Despite the use of aggressive therapeutic modalities, including
surgical resection, radiation and chemotherapy, the prognosis for
patients with malignant gliomas remains poor (2–4). Advances
in increasing the survival rate of glioma patients have been
limited, as the pathophysiological mechanisms of glioma remains
unclear. Therefore, a deeper understanding of the pathways involved
in the development of glioma is required to develop a novel
therapeutic target for glioma.
Recent improvements in high-resolution microarray
and genome-wide sequencing analysis have revealed that human genome
includes ~20,000 protein-coding genes, however, most of the human
genome is pervasively transcribed into ncRNA (5,6). ncRNAs
are divided into two groups, according to their size, long ncRNAs
(lncRNA) and small ncRNAs. Of the ncRNAs, microRNAs have attracted
considerable attention as they serve a number of pivotal roles in
cancer via the silencing of target genes (7,8). However,
the field of lncRNAs is an emerging area in ncRNAs study. lncRNAs
have been reported to regulate a wide of biological processes as
they are involved in every step of mRNA biology, including
transcription, mRNA splicing, RNA decay and translation (9–11).
lncRNAs are a class of non-coding RNA transcript
>200 nucleotides in length that have no protein-coding
potential. Recent studies have demonstrated that lncRNAs serve a
vital role in epigenetic modification. lncRNAs participate in
various biological and pathological processes by controlling gene
expression via diverse mechanisms including transcription,
post-transcriptional processing, genomic imprinting, chromatin
modification and the regulation of protein function (12–14). For
example, Wang et al (15)
demonstrated that the ectopic expression of maternally expressed 3
(MEG3), a lncRNA that is markedly downregulated in astrocytoma,
could inhibit cell proliferation and promote cell apoptosis in
astrocytoma cells, indicating the potential tumor-suppressive
function of MEG3. In addition, the expression of HOX transcript
antisense RNA (HOTAIR) was closely associated with glioma grade and
poor prognosis, and an independent prognostic factor in GBM
patients (16). Knockdown of HOTAIR
inhibited colony formation and cell cycle
G0/G1 arrest (16). These findings indicate that lncRNAs
are involved in the development of gliomas. lncRNA SPRY4-intronic
transcript 1 (SPRY4-IT1) is a 687-nucleotide unspliced,
polyadenylated transcript that is transcribed from the second
intron of the SPRY4 gene. The upregulation of SPRY4-IT1 expression
has been observed in melanoma cells, whereas the knockdown of
SPRY4-IT1 inhibited invasion, inducing cell growth arrest and
apoptosis (17). However, the role of
lncRNA SPRY4-IT1 in glioma is unclear.
Spindle and kinetochore associated complex subunit 2
(SKA2) is encoded by an 831-nucleotide cDNA sequence and is located
on human chromosome 17q 23.2. SKA2 is required for the assembly of
condensed chromosomes on the metaphase plate and is involved in the
maintenance of the metaphase plate and/or spindle checkpoint
silencing (18). Rice et al
(19) demonstrated that the enforced
expression of SKA2 induced glucocorticoid transactivation in HepG2
cells, whereas knockdown of SKA2 in A549 human lung epithelial
cells reduced transactivation and suppressed dexamethasone
inhibition of proliferation.
The objective of the present study was to explore
the role of SPRY4-IT1 in glioma. SPRY4-IT1 expression was examined
in glioma tissues and U251 cell lines using reverse
transcription-quantitative polymerase chain reaction (RT-qPCR). and
the biological functions of SPRY4-IT1 in U251 cells. Additionally,
it was observed that SKA2 was a downstream target gene of SPRY4-IT1
and that SKA2 promoted U251 cells proliferation and invasion. The
present study promotes the understanding of the role of SPRY4-IT1
as regulators of glioma pathogenesis, and contributes to the
development of lncRNA-directed diagnostics and therapeutics.
Materials and methods
Materials
Fetal bovine serum (FBS) and Dulbecco's modified
Eagle's medium (DMEM) were purchased from Hyclone; GE Healthcare
Life Sciences (Logan, UT, USA). Lipofectamine 2000 and TRIzol
reagent was purchased from Invitrogen; Thermo Fisher Scientific,
Inc. (Waltham, MA, USA). M-MLV Reverse Transcriptase was obtained
from Promega Corporation (Madison, WI, USA). All other chemicals
were purchased from Sigma-Aldrich; Merck KGaA (Darmstadt, Germany).
The antibodies used were as follows: Anti-proliferating cell
nuclear antigen (PCNA; cat. no. bs-0754R), anti-cyclin D1 (cat. no.
bs-0623R); anti-matrix metalloproteinase-2 (MMP2; cat. no.
bs-4605R); and anti-MMP9 (1:400 dilution, cat. no. bs-4593R; BIOSS,
Beijing, China), anti-β-actin (cat. no. ab8226), anti-SKA2 (cat.
no. ab91551) (1:1,000 dilution; Abcam, Cambridge, UK).
Patients and tissue samples
Tissue samples from glioma tumors and normal brain
tissues were collected from the neurosurgery department of the
Second Affiliated Hospital of Anhui Medical University (Hefei,
China) between May 2014 and March 2015. Samples were collected and
preserved at −80°C and their histological type was further
confirmed according to the World Health Organization (WHO)
criteria. A total of 64 glioma samples (WHO I/II, n=27 WHO III/IV,
n=37) and normal brain tissues (n=9) were used in the present study
(20). Patients selected included 50
males and 23 females, with an age range between 16 and 64 years
(median age, 46 years). The present study was approved by the
Biomedical Ethics Committee of Anhui Medical University and
patients provided written informed consent.
