Introduction
Cervical cancer is one of the most common types of
malignant tumour and the fourth leading cause of cancer-associated
mortality in women worldwide (1,2). Although
a cervical cancer vaccine has been successfully developed, cervical
cancer is still associated with high worldwide mortality and
morbidity rates. At presentation, the majority of cases involve the
advanced stage. Cervical carcinogenesis is a multistep process that
involves genetic and epigenetic alterations of protein-coding
oncogenes, and suppressor genes. A previous study has revealed that
persistent infections with high-risk human papillomaviruses (HPVs)
are the main causes of almost all cervical cancer cases (3). However, a substantial body of evidence
has revealed that HPV infections alone are insufficient to induce
malignant transformation, and that other genetic alterations may be
involved in tumorigenesis and tumour progression (4). Based on this hypothesis, our previous
study indicated that the overexpression of ribonucleotide reductase
subunit M2 (RRM2), a downstream target gene of HPVE7, promotes
cervical carcinogenesis (5).
Human ribonucleotide reductase (RNR) is the
rate-limiting enzyme in the production of 2′-deoxyribonucleoside
5′-diphosphates, and is required for DNA synthesis and repair
(6). RNR consists of three subunits,
R1, R2 and p53R2. RRM1 contains enzymatically active sites and
binding sites for allosteric effectors. RRM2 and p53R2 are 80%
homologous, and both possess a diiron-tyrosyl radical cofactor that
is essential for enzyme activity (7).
RRM2 is expressed only during the late G1/early S phase and is
degraded in the late S phase (8).
RRM1 interacts with either RRM2 or p53R2 to become the
catalytically active form of eukaryotic RR. Therefore, the
catalytic activity of RNR is tightly controlled during the cell
cycle according to the level of RRM2. Previous studies have
demonstrated that the overexpression of RRM2 correlates with
cellular invasiveness, metastasis, tumorigenesis and poor patient
outcome (9–11). The characteristics of RRM2 that
promote tumour progression are associated with its capability to
induce the activities of various oncogenes, including those
encoding nuclear factor-kB, Myc proto-oncogene protein (c-Myc),
tyrosine-protein kinase transforming protein Fes and ornithine
decarboxylase (12,13). RRM2 has been reported to be regulated
by cell cycle-associated factors, including transcription factor
E2F (14). Inhibition of RRM2
expression induces apoptosis and G1/S-phase cell cycle arrest in
ovarian cancer (15). Elevated RRM2
expression has been reported to be associated with poor prognosis
of cervical cancer. However, the mechanisms by which RRM2 regulates
biological functions remain unknown.
Tumorigenesis is the result of uncontrollable cell
proliferation, which may be caused by various carcinogenic factors.
Several studies have demonstrated that inhibition of cell apoptosis
significantly promotes tumorigenesis (16,17).
Therefore, understanding the molecular mechanisms that underlie
cell apoptosis and cell cycle progression is critical for cancer
prevention. Cell cycle progression is an important component of
controlling cell proliferation (18).
The G1- to S-phase transition is a critical stage for cancer
formation (19).
To date, few studies have investigated the functions
of RRM2 in cervical cancer (5,20). The
results of the present study revealed that downregulation of RRM2
in cervical cancer cells increase apoptosis and G1-phase arrest. In
addition, RRM2 inhibits cell proliferation. To the best of our
knowledge, this study is the first to indicate that RRM2 has a
critical role in regulating apoptosis in cervical cancer cells.
Materials and methods
Cell culture, patient samples and
reagents
The human cervical cancer cell lines HeLa, SiHa,
CaSki and C33A were obtained from the Type Culture Collection of
the Chinese Academy of Sciences (Shanghai, China) and cultured in
Dulbecco's modified Eagle's medium (DMEM) supplemented with 10%
fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc., Waltham,
MA, USA) at 37°C in a humidified 5% CO2 atmosphere. Low
melting temperature agarose were purchased from BD Biosciences,
(Franklin Lakes, NJ, USA). Samples from 60 cases of cervical cancer
were collected from the Hubei Tumour Hospital (Wuhan, China). All
carcinoma tissues were confirmed as invasive cervical cancer and
the molecular subtypes of cervical carcinoma were identified based
on the 2009 International Federation of Gynaecology and Obstetrics
(FIGO) Classification (21).
Paracarcinoma tissues were also collected. The present study was
approved by the Institutional Review Board of Jiaying University on
human subject research and conducted in accordance with the
Declaration of Helsinki. Written informed consent was obtained from
each study participant. The mouse monoclonal antibodies against
RRM2 were purchased from Epitomics (cat. no. 8029-1; Abcam,
Cambridge, UK). GAPDH (cat. no. 97166), cyclin-dependent kinase 4
(CDK4; cat. no. 12790), cyclin D1 (cat. no. 2978), phospho-AKT
(S473) (cat. no. 4058), AKT (cat. no. 9272), c-Myc (cat. no.
