Introduction
Acute lymphoblastic leukemia (ALL) is the most
common type of leukemia in children, with a low morbidity in adults
(1). The efficacy of treatment is
significantly increased by optimization of chemotherapy, improved
treatment conditions and risk stratification (2). Factors, including age, white blood cell
count, genetic characteristics and treatment response, determine
the prognosis of adults with ALL (3).
The genetic characteristics encompass genomic mutations and gene
variations, and genomic analysis proposes a novel perspective on
the pathogenesis and prognosis of ALL (4). The association between gene copy number
variations (CNVs) and prognosis in adults with ALL has been
investigated, but remains inconclusive.
Multi-link probe amplification (MLPA) was initially
reported by Schwab et al (5)
and Schouten et al (6). This
method permits detection of multiple minor CNVs in the human genome
and differences in the relative copy number of the target
sequences. The method is commonly used to analyze the multiple gene
polymorphisms underlying the disease, particularly for the analysis
of large samples.
The present study used MLPA to analyze the gene CNVs
in 87 adults with ALL treated between July 2009 and March 2015 at
the Institute of Hematology and Blood Diseases Hospital (Tianjin,
China). The aim of the present study was to determine the
association between gene CNVs and the prognosis of a Chinese
population of adults with ALL.
Materials and methods
Patients and samples
A total of 87 adult patients with ALL that were
diagnosed and treated at the Leukemia department, Institute of
Hematology and Blood Diseases Hospital between July 2009 and March
2015 were enrolled in the present study. The inclusion criteria was
patients who were diagnosed with ALL aged >14 years. Individuals
who had received treatment in other hospitals or were unable to
afford regular chemotherapy were excluded. All the patients
enrolled in the present study provided written informed consent and
the study was approved by Ethics Committee of the Institute of
Hematology and Blood Diseases Hospital (Tianjin, China). The
diagnosis was based on the morphology, immunophenotype, and
molecular and cytogenetic analysis. The median follow-up time was
12.12 months (range, 1.25–63 months) and the rate of loss to
follow-up was 5.7% (5/87). The patients were treated with regimens
prescribed by ChiCTR-TRC-00000397 as described in Zhao et al
(7), Bone marrow (BM) mononuclear
cells (MNC) were collected prior to the induction of treatment and
a QIAamp DNA Blood Mini kit (cat. no. 51104; Qiagen GmbH, Hilden,
Germany) was used for DNA extraction, according to the
manufacturer's protocols. TRIzol™ (Life Technologies;
Thermo Fisher Scientific, Inc., Waltham, MA, USA) was used to
extract RNA, RNA was also extracted from the MNCs of 50 patients
(MNCs <106, as dictated by the TRIzol protocol) and
was synthesized into cDNA, as previously described (8). Nested reverse transcription polymerase
chain reaction (RT-PCR) was performed, as previously described (PCR
Master mix; Takara Biotechnology Co., Ltd., Dalian, China)
(8).
The present study investigated 87 adults with ALL,
including 54 males and 33 females, with a median age of 19 years
(range, 14–61 years). Of these patients, 69 presented with B-ALL
and 18 with T-ALL. Among the patients with B-ALL, 29 patients
exhibited abnormal t(9;22)/BCR-ABL1, which is also described as Ph
positive chromosome (Ph+ ALL) and 40 exhibited the Ph
negative chromosome (Ph− ALL).
Subgroups included 53 patients in the high-risk
group (HR) and 16 in the low-risk group (SR). The T-ALL group
included 15 cases of HR and 3 cases of SR. The prognosis was based
on the guidelines by Gökbuget and Hoelzer (9). The age of the SR group was ≤35 years and
the white blood cell count was <30×109/l; and
TEL-AML1, HOX11, NOTCH1, 9p and polyploidy were observed. In
contrast, the HR group included patients aged ≥35 years with a
white blood cell count of >30×109/l in B-ALL
(>100×109/l in T-ALL), diagnosed with pro B-ALL and
exhibiting a complex and hypodiploid karyotype. Furthermore, DNA of
10 healthy people were extracted as normal control, including 6
males and 4 females (age range, 22–45 years). The samples from
volunteers were collected from January to April 2015 at the
Institute of Hematology and Blood Diseases Hospital.