Cell culture procedures
Human astrocytoma U251 cells were purchased from the
American Type Culture Collection (ATCC; Manassas, VA, USA). Cells
were cultured in DMEM, supplemented with 10% heat-inactivated FBS
and 100 U/ml penicillin/streptomycin (Thermo Fisher Scientific,
Inc.). Cells cultures were maintained at 37°C in humidified
atmosphere of 5% CO2. Small interfering RNAs (siRNAs)
were chemically synthesized by GenePharma (Shanghai, China). The
sequences are: SPRY4-IT1 siRNA, GCT TTC TGA TTC CAA GGC CTA TTA A;
SKA2 siRNA, AAG AAA TCA AGA CTA ATC ATC TT; Si-NC, UUC UCC GAA CGU
GUC ACG UTT. U251 cells (2×105) were transfected with
siRNA using Lipofectamine 2000 transfection reagent (Life
Technologies; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. After 48 h, cells transfected with
siRNA were harvested for RT-qPCR to determine the transfection
efficiency.
RT-qPCR analysis
Total RNA was extracted from U251 cells and patient
samples using TRIzol reagent. The first-strand cDNA was synthesized
from total RNA using the Thermoscript RT-PCR system (Thermo Fisher
Scientific, Inc.) at 65°C for 5 min. RT-qPCR analysis was performed
using SYBR Green Master Mix kit on Thermo Fisher connect Real-Time
PCR platform (Thermo Fisher Scientific, Inc.). In brief, each PCR
reaction mixture containing 10 µl of 2X SYBR GreenMaster Mix, 1 µl
of sense and antisense primers (5 µmol/µl) and 1 µl of cDNA (10
ng), was run for 45 cycles with denaturation at 95°C for 15 sec,
annealing at 60°C for 30 sec and extension at 72°C for 30 sec in a
total volume of 20 µl. For relative quantification,
2−∆∆Cq was calculated and used as an indication of the
relative expression levels (21),
which was calculated by subtracting quantification cycle (Cq)
values of the control gene from the Cq values of SPRY4-IT1. The
primer sequences were as follows: SPRY4-IT1 forward,
5′-AGCCACATAAATTCAGCAGA-3′ and reverse,
5′-CGATGTAGTAGGATTCCTTTCA-3′; SKA2 forward,
5′-CCGCTTTAAACCAGTTGCTG-3′ and reverse, 5′-CTCTGCCGCAGTTTTCTCTT-3′.
GAPDH was applied as an internal control. The primer sequences of
GAPDH were: Forward, 5′-AGCAAGAGCACAAGAGGAAG-3′ and reverse,
5′-GGTTGAGCACAGGGTACTTT-3′.
MTT assay
U251 cells were trypsinized, resuspended and seeded
into 96-well plates at a concentration of 2,000 cells/well, and
incubated at 37°C 48 h after siRNA treatment. The number of viable
cells was measured at daily intervals (0,12, 24, and 48 h). At each
time-point, 10 µl of 5 mg/ml MTT (Beijing Dingguo Biotechnology
Co., Ltd., Beijing, China) was added and incubated for 4 h. Then
the medium was removed carefully and 100 µl dimethyl sulfoxide was
added at the end of incubation. The absorbance was measured at 592
nm on the spectrophotometer.
Colony formation assay
A total of 200 U251 cells were seeded in 6-well
plates following a 48-h siRNA treatment. The medium was changed at
regular time intervals. After 7 days of culture at 37°C, the
natural colonies were washed with PBS and fixed with 4%
paraformaldehyde for 30 min at room temperature. The colonies were
then stained with methylene blue (1%) for 10 min at room
temperature, washed with water and air-dried. The total number of
colonies with more than 50 cells was counted using a fluorescent
microscope (×100 magnification).
Scratch wound assay
U251 astrocytoma cells were transfected with siRNA
targeted at SPRY4-IT1 (si-SPRY4-IT1) or si-negative control
(si-NC). Cells (5×105) were cultured in 6-well plates.
Wounds were created in adherent cells using a 20 µl pipette tip at
48 h after transfection. The cells were then washed three times
with PBS to remove any free-floating cells and debris. Serum-free
DMEM was added, and the cells were incubated under normal
conditions. Wound healing was observed after 24 h using a light
microscope. Images of representative scratch lines were captured
using digital microscopy once culture inserts were removed. Each
experiment was repeated in triplicate.
In vitro migration and invasion
assays
U251 cells (5×105) were transfected with
si-SPRY4-IT1 after 48 h, and then plated into the upper chamber of
polycarbonate transwell filters (without Matrigel for the transwell
assay) or plated on the top side of polycarbonate transwell filter
coated with Matrigel (for the invasion assay) in the upper chamber
of the QCM™ 24-Well Cell Invasion Assay (Cell Biolabs, Inc., Sand
Diego, CA, USA). For transwell migration assays, cells were
suspended in 150 µl DMEM without serum, and DMEM supplemented with
10% FBS was used in the lower chamber. For the invasion assay,
cells were suspended in medium without serum, and DMEM supplemented
with 10% FBS was used as a chemoattractant in the lower chamber.
The cells were incubated at 37°C for 24 h for each assay. The
non-migratory or non-invasive cells in the top chambers were
removed with cotton swabs. The migrated and invaded cells on the
lower membrane surface were fixed with methanol and stained with
crystal violet. Cells were counted visually in 5 random fields
using a light microscope (×100). In addition, migrated and invaded
cells were dissociated, lysed and quantified at 570 nm using
spectrophotometer.