13987), β-actin (cat. no. 3700), extracellular signal-regulated
kinase (ERK; cat. no. 9122) and phospho-ERK1/2 (Thr202/Tyr204)
(cat. no. 9101) antibodies were all purchased from Cell Signalling
Technology, Inc. (Danvers, MA, USA). Horseradish
peroxidase-conjugated secondary antibodies were from Sigma-Aldrich
(Merck KGaA; Darmstadt, Germany). The negative control (NC) small
interfering siRNA was chemically synthesised from Guangzhou
RioboBio Co., Ltd. (Guangzhou, China).
siRNA transfection
The human cervical cancer cells were seeded at
40–50% confluency and transfected with siRNA or negative control
siRNA using Lipofectamine® 2000 in Opti-MEM (both from
Invitrogen; Thermo Fisher Scientific, Inc.) following the
manufacturer's instructions. siRNA was transfected at a
concentration of 100 nM, while negative control siRNA was
transfected at 30 nM.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from cells using the TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, Inc.), and cDNA was
synthesized using the RevertAid First Strand cDNA Synthesis kit
K1622 (Thermo Fisher Scientific, Inc.). RT-qPCR was performed using
the FastStart Universal SYBR Green Master kit (Roche Diagnostics,
Basel, Switzerland) and analysed using an Applied Biosystems 7900
Real-Time PCR system. The concentrations of RNA were determined
using a NanoDrop ND-1000 instrument (Thermo Fisher Scientific,
Inc.), and aliquots of the samples were stored at −80°C. Primer
sequences were as follows: GAPDH forward, 5′-TGAACGGGAAGCTCACTGG-3′
and reverse, 5′-TCCACCACCCTGTTGCTGT-3′; RRM2 forward,
5′-TTTAGTGAGCTTAGCACAGCGGGA-3′ and reverse,
5′-AAATCTGCGTTGAAGCAGTGAGGC-3′. Data analysis was performed using
the 2−∆∆Cq relative expression quantity method (22).
Protein extraction and western blot
analysis
All cells were washed with PBS and lysed with RIPA
lysis buffer (Beyotime Institute of Biotechnology, Haimen, China)
supplemented with a protease and phosphatase inhibitor cocktail
(cat. no. 78440; Thermo Fisher Scientific, Inc.) on ice for 30 min.
Cell lysates were centrifuged for 10 min (12,000 × g, 4°C), the
precipitates were collected and the total protein concentrations
were determined using the BCA Protein assay kit (Beyotime Institute
of Biotechnology). Proteins (40 µg/well) were separated by 12%
SDS-PAGE and transferred to polyvinylidene fluoride membranes (EMD
Millipore, Billerica, MA, USA). The membranes were blocked in 5%
non-fat milk for 2 h at room temperature, then the membranes were
cultured at 4°C overnight with the primary antibodies CDK4
(1:1,000), GAPDH, cyclin D1, phospho-AKT (S473), AKT, c-Myc,
β-actin, ERK and phospho-ERK1/2 (Thr202/Tyr204) (1:2,000), rinsed
with Tris-buffered saline with Tween-20 (10 mM Tris-Hcl, 150 mM
NaCl, 0.1% Tween-20) for three times. Subsequently, membranes were
incubated with rabbit anti-mouse (cat. no. A9044) or goat
anti-rabbit (cat. no. A0545) secondary antibody (1:20,000;
Sigma-Aldrich; Merck KGaA) for 2 h at room temperature. The signals
were detected with the ECL Western Blotting substrate (Thermo
Fisher Scientific, Inc.), and protein band intensity was measured
using Quantity One software (version 4.52; Bio-Rad Laboratories,
Inc., Hercules, CA, USA). Three independent experiments were
performed.
Cell cycle analysis
Cervical cancer cells were cultured for 48 h,
digested by trypsinization, washed twice in PBS, and fixed with
cold 75% ethanol for 2 h at −2°C. The cells were washed twice in
PBS and resuspended in PBS containing DNase-free RNase A (20 µg/ml)
and propidium iodide (PI; 50 µg/ml) (BD Biosciences) in the dark
for 30 min at room temperature. The PI signal was determined by
flow cytometry at an excitation wavelength of 488 nm. The results
were analysed using ModFit LT™ software (version 4.1; Verity
Software House, Inc., Topsham, ME, USA).
Apoptosis assay
After 48 h transfections, an Annexin V-fluorescein
isothiocyanate (FITC) kit (Nanjing KeyGen Biotech, Co., Ltd.,
Nanjing, China) was used to assess apoptosis by flow cytometry.
Briefly, cells transfected with the siRNA or siRNA mock were
resuspended in 100 ml of binding buffer at a density of
1×106 cells/ml, and incubated with Annexin V-FITC for 15
min at room temperature away from bright light, then PI was added
for 5 min at room temperature. The cells were analysed with the
Beckman CXP software 2.1 on an FC-500 flow cytometer (both from
Beckman Coulter, Inc., Brea, CA, USA) within 1 h of the cell
collection. Apoptosis determination included early apoptotic cells
(Annexin V-positive, PI-negative) and late apoptotic cells Annexin
V-positive, PI-positive).
Cell proliferation assay
The cervical cancer cells were seeded in 96-well
plates at 5×103 cells/well and transfected on the
following day. Cell proliferation was determined at 24, 48, 72 and
96 h using the CellTiter 96 AQueousOne Solution Cell Proliferation
Assay kit (Promega Corporation, Madison, WI, USA), according to the
manufacturer's instructions as we previously described (23). The assays were performed in triplicate
and were repeated three times.
Colony formation assay
In order to perform the colony formation assay, 1 ml
of 0.6% agarose + 10% fetal calf serum-DMEM solution (Gibco; Thermo
Fisher Scientific, Inc.) was added to six-well plates. Cells
transfected with siRRM2 or siNC for 48 h were trypsinised and
seeded into the six-well plates at a density of 5×102
cells/well to form natural colonies. After culturing for 14 days at
37°C, cells were washed twice with PBS, fixed with 4% methanol for
20 min at room temperature and stained with haematoxylin for 20 min
at room temperature. Then, the colonies were statistically
analysed. The colony formation efficiency (%) was calculated as the
(number of colonies/number of cells seeded) × 100. The experiment
was performed in triplicate and repeated at least three times to
ensure data reproducibility.
Immunohistochemical staining
Immunohistochemical staining was performed as
described in our previous study (21). Briefly, formaldehyde-fixed
paraffin-embedded tissue sections were dewaxed in xylene solution
and rehydrated using graded ethanol solutions. Antigen retrieval
was performed using citrate buffer at 90°C for 30 min. The slides
were then incubated with RRM2 antibody (1:200; Epitomics,
Burlingame, CA, USA) at 4°C overnight, and horseradish
peroxidase-conjugated secondary antibody (rabbit anti-mouse IgG)
was added for 30 min at 37°C. The colour reaction was developed
with 3,3′-diaminobenzidine tetrahydrochloride/0.03%
H2O2 at room temperature without light for 10
min, followed by counter staining with haematoxylin for 2–4 min at
room temperature.
In vivo xenografts
Four-week-old female BALB/c nude mice (n=10; mean
weight was 18 g) were purchased from Shanghai SLAC Laboratory
Animal Co., Ltd. (Shanghai, China). The animals were housed in a
specific pathogen-free environment, and provided sterile food and
water ad libitum. Mice were divided into two groups (n=5),
either transfected with siRRM2 or negative control siRNA. A total
of 5×106 HeLa cells suspended in 200 µl PBS were
subcutaneously injected into the right flanks of the mice. Tumour
xenograft diameters were measured using digital calipers twice/week
for 35 days, and the tumour volume was calculated using the
following formula: Width2 × length × 0.5. On day 35, the
mice were sacrificed and the tumours were excised, fixed in 10%
formalin at 4°C for 24 h, and then washed with PBS twice and stored
in 0.1% sodium azide (http://www.bszh.com; Beijing, China) solution for
further paraffin embedded slide analysis. All animal experiments
were approved by the JiaYing University Animal Care and Use
Committee.
Statistical analysis
All data were obtained from at least three
independent experiments and are presented as the mean ± standard
deviation. Student's t-test was used to analyse the differences in
the means between the two different groups. Data were analysed
using GraphPad Prism software (version 6.0; GraphPad Software,
Inc., La Jolla, CA, USA). P<0.05 was considered to indicate a
statistically significant difference.
Results
RRM2 expression is associated with
cervical carcinogenesis
RRM2 contributes to the malignant cellular phenotype
of multiple human cancer types and its overexpression plays a
critical role in tumour invasion (10,12).
However, little is known regarding its functions in cervical
carcinogenesis. To identify the association between RRM2 expression
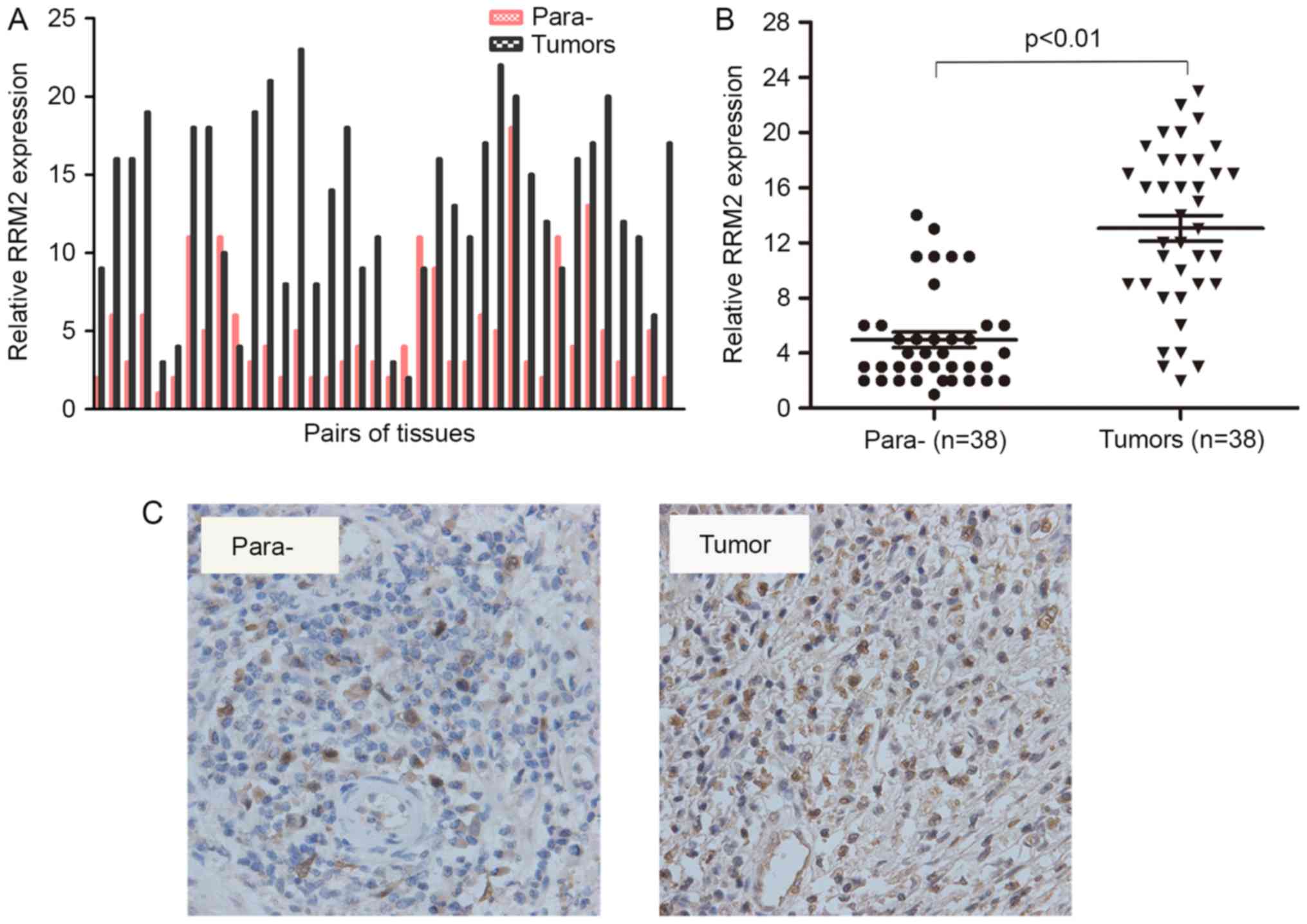
and cervical cancer carcinogenesis, the expression of RRM2 in 38
pairs of specimens was assessed using RT-qPCR. Each bar represents
a single patient. The RT-PCR results revealed that the RNA levels
of RRM2 were significantly increased in cervical cancer tissues
compared with paracarcinoma tissues (Fig.