Western blotting
U251 astrocytoma cells transfected with si-SPRY4-IT1
were lysed with RIPA lysis buffer (Beyotime Institute of
Biotechnology, Haimen, China). Whole extracts were prepared, and
protein concentrations were determined using the BCA protein assay
kit (Boster Biological Technology, Pleasanton, CA, USA). Whole-cell
extracts (20 or 40 µg) were then fractionated using 8 or 12%
SDS-PAGE. Gels were run at a 120 V for 2 h then transfer onto a
polyvinylidene fluoride membrane (EMD Millipore, Billerica, MA,
USA). Nitrocellulose blots were incubated for 1 h with primary
antibodies diluted in TBS/Tween-20 (0.075% Tween-20) containing 3%
skimmed milk powder. Antibody was diluted. Following incubation
with the afore mentioned primary antibodies at 4°C overnight, blots
were washed three times in TBS/Tween-20 prior to incubation at room
temperature for 1 h with goat anti-mouse (cat. no. ZB-2305) or
anti-rabbit (cat. no. ZB-2301) (both from ZSGB-BIO; Origene,
Beijing, China) horseradish peroxidase conjugated antibody at a
1:10,000 dilution in TBS/Tween-20 containing 5% milk. Following
extensive washing in TBS/Tween-20, the blots were rinsed with
distilled water and proteins were detected using the enhanced
chemiluminescence system. Proteins were visualized with an enhanced
chemiluminescence kit (ECL-plus; Thermo Fisher Scientific, Inc.).
The quantification of the bands was performed using ImageJ software
(version 1.48; National Institutes of Health, Bethesda, MD,
USA).
Statistical analysis
All data were expressed as mean ± standard deviation
of three independent experiments, in which each assay was performed
in triplicate. Data were analyzed with SPSS 16.0 software (SPSS,
Inc., Chicago, IL, USA). Evaluation of the data was performed by
t-test (two-sided) or one-way analysis of variance, followed by
Tukey's post hoc test. Pearson's test was performed to calculate
the association between SPRY-IT1 and SKA2 expression. P<0.05 was
considered to indicate a statistically significant difference.
Results
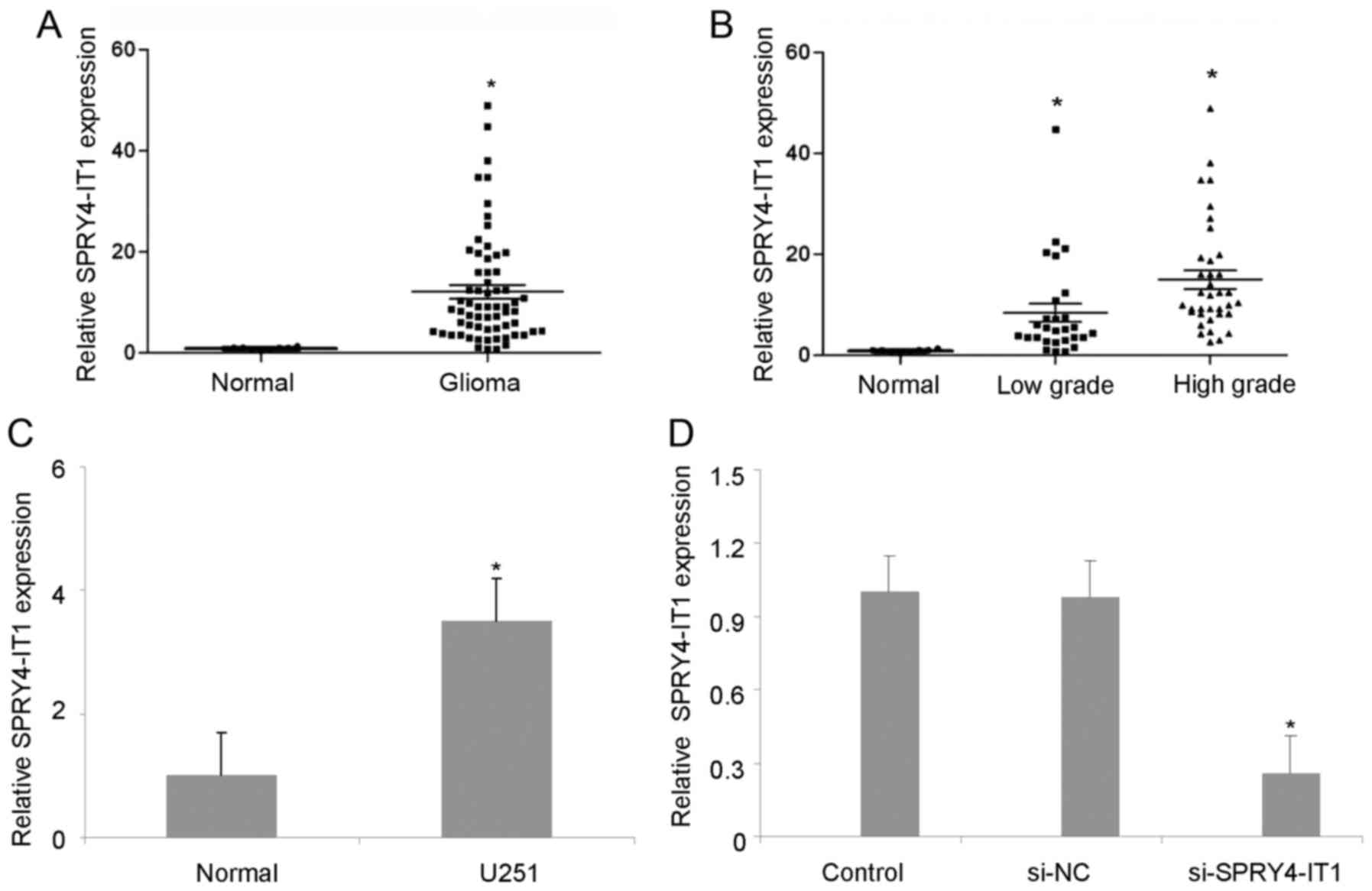
Expression of SPRY4-IT1 was
upregulated in glioma
To determine the association between SPRY4-IT1
expression and glioma with different degrees, RT-qPCR was performed
to evaluate the expression levels of SPRY4-IT1 in 9 normal brain
tissue samples and 64 freshly dissected glioma samples. As shown in
Fig. 1A, levels of SPRY4-IT1 were
upregulated in gliomas tissues compared with normal brain tissues.