1A and B). Furthermore, an immunohistochemical analysis of 5
other pairs of cervical cancer samples showed positive staining for
RRM2 in all tumour tissues (Fig. 1C).
These results demonstrated that the mRNA and protein levels of RRM2
were higher in tumours compared with in paracarcinoma tissues.
RRM2 knockdown inhibits the
proliferation of cervical cancer cells in vitro
Our previous study revealed that overexpression of
RRM2 induces angiogenesis in cervical cancer (5), but its involvement in other aspects of
cervical cancer development remains unclear. To elucidate the
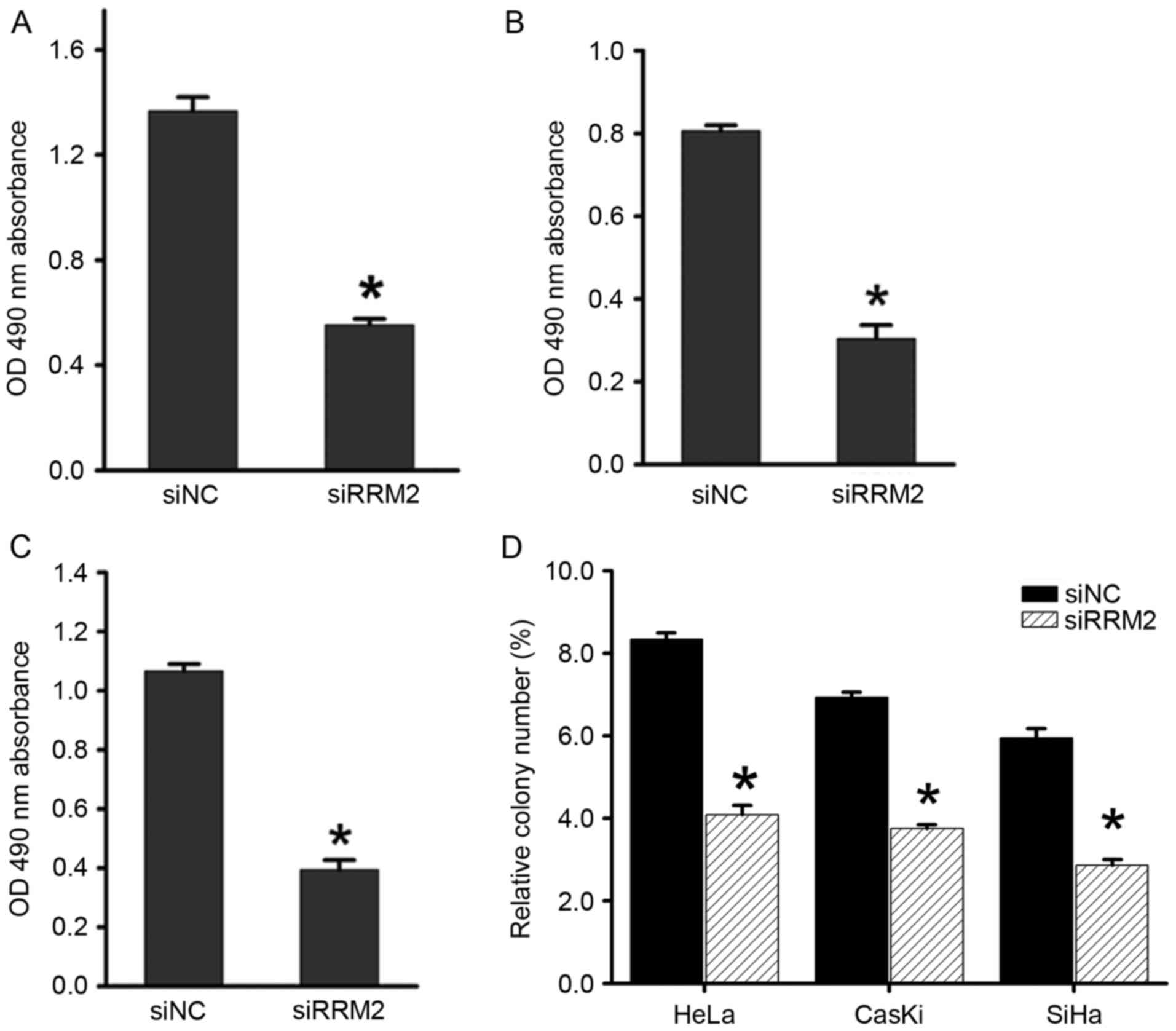
biological function of RRM2 in cervical tumorigenesis, RRM2
expression was transiently attenuated using siRNA in cervical
cancer cell lines (HeLa, SiHa and CaSki) and the most effective
siRNA was chosen for further experiments. The Cell Titer 96
AQueousOne Solution Cell Proliferation assay showed that
downregulation of RRM2 significantly inhibited cell proliferation
in all three cell lines compared with the control groups (Fig. 2A-C). Similarly, in an
anchorage-dependent monolayer colony formation assay, a significant
reduction in the colony number of each cell line transfected with
siRRM2 compared with the negative control siRNA group was observed
(Fig. 2D).
Downregulation of RRM2 inhibits
tumorigenesis in vivo
To further investigate the effects of RRM2 on the
tumorigenicity of cervical cancer, in vivo tumour formation
assays were performed. HeLa cervical cancer cells, which were
transfected with siRRM2 or negative control siRNA, were injected
into nude mice. Thirty-five days later, the mice were sacrificed,
and the tumours were removed and weighed. A significant decrease in
xenograft weight and volume was observed in the siRRM2 group
(Fig. 3A-C). To further examine the
expression of RRM2 in mice treated with siRRM2 or negative control
siRNA, total RNA was extracted from solid tumours and RT-qPCR was
performed. The results showed that RRM2 was significantly reduced
by siRRM2 (Fig. 3D), which suggested
that the downregulation of RRM2 inhibited tumorigenesis in
vivo. Therefore, these findings suggest that RRM2 plays an
important role in promoting the malignant growth of cervical cancer
cells in vivo.
Downregulation of RRM2 causes cell
cycle arrest at the G1 phase
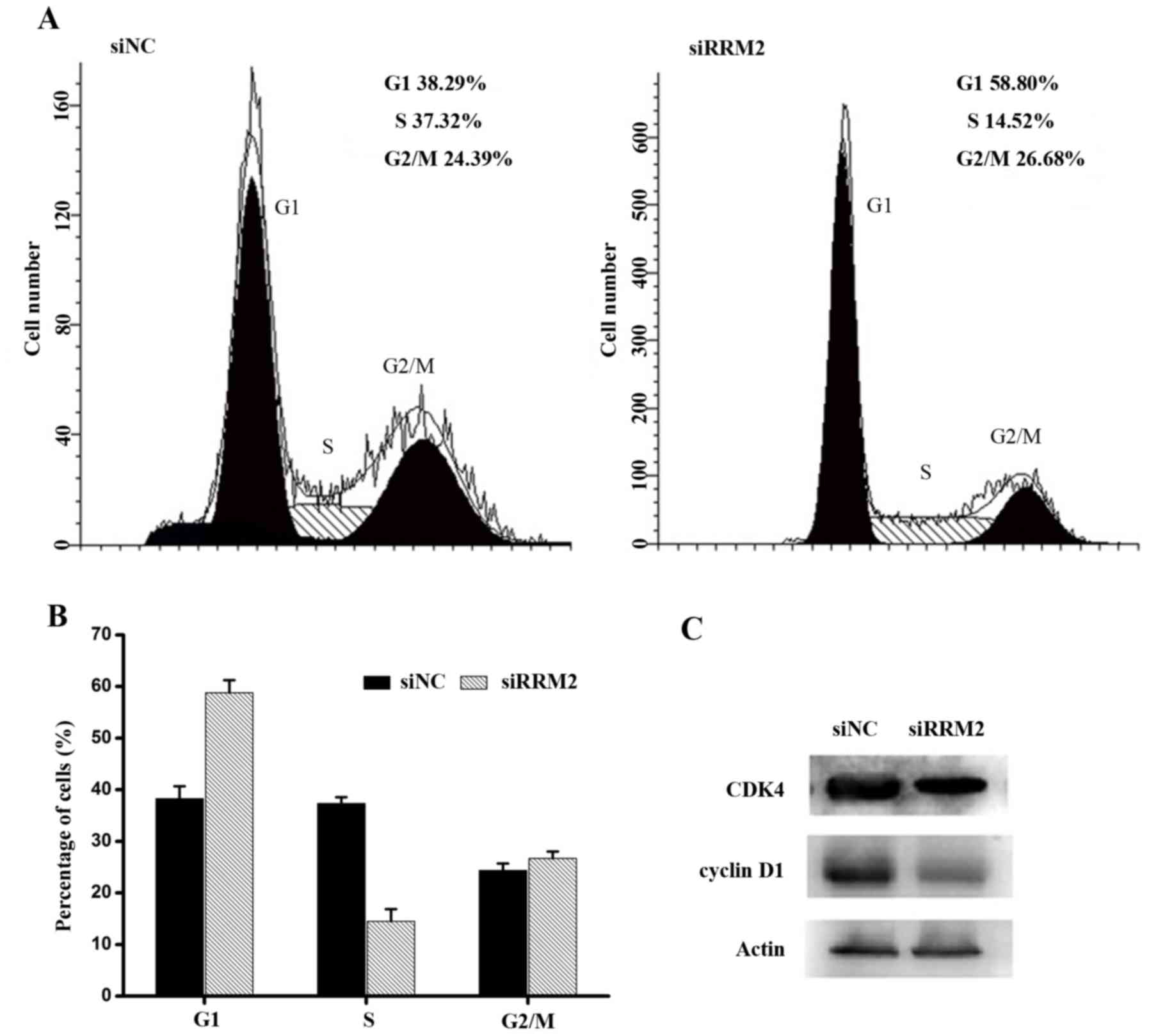
To better understand the growth inhibitory effect of
siRRM2 on cervical cancer cell lines, the cell cycle distribution
was analysed by flow cytometry. Cervical cancer cell lines were
transfected with siRRM2 or negative control siRNA, as shown in
Fig. 4A and B. Forty-eight hours
after the transient attenuation of RRM2 by siRNA, the percentage of
G1-phase cells increased, and the percentage of S-phase cells
decreased compared with the percentage of cells from the siRNA
control group. This result indicated that the downregulation of
RRM2 resulted in cell cycle arrest. To further evaluate the
mechanisms underlying cell cycle arrest after the siRRM2
transfection, cell cycle-associated proteins were analysed using
western blot analysis. It was demonstrated that active CDK4 and
cyclin D1 proteins were markedly downregulated compared with the
control group (Fig. 4C). Therefore,
it was concluded that the downregulation of RRM2 inhibited the
expression of active cyclin D1 and CDK4, leading to cell cycle
arrest at the G1 phase in cervical cancer cells.