Additionally, expression of SPRY4-IT1 was markedly upregulated in
high-grade glioma samples (WHO tumor grades III and IV) and, to a
lesser degree, increased in WHO tumor grades I and II glioma
samples, compared with normal brain tissues (Fig. 1B). SPRY4-IT1 expression was also
examined in U251 cell lines and normal brain tissues. As shown in
Fig. 1C, SPRY4-IT1 expression was
higher in U251 cell lines than in normal brain tissues. Therefore,
these data clearly indicate that high levels of SPRY4-IT1
expression are present in glioma tissues and indicate that this
high expression may contribute to glioma pathogenesis.
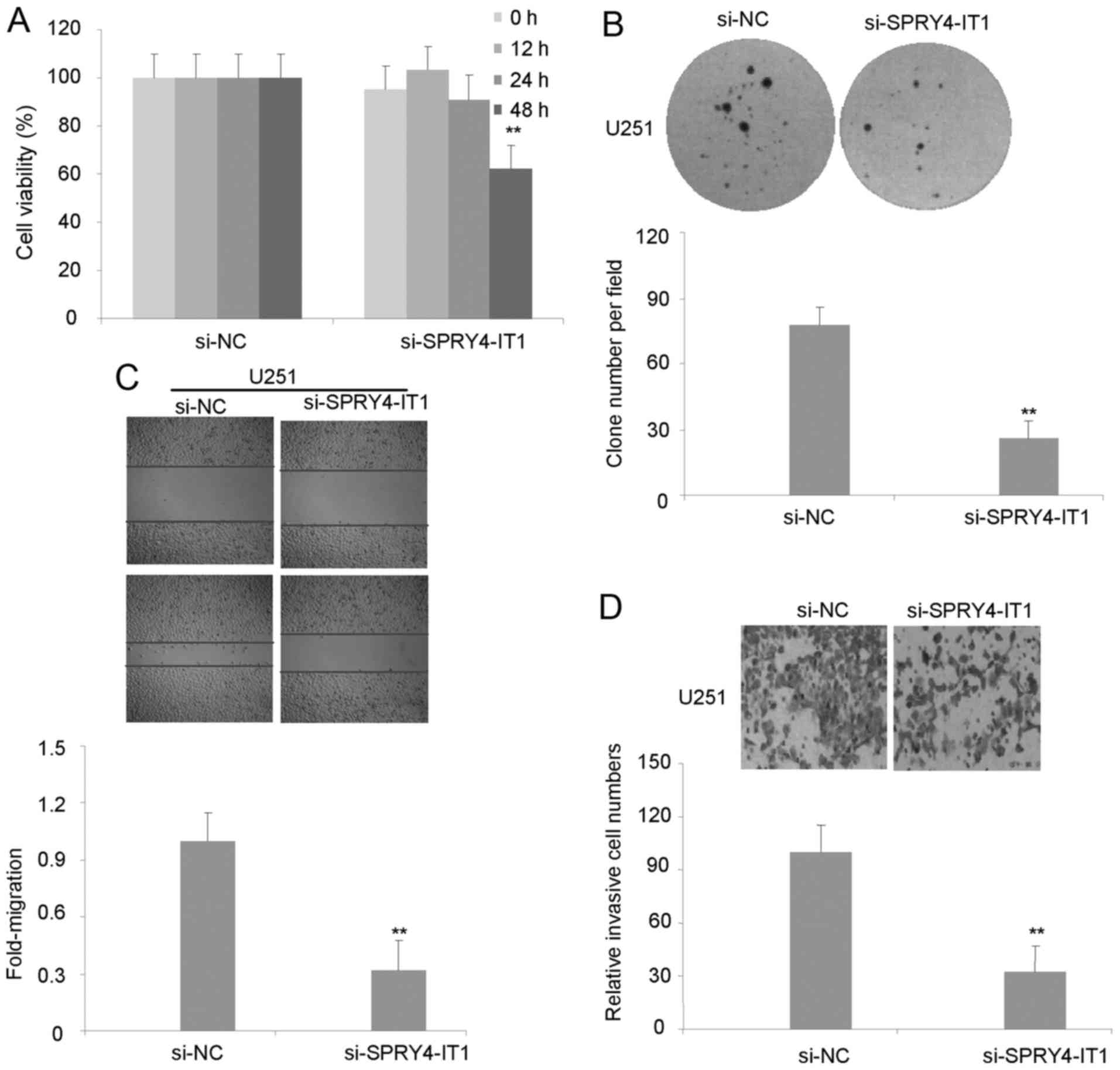
Knockdown of SPRY4-IT1 inhibited U251
cells proliferation
Astrocytoma is a glioma that originates from
astrocytes (20). U251 astrocytoma
cells are frequently used to study glioma malignant biological
characteristics, so U251 cells were selected for use in the present
study. To determine whether siRNAs were able to be transfected into
U251 cells, cells transfected with si-SPRY4-IT1 and si-NC were
assessed by RT-qPCR. Cells transfected with si-SPRY4-IT1 exhibited
significantly lower expression of SPRY4-IT1 mRNA in astrocytoma
cells compared with si-NC cells (Fig.
1D). To investigate the role of SPRY4-IT1 in U251 cells
proliferation, an MTT assay was performed. Knockdown of SPRY4-IT1
in U251 cells markedly suppressed cellular proliferation (Fig. 2A). Similarly, the size of colonies
formed by si-SPRY4-IT1 cells were markedly decreased in U251 cells
compared with si-NC cells, indicating that the decrease of
SPRY4-IT1 expression significantly inhibits colony formation by
U251 cells (Fig. 2B).
SPRY4-IT1 knockdown suppressed U251
cells migration and invasion
To investigate whether SPRY4-IT1 serves a direct
functional role in facilitating U251 cell migration, U251 cell
migration was assessed using a scratch wound assay. Data from this
assay revealed that transfection with si-SPRY4-IT1 but not si-NC
reduced the cell migration of U251 cells (Fig. 2C). As shown in Fig. 2D, invasion of U251 cells was reduced
following inhibition of SPRY4-IT1. These results indicate that
SPRY4-IT1 can promote the U251 cell migratory and invasive
phenotype.