Downregulation of RRM2 increases
apoptosis in cervical cancer cells
Next, the present study focused on the role of RRM2
in regulating cell apoptosis. An Annexin V-FITC apoptosis assay was
performed to evaluate the effects of RRM2-mediated apoptosis
regulation on cervical cancer tumorigenesis. As shown in Fig. 5A and B, there was an increase in the
early and late apoptotic cell populations of HeLa cells after the
siRRM2 transfection. The combined percentage of early and late
apoptotic siRRM2-treated cells was ~40%, which was significantly
increased compared with the control group (Fig. 5B). These results suggested that RRM2
suppresses cervical cancer cell growth by inducing G1-phase arrest
and apoptosis.
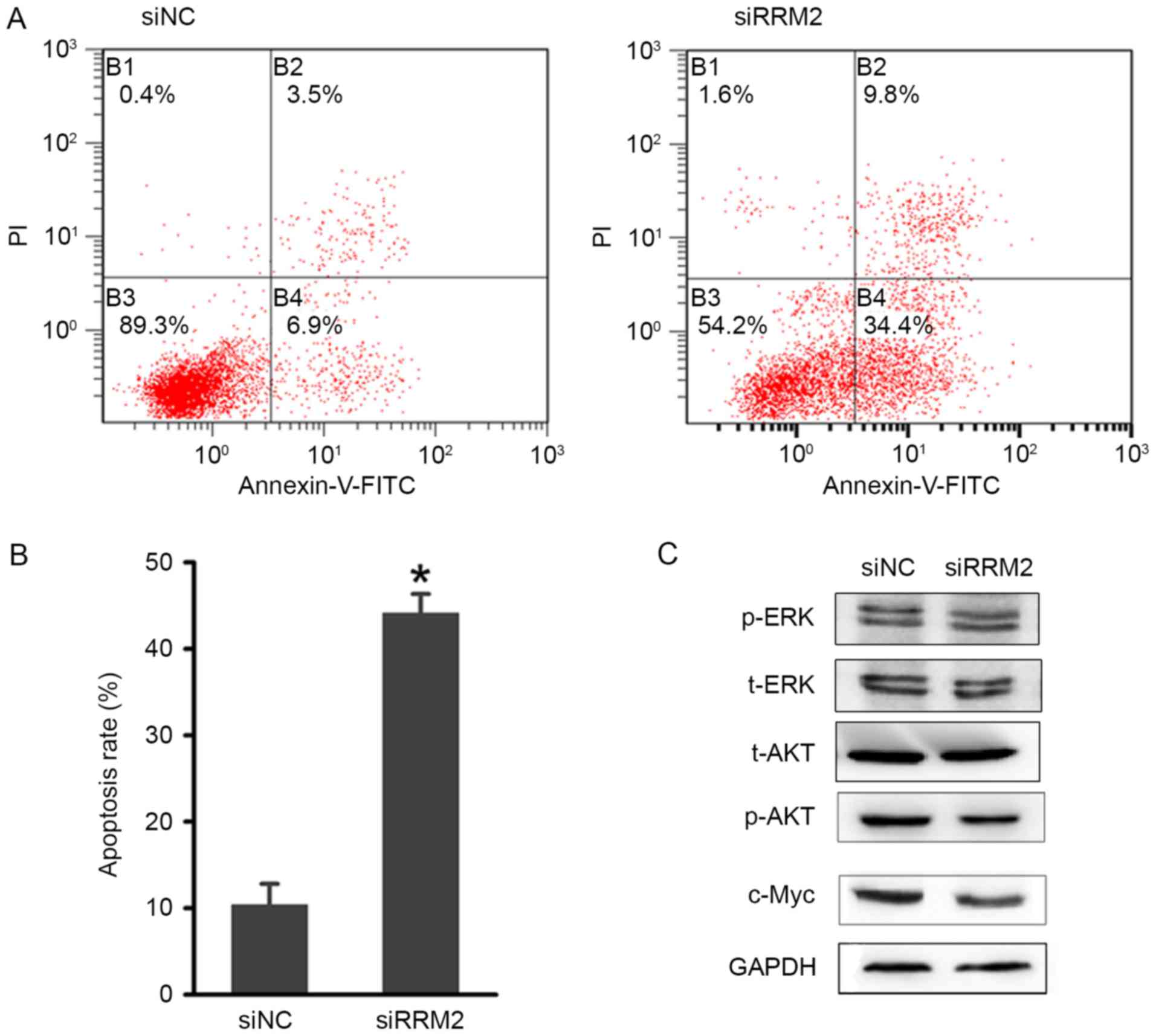 | Figure 5.Effect of RRM2 downregulation on
cervical cancer cell apoptosis. (A) Flow cytometric analysis of
HeLa cell apoptosis with RRM2 siRNA and NC siRNA. (B) Quantitative
analysis of the apoptosis rate following transfection. (C)
Representative blots demonstrating that RRM2 downregulation reduced
p-AKT and c-Myc protein expression. The protein expression level of
t-AKT, t-ERK and p-ERK were also analysed by western blotting, but
no evident changes were observed. *P<0.05. RRM2, ribonucleotide
reductase subunit M2; si, small interfering RNA; NC, negative
control; p, phosphorylated; t, total; AKT, AKT serine/threonine
kinase; ERK, extracellular signal-regulated kinase; c-Myc, Myc
proto-oncogene protein. |
To investigate the potential involvement of
signalling mechanisms in the apoptosis and cell cycle arrest that
was induced by RRM2 downregulation, western blot analysis of
associated proteins was performed. Based on previous studies
(24,25), several phosphorylated proteins
involved in cell cycle regulation were examined. As shown in
Fig. 5C, the phosphorylation of the
AKT protein was markedly reduced, while little variation in the
phosphorylation of the ERK1/2 protein was observed. The expression
of c-Myc was also markedly downregulated. Considering the
association between AKT and c-Myc in regulating tumorigenesis, the
AKT signalling pathway may be influenced by RRM2.
Discussion
Accumulating evidence has suggested that RRM2
upregulation is associated with cellular invasiveness (12), metastasis (26), tumorigenesis (10) and poor prognosis (27) in several types of cancer, including
colon cancer, pancreatic adenocarcinoma and lung adenocarcinoma. It
has been suggested that RRM2 may be a diagnostic and prognostic
molecular biomarker, and therapeutic target for cancer in the
future. However, the role of RRM2 in the regulation of apoptosis
and the cell cycle in cervical cancer has not been elucidated. To
the best of our knowledge, the results of the present study
presented a previously unknown role for RRM2 in cervical
carcinogenesis. The results of this study suggest that RRM2 may be
a promising target for the treatment of cervical cancer.
In the present study, it was demonstrated that RRM2
mRNA was significantly upregulated in cervical cancer compared with
paracarcinoma tissues. RRM2 knockdown by siRNA significantly
inhibited cervical cancer cell proliferation and induced cell
apoptosis in vitro. Based on the in vitro results,
the role of siRRM2 in tumorigenesis in nude mice was examined. It
was demonstrated that downregulation of RRM2 prevented xenograft
tumour growth in vivo, suggesting that decreased RRM2
expression inhibited tumorigenesis in cervical cancer. In line with
these results, the cell cycle distribution was further analysed to
investigate the mechanisms underlying the growth suppressive role
of siRRM2. It was revealed that RRM2 knockdown inhibited cell cycle
progression at the G1 phase through the AKT signalling pathway in
cervical cancer cells, and CDK4 and cyclin D1 protein expression
was downregulated. In addition, c-Myc expression was markedly
downregulated. Considering the association between AKT and c-Myc in
regulating tumorigenesis, further studies are needed to provide
further insights into the detailed molecular mechanisms by which
RRM2 decreases activation of the AKT signalling pathway.
RRM2 knockdown was shown to reduce cell
proliferation and the invasive ability in gastric cancer (28). In addition, silencing of RRM2
expression in pancreatic adenocarcinoma was demonstrated to
attenuate cellular invasiveness (12). In the present study, it was revealed
that the downregulation of RRM2 significantly induced apoptosis and
prevented cell cycle progression at the G1 phase in cervical cancer
cells. These results suggest that RRM2 has a significant role in
driving tumour cell invasion and metastasis. However, whether the
effect of RRM2 in regulating cervical cancer cell apoptosis, and
cell cycle arrest is time- and concentration-dependent will need to
be elucidated. Assays that assess the effect of RRM2 in metastasis
in vivo should be addressed in future studies to fully
understand its biological functions in cervical cancer
progression.
Several studies have shown that RRM2 is an
independent prognostic factor and may predict poor survival for
certain cancer types (29–31). However, few studies have elucidated
the underlying mechanisms of RRM2 in cervical cancer. To the best
of our knowledge, the results of the present study are the first to
demonstrate that the downregulation of RRM2 inhibits cervical
cancer tumorigenesis and progression. This may aid in improving the
understanding of the molecular mechanisms underlying cervical
cancer. It is of interest to further investigate whether RRM2 is
able to act as a biomarker for predicting the cervical cancer
prognosis and patient survival in the future.
Acknowledgements
The present study was supported by the Chinese
National Natural Science Foundation (grant no. 81502257) and by the
Initial Scientific Research Fund of Young Teachers in Jiaying
University (grant no. 311A0409).
References
|
1
|
Bodily JM, Mehta KP and Laimins LA: Human
papillomavirus E7 enhances hypoxia-inducible factor 1-mediated
transcription by inhibiting binding of histone deacetylases. Cancer
Res. 71:1187–1195. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hildesheim A and Wang SS: Host and viral
genetics and risk of cervical cancer: A review. Virus Res.
89:229–240. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mansur CP and Androphy EJ: Cellular
transformation by papillomavirus oncoproteins. Biochim Biophys
Acta. 1155:323–345. 1993.PubMed/NCBI
|
|
5
|
Wang N, Zhan T, Ke T, Huang X, Ke D, Wang
Q and Li H: Increased expression of RRM2 by human papillomavirus E7
oncoprotein promotes angiogenesis in cervical cancer. Br J Cancer.