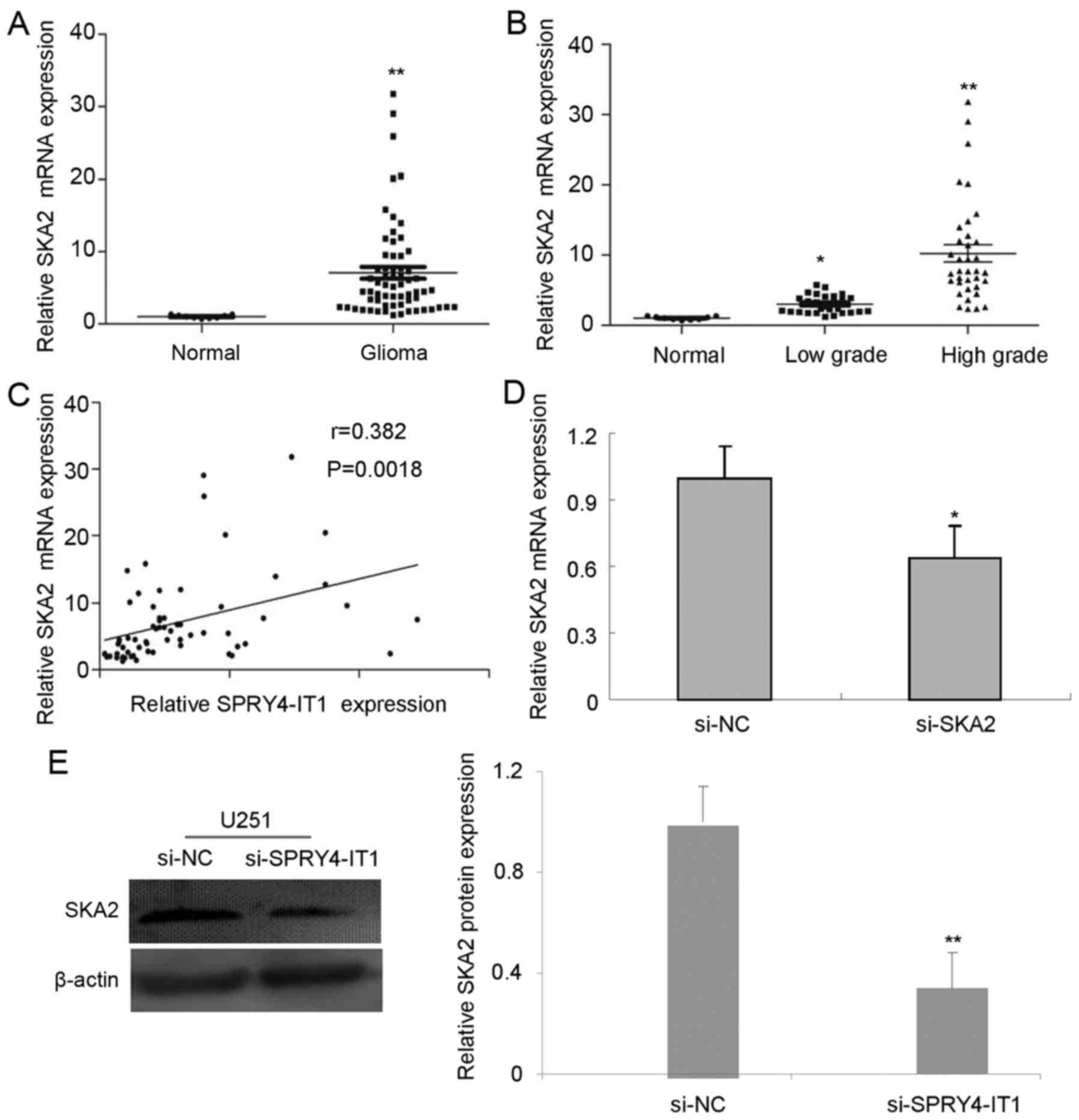
Expression of SKA2 is upregulated, and
positively correlated with SPRY4-IT1 in glioma tissues
The correlation of SPRY4-IT1 and SKA2 expression was
assessed in glioma samples. To identify the aberrant expression of
SKA2 in glioma tissues further, RT-qPCR was used. Similar to the
expression of SPRY4-IT1, the expression of SKA2 was much higher in
glioma tissues than in normal brain tissues (Fig. 3A). The expression of SKA2 was higher
in high-grade glioma compared with low-grade glioma (Fig. 3B). Notably, Pearson's correlation
analysis demonstrated that the expression of SPRY4-IT1 was
positively correlated with that of SKA2 in glioma tissues, which
further indicates that SPRY4-IT1 is associated with the expression
of SKA2 (Fig. 3C).
To determine the role of SPRY4-IT1 in SKA2
expression, U251 cells were transfected with si-SPRY4-IT1. U251
cells transfected with si-SPRY4-IT1 expressed lower levels of SKA2
mRNA than cells transfected with si-NC (Fig. 3D). Similarly, the protein expression
of SKA2 was significantly reduced in si-SPRY4-IT1 cells compared
with si-NC U251 cells (Fig. 3C).
These data indicated that SPRY4-IT1 promotes SKA2 expression in
U251 cells.