110:1034–44. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nordlund P and Reichard P: Ribonucleotide
reductases. Annu Rev Biochem. 75:681–706. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tanaka H, Arakawa H, Yamaguchi T,
Shiraishi K, Fukuda S, Matsui K, Takei Y and Nakamura Y: A
ribonucleotide reductase gene involved in a p53-dependent
cell-cycle checkpoint for DNA damage. Nature. 404:42–49. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Engström Y, Eriksson S, Jildevik I, Skog
S, Thelander L and Tribukait B: Cell cycle-dependent expression of
mammalian ribonucleotide reductase. Differential regulation of the
two subunits. J Biol Chem. 260:9114–9116. 1985.PubMed/NCBI
|
|
9
|
Zhou BS, Tsai P, Ker R, Tsai J, Ho R, Yu
J, Shih J and Yen Y: Overexpression oftransfected human
ribonucleotide reductase M2 subunitin human cancer cells enhances
their invasive potential. Clin Exp Metastasis. 16:43–49. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang K, Hu S, Wu J, Chen L, Lu J, Wang X,
Liu X, Zhou B and Yen Y: Overexpression of RRM2 decreases
thrombspondin-1 and increases VEGF production in human cancer cells
in vitro and in vivo: Implication of RRM2 in angiogenesis. Mol
Cancer. 8:11–22. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu X, Zhang H, Lai L, Wang X, Loera S,
Xue L, He H, Zhang K, Hu S, Huang Y, et al: Ribonucleotide
reductase small subunit M2 serves as a prognostic biomarker and
predicts poor survival of colorectal cancers. Clin Sci (Lond).
124:567–578. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Duxbury MS and Whang EE: RRM2 induces
NF-kappaB-dependent MMP-9 activation and enhances cellular
invasiveness. Biochem Biophys Res Commun. 354:190–196. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fan H, Villegas C, Huang A and Wright JA:
The mammalian ribonucleotide reductase R2 component cooperates with
a variety of oncogenes in mechanisms of cellular transformation.
Cancer Res. 58:1650–1653. 1998.PubMed/NCBI
|
|
14
|
Chabes AL, Björklund S and Thelander L: S
Phase-specific transcription of the mouse ribonucleotide reductase
R2 gene requires both a proximal repressive E2F-binding site and an
upstream promoter activating region. J Biol Chem. 279:10796–10807.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang M, Wang J, Yao R and Wang L: Small
interfering RNA (siRNA)-mediated silencing of the M2 subunit of
ribonucleotide reductase: A novel therapeutic strategy in ovarian
cancer. Int J Gynecol Cancer. 23:659–666. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hanahan D and Weinberg RA: The hallmarks
of cancer. Cell. 100:57–70. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fulda S and Vucic D: Targeting IAP
proteins for therapeutic intervention in cancer. Nat Rev Drug
Discov. 11:109–124. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Vermeulen K, Van Bockstaele DR and
Berneman ZN: The cell cycle: A review of regulation, deregulation
and therapeutic targets in cancer. Cell Prolif. 36:131–149. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen HZ, Tsai SY and Leone G: Emerging
roles of E2Fs in cancer: An exit from cell cycle control. Nat Rev
Cancer. 9:785–797. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Su YF, Wu TF, Ko JL, Tsai HT, Tee YT,
Chien MH, Chou CH, Lin WL, Low HY, Chou MY, et al: The expression
of ribonucleotide reductase M2 in the carcinogenesis of uterine
cervix and its relationship with clinicopathological
characteristics and prognosis of cancer patients. PLoS One. 9:e
916442014. View Article : Google Scholar
|
|
21
|
Pecorelli S: Revised FIGO staging for
carcinoma of the vulva, cervix, and endometrium. Int J Gynaecol
Obstet. 105:103–104. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang N, Zhou Y, Zheng L and Li H: miR-31
is an independent prognostic factor and functions as an oncomir in
cervical cancer via targeting ARID1A. Gynecol Oncol. 134:129–137.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Molina JR and Adjei AA: The Ras/Raf/MAPK
pathway. J Thorac Oncol. 1:7–9. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li XD, Zhang YJ and Han JC: Betulin
inhibits lung carcinoma proliferation through activation of AMPK
signaling. Tumour Bio. l35:1–11158. 2014.
|
|
26
|
Liu X, Zhou B, Xue L, Yen F, Chu P, Un F
and Yen Y: Ribonucleotide reductase subunits M2 and p53R2 are
potential biomarkers for metastasis of colon cancer. Clin
Colorectal Cancer. 6:374–381. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Souglakos J, Boukovinas I, Taron M, Mendez
P, Mavroudis D, Tripaki M, Hatzidaki D, Koutsopoulos A,
Stathopoulos E, Georgoulias V and Rosell R: Ribonucleotide
reductase subunits M1 and M2 mRNA expression levels and clinical
outcome of lung adenocarcinoma patients treated with
docetaxel/gemcitabine. Br J Cancer. 98:1710–1715. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kang W, Tong JH, Chan AW, Zhao J, Wang S,
Dong Y, Sin FM, Yeung S, Cheng AS, Yu J and To K: Targeting
ribonucleotide reductase M2 subunit by small interfering RNA exerts
anti-oncogenic effects in gastric adenocarcinoma. Oncol Rep.
31:2579–2586. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ferrandina G, Mey V, Nannizzi S, Ricciardi
S, Petrillo M, Ferlini C, Danesi R, Scambia G and Del Tacca M:
Expression of nucleoside transporters, deoxycitidine kinase,
ribonucleotide reductase regulatory subunits, and gemcitabine
catabolic enzymes in primary ovarian cancer. Cancer Chemother
Pharmacol. 65:679–686. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kretschxmer C, Sterner-Kock A, Siedentopf
F, Schoenegg W, Schlag PM and Kemmner W: Identification of early
molecular markers for breast cancer. Mol Cancer. 10:152011.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Morikawa T, Maeda D, Kume H, Homma Y and
Fukayama M: Ribonucleotide reductase M2 subunit is a novel
diagnostic marker and a potential therapeutic target in bladder
cancer. Histopathology. 57:885–892. 2010. View Article : Google Scholar : PubMed/NCBI
|