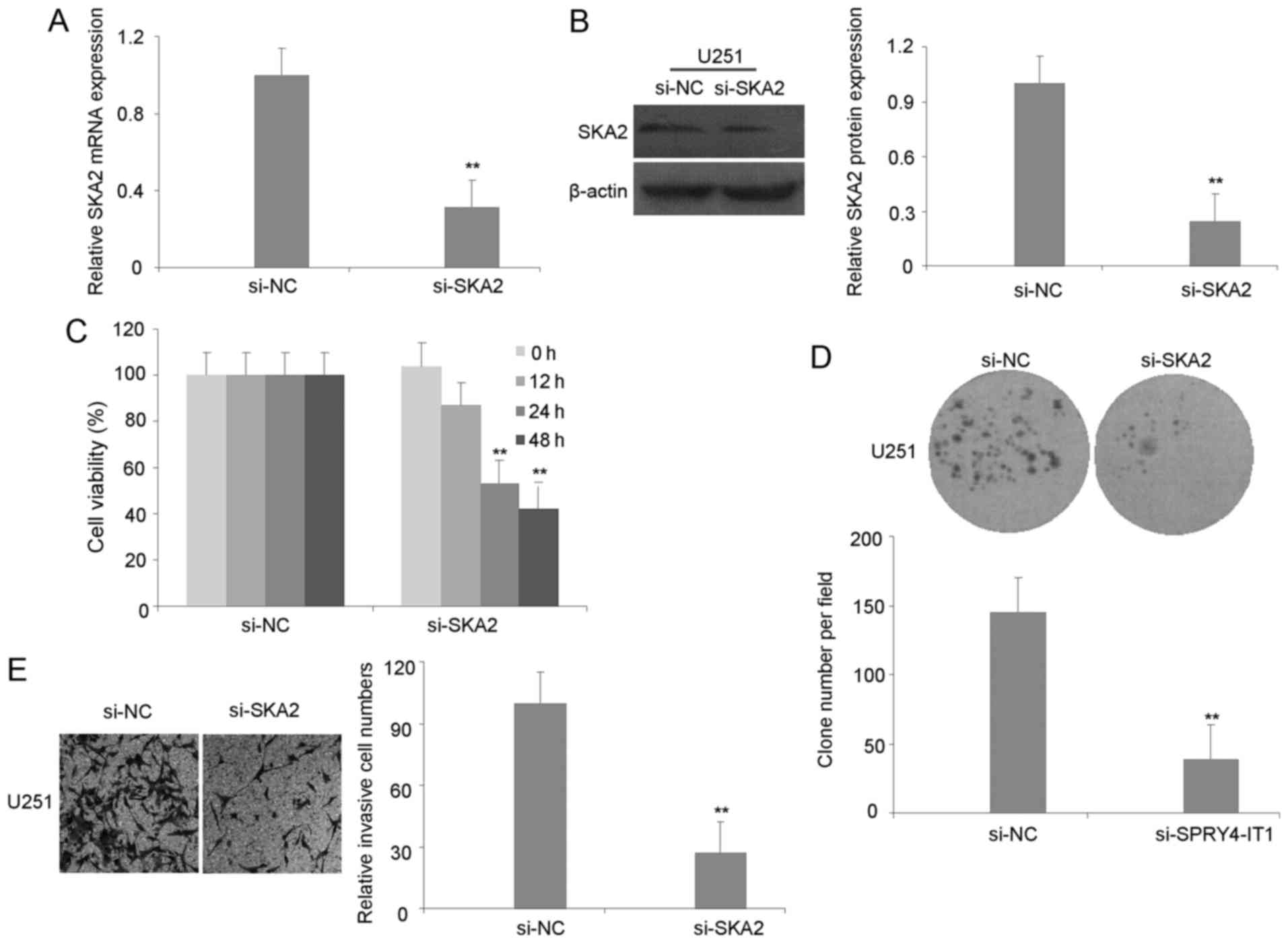
Knockdown of SKA2 inhibits the
biological behavior of U251 cells
To investigate the role of SKA2 on U251 cells, SKA2
expression was knocked down. SKA2 expression was markedly decreased
in U251 cells following transfection of si-SKA2 compared with si-NC
(Fig. 4A and B). Similar to results
observed with SPRY4-IT1-knockdown, knockdown of SKA2 significantly
inhibited U251 cell proliferation and colony formation (Fig. 4C and D). Additionally, the invasion of
U251 cells was inhibited in si-SKA2 cells compared with si-NC cells
(Fig. 4E). To investigate the
mechanism by which SKA2-knockdown inhibited formation of the glioma
malignant phenotype, investigated the effect of SKA2 on the
expression of proteins involved in cellular proliferation,
including cyclin D1 and PCNA, and invasion, including MMP2 and
MMP9. Knockdown of SKA2 reduced the expression of cyclin D1 and
PCNA in U251 cells compared with si-NC cells, whereas the
expression of MMP2 and MMP9 was significantly reduced in the cells
transfected with si-SKA2 siRNA (Fig.
5A). Together, these results indicate that the knockdown of
SKA2 inhibits cell proliferation, which is associated with the
decreased expression of cyclin D1 and PCNA, and suppresses the
invasive phenotype associated with the reduced expression levels of
MMP2 and MMP9 in U251 cells.
Discussion
Glioma is the most common primary malignancy of the
human brain (22). Despite the use of
aggressive therapeutic strategies, including surgical resection,
radiotherapy and chemotherapy, the prognosis of glioma patients
remains poor (23). Previous studies
demonstrated that glioma development is associated with rates of
cellular proliferation, migration, apoptosis and invasion (1,24). It is
therefore vital to understand the major regulatory mechanisms of
this malignancy, which is key to the development of novel and
effective therapeutic interventions for gliomas.
lncRNAs are a class of RNA molecules that regulate
the transcription of target genes, meaning that the difference in
lncRNA profiling between cancer and normal cells may indicate the
mechanisms of cancer transformation. Evidence indicates that
lncRNAs may serve pivotal roles in a range of cancer-associated
biological processes, including in glioma (15,16). By
acting as oncogenes or tumor suppressors, lncRNAs have the
potential to contribute to glioma initiation, progression and other
malignant phenotypes. SPRY4-IT1 is a lncRNA that was initially
associated with melanoma, and is upregulated in melanoma patient
samples (17). The present study
investigated SPRY4-IT1 expression in gliomas patients for the first
time. SPRY4-IT1 expression was upregulated in gliomas tissues and
cell lines compared with normal brain tissues. However, SPRY4-IT1
expression was markedly downregulated in non-small cell lung cancer
tissues, compared with normal tissues (25). These findings indicate that SPRY4-IT1
expression may be tissue- and cell-specific, and that SPRY4-IT1 may
serve different roles in different tissues and stages of life
process.
A previous study demonstrated that SPRY4-IT1
regulated the cell growth in melanoma cells; siRNA-mediated
SPRY4-IT1-knockdown in melanoma cells resulted in the inhibition of
cell growth, and an increase in the rate of apoptosis (17). Xie et al (26) demonstrated that knockdown of SPRY4-IT1
decreased the growth of esophageal squamous cell carcinoma in a
xenograft mice model. The present study demonstrated that knockdown
of SPRY4-IT1 significantly decreased the growth of U251 cells in
vitro. Overexpression of SPRY4-IT1 has been shown to promote
melanoma cell migration and invasiveness, whereas knockdown of
SPRY4-IT1 inhibits melanoma cell invasion (17). The present study revealed that
knockdown of SPRY4-IT1 inhibited the migration and invasion of U251
cells; however, this phenomenon was not consistent with the study
reported by Zou et al conducted in trophoblast cells
(27). Knockdown of SPRY4-IT1 induced
the migration of HTR-8/SVneo cells, with overexpression of
SPRY4-IT1 resulting in the inhibition of this migration (28). These findings indicate that lncRNA may
present different biological functions in different cancer cells.
For example, overexpression of HOTAIR results in an increase in the
invasiveness and metastasis in primary breast tumors (29). Accordingly, knockdown of HOTAIR
inhibited cell proliferation and induced apoptosis in pancreatic
cancer cells (28). Although only a
few functional lncRNAs have been well identified, they have been
found to regulate gene expression at the levels of transcription,
post-transcription and chromatin modification (30,31).
SKA2 has been reported to locate to the
kinetochore-microtubule interface during mitosis, whereas
SKA-depleted cells cause chromosome congression defects and
subsequently cell death (18,32). A prior study revealed that A549 cells
were treated with a microRNA-301 inhibitor or SKA2-specific siRNA,
the mitotic index of the cells significantly increased, with a
corresponding decrease in the colony formation ability (33). Overexpression of SKA2 appears to
promote proliferation and human breast cancer progression (34). Recently, microarray analysis to
identify genes that exhibited a change in expression following
SPRY4-IT1 knockdown in MDA-MB-231 cells, indicating that SKA2 may
represent a notable downstream effector of SPRY4-IT1 (35). The present study revealed that SKA2
expressions were elevated in glioma tissues; a significant positive
correlation was also observed between SPRY4-IT1 and SKA2 in human
glioma tissues. An in vitro experiment confirmed that
SPRY4-IT1 knockdown markedly reduced SKA2 expression in U251 cells.
However, the biological function of SKA2 in glioma remains unknown.
The present study provides evidence that SKA2 serves an oncogenic
role in U251 cells. SKA2 knockdown suppressed cellular
proliferation and the invasiveness of U251 cells. The mechanism
behind these behaviors may be associated with the decreased
expression of proteins associated with proliferation and invasion.
However, the precise molecular mechanism of how SPRY4-IT1 regulates
SKA2 expression remains unclear and requires further study.
In conclusion, to the best of our knowledge for the
first time, the present study demonstrated that SPRY4-IT1
expression was increased in glioma tissues, whereas the knockdown
of SPRY4-IT1 inhibited U251 cells growth, migration and invasion.
SPRY4-IT1 may control the glioma malignant phenotype via regulation
of SKA2 expression. Further investigation of the functional and
clinical implications of SPRY4-IT1 and its target SKA2 may
facilitate the identification of novel therapeutic targets for
glioma.
Acknowledgements
This study was supported by grants from the National
Natural Science Foundation of China (nos. 81402078 and 81502149)
and the Natural Science Foundation of Anhui Province (nos.
1608085MH225 and 1508085MH194).
Glossary
Abbreviations
Abbreviations:
|
CNS
|
central nervous system
|
|
lncRNA
|
long non-coding RNAs
|
|
SKA2
|
spindle and kinetochore associated
complex subunit 2
|
|
DMEM
|
Dulbecco's modified Eagle's medium
|
|
FBS
|
fetal bovine serum
|
|
SPRY4-IT1
|
SPRY4-intronic transcript 1
|
|
NC
|
negative control
|
References
|
1
|
Hamza MA and Gilbert M: Targeted therapy
in gliomas. Curr Oncol Rep. 16:3792014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
De Witt Hamer PC, Robles SG, Zwinderman
AH, Duffau H and Berger MS: Impact of intraoperative stimulation
brain mapping on glioma surgery outcome: A meta-analysis. J Clin
Oncol. 30:2559–2565. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Smith JS, Chang EF, Lamborn KR, Chang SM,
Prados MD, Cha S, Tihan T, Vandenberg S, McDermott MW and Berger
MS: Role of extent of resection in the long-term outcome of
low-grade hemispheric gliomas. J Clin Oncol. 26:1338–1345. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
van den Bent MJ, Afra D, de Witte O, Ben
Hassel M, Schraub S, Hoang-Xuan K, Malmström PO, Collette L,
Piérart M, Mirimanoff R, et al: Long-term efficacy of early versus
delayed radiotherapy for low-grade astrocytoma and
oligodendroglioma in adults: The EORTC 22845 randomised trial.
Lancet. 366:985–990. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Carninci P, Kasukawa T, Katayama S, Gough
J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al:
The transcriptional landscape of the mammalian genome. Science.
309:1559–1563. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Trapnell C, Williams BA, Pertea G,
Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ and Pachter
L: Transcript assembly and quantification by RNA-Seq reveals
unannotated transcripts and isoform switching during cell
differentiation. Nat Biotechnol. 28:511–515. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Farazi TA, Spitzer JI, Morozov P and
Tuschl T: miRNAs in human cancer. J Pathol. 223:102–115. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Louro R, Smirnova AS and Verjovski-Almeida
S: Long intronic noncoding RNA transcription: Expression noise or
expression choice? Genomics. 93:291–298. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nagano T and Fraser P: No-nonsense
functions for long noncoding RNAs. Cell. 145:178–181. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wapinski O and Chang HY: Long noncoding
RNAs and human disease. Trends Cell Biol. 21:354–361. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pandey RR, Mondal T, Mohammad F, Enroth S,
Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D and Kanduri C:
Kcnq1ot1 antisense noncoding RNA mediates lineage-specific
transcriptional silencing through chromatin-level regulation. Mol
Cell. 32:232–246. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang X, Arai S, Song X, Reichart D, Du K,
Pascual G, Tempst P, Rosenfeld MG, Glass CK and Kurokawa R: Induced
ncRNAs allosterically modify RNA-binding proteins in cis to inhibit
transcription. Nature. 454:126–130. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhao J, Sun BK, Erwin JA, Song JJ and Lee
JT: Polycomb proteins targeted by a short repeat RNA to the mouse X
chromosome. Science. 322:750–756. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang P, Ren Z and Sun P: Overexpression of
the long non-coding RNA MEG3 impairs in vitro glioma cell
proliferation. J Cell Biochem. 113:1868–1874. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang JX, Han L, Bao ZS, Wang YY, Chen LY,
Yan W, Yu SZ, Pu PY, Liu N, You YP, et al: HOTAIR, a cell
cycle-associated long noncoding RNA and a strong predictor of
survival, is preferentially expressed in classical and mesenchymal
glioma. Neuro Oncol. 15:1595–1603. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Khaitan D, Dinger ME, Mazar J, Crawford J,
Smith MA, Mattick JS and Perera RJ: The melanoma-upregulated long
noncoding RNA SPRY4-IT1 modulates apoptosis and invasion. Cancer
Res. 71:3852–3862. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hanisch A, Sillje HH and Nigg EA: Timely
anaphase onset requires a novel spindle and kinetochore complex
comprising Ska1 and Ska2. EMBO J. 25:5504–5515. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Rice L, Waters CE, Eccles J, Garside H,
Sommer P, Kay P, Blackhall FH, Zeef L, Telfer B, Stratford I, et
al: Identification and functional analysis of SKA2 interaction with
the glucocorticoid receptor. J Endocrinol. 198:499–509. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Louis DN, Ohgaki H, Wiestler OD, Cavenee
WK, Burger PC, Jouvet A, Scheithauer BW and Kleihues P: The 2007
WHO classification of tumours of the central nervous system. Acta
Neuropathol. 114:97–109. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)). Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Furnari FB, Fenton T, Bachoo RM, Mukasa A,
Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, et al:
Malignant astrocytic glioma: Genetics, biology, and paths to
treatment. Genes Dev. 21:2683–2710. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
He J, Zhang W, Zhou Q, Zhao T, Song Y,
Chai L and Li Y: Low-expression of microRNA-107 inhibits cell
apoptosis in glioma by upregulation of SALL4. Int J Biochem Cell
Biol. 45:1962–1973. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Diamandis P and Aldape KD: Insights from
molecular profiling of adult glioma. J Clin Oncol. 35:2386–2393.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sun M, Liu XH, Lu KH, Nie FQ, Xia R, Kong
R, Yang JS, Xu TP, Liu YW, Zou YF, et al: EZH2-mediated epigenetic
suppression of long noncoding RNA SPRY4-IT1 promotes NSCLC cell
proliferation and metastasis by affecting the
epithelial-mesenchymal transition. Cell Death Dis. 5:e12982014.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xie HW, Wu QQ, Zhu B, Chen FJ, Ji L, Li
SQ, Wang CM, Tong YS, Tuo L, Wu M, et al: Long noncoding RNA
SPRY4-IT1 is upregulated in esophageal squamous cell carcinoma and
associated with poor prognosis. Tumour Biol. 35:7743–7754. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zou Y, Jiang Z, Yu X, Sun M, Zhang Y, Zuo
Q, Zhou J, Yang N, Han P, Ge Z, et al: Upregulation of long
noncoding RNA SPRY4-IT1 modulates proliferation, migration,
apoptosis, and network formation in trophoblast cells HTR-8SV/neo.
PLoS One. 8:e795982013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kim K, Jutooru I, Chadalapaka G, Johnson
G, Frank J, Burghardt R, Kim S and Safe S: HOTAIR is a negative
prognostic factor and exhibits pro-oncogenic activity in pancreatic
cancer. Oncogene. 32:1616–1625. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tsai MC, Manor O, Wan Y, Mosammaparast N,
Wang JK, Lan F, Shi Y, Segal E and Chang HY: Long noncoding RNA as
modular scaffold of histone modification complexes. Science.
329:689–693. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gaitanos TN, Santamaria A, Jeyaprakash AA,
Wang B, Conti E and Nigg EA: Stable kinetochore-microtubule
interactions depend on the Ska complex and its new component
Ska3/C13Orf3. EMBO J. 28:1442–1452. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cao G, Huang B, Liu Z, Zhang J, Xu H, Xia
W, Li J, Li S, Chen L, Ding H, et al: Intronic miR-301 feedback
regulates its host gene, ska2, in A549 cells by targeting MEOX2 to
affect ERK/CREB pathways. Biochem Biophys Res Commun. 396:978–982.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Shi W, Gerster K, Alajez NM, Tsang J,
Waldron L, Pintilie M, Hui AB, Sykes J, P'ng C, Miller N, et al:
MicroRNA-301 mediates proliferation and invasion in human breast
cancer. Cancer Res. 71:2926–2937. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Shi Y, Li J, Liu Y, Ding J, Fan Y, Tian Y,
Wang L, Lian Y, Wang K and Shu Y: The long noncoding RNA SPRY4-IT1
increases the proliferation of human breast cancer cells by
upregulating ZNF703 expression. Mol Cancer. 14:512015. View Article : Google Scholar : PubMed/NCBI
|