Analysis of copy number alterations
(CNAs)
The SALSA MLPA P335 ALL-IKZF1 kit (MRC Holland,
Amsterdam, the Netherlands) was applied to detect the gene CNAs,
according to the manufacturer's protocol. This kit was able to
detect the deletions of IKAROS family zinc finger 1 (IKZF1),
purinergic receptor P2Y8 (P2RY8), zinc finger protein,
Y-linked (ZFY), Janus kinase 2 (JAK2), paired box 5
(PAX5), ETS variant 6 (ETV6), RB transcriptional
corepressor 1 (RB1), BTG anti-proliferation factor 1
(BTG1), early B-cell factor 1 (EBF1), cyclin
dependent kinase inhibitor 2A/2B (CDKN2A/2B), cytokine
receptor like factor 2 (CRLF2), interleukin 3 receptor
subunit α (IL3RA), colony-stimulating factor 2 receptor α
subunit (CSF2RA) and short stature homeobox (SHOX)
genes. Electrophoresis (pop7 polymer used as supplied) and
quantification of fluorescein amidite-labeled amp (4 nmol/ml) icons
were performed on an ABI-3730 genetic analyzer (Applied Biosystems;
Thermo Fisher Scientific, Inc.), 80°C, 2 min. The resulting peak
intensities were normalized to the manufacturer's control probes
and the DNA from the normal control was used as a reference.
Statistical analysis
All statistical analyses were performed using SPSS
21.0 software (IBM Corp., Armonk, NY, USA). The data are presented
as median ± quartile. Relapse-free survival (RFS; defined as the
time between diagnosis and relapse) and overall survival (OS;
defined as the time between diagnosis and mortality or last
follow-up) were analyzed using the Kaplan-Meier method and the
differences between multiple groups were analyzed using the
log-rank test. Cox proportional hazards regression models were used
to assess the prognostic relevance of different factors. Other
comparisons were performed using the X2, Fisher exact, as
appropriate. P<0.05 was considered to indicate a statistically
significant difference.
Results
MLPA
Analysis of gene deletions
Gene deletions were detected in 58/87 (66.7%) cases
of ALL. The common deletions included those in the IKZF1
32.2% (28/87), CDKN2A 35.6% (31/87), CDKN2B 29.9%
(26/87), PAX5 18.4% (16/87) and RB1 13.8% (12/87)
genes. Deletions in the genes, EBF1 (8/87, 9.2%),
BTG1 (8/87, 9.2%) and ETV6 (7/87, 8%), while
deletions in the genes, IL3RA-1, JAK2, CSF2RA-1 and
P2RY8, accounted for <5%, and no gene deletions were
observed in 29 patients (33.3%; Fig.
1A).
 | Figure 1.Frequencies of copy number
alterations. (A) Gene deletions in the entire cohort (n=87). (B)
Gene deletions in patients with B-ALL or T-ALL. (C) Gene
amplification in the whole cohort; B-ALL, B-cell acute
lymphoblastic leukemia; T-ALL, T-cell acute lymphoblastic leukemia;
IKZF1, IKAROS family zinc finger 1; CDKN2A/2B, cyclin dependent
kinase inhibitor 2A/2B; C&I, CDKN2A/2B and IKZF1; RB1, RB
transcriptional corepressor 1; ETV6, ETS variant 6; PAX5, paired
box 5; BTG1, BTG anti-proliferation factor 1; EBF1, early B-cell
factor 1; JAK2, Janus kinase 2; CSF2RA, colony-stimulating factor 2
receptor α subunit; IL3RA-1, interleukin 3 receptor subunit α;
P2RY8, purinergic receptor P2Y8. |
In the B-ALL group, gene deletions were detected in
45/69 patients (65.2%). The commonly deleted genes were
IKZF1 (28/69, 40.6%), CDKN2A (22/69, 31.9%),
CDKN2B (20/69, 29%), PAX5 (15/69, 21.7%), RB1
(10/69, 14.5%), BTG1 (7/69, 10.1%), EBF1 (8/69, 9.2%)
and ETV6 (7/69, 8%). No gene deletions were observed in 24
patients (34.8%; Fig. 1B). Among
those with gene deletions, one single gene deletion was observed in
16 patients (16/45, 35.6%), two were observed in 11 patients
(11/45, 24.4%) and ≥3 were observed in 18 patients (18/45, 40%).
The loss of CDKN2A and/or CDKN2B (CDKN2A/2B)
was reported in 25 patients (25/45, 55.6%). The loss of
IKZF1 and CDKN2A/2B was observed in 11 patients
(11/45, 22.4%). Simultaneous deletions of IKZF1 and other
genes were reported in 23 patients (23/28, 82.1%). A total of 14
(14/15, 93.3%) cases of PAX5 deletions and 16 (17/25, 64%)
of CDKN2A/2B deletions were accompanied by the deletion of
other genes. All patients with BTG1 and RB1 deletions
exhibited deletions in other genes. At the end of the follow-up
period, 22 cases of recurrence were observed in patients with
B-ALL. Of these, 15 exhibited gene deletions, including 9 (9/28,
32.1%) with the loss of IKZF1, 9 (9/25, 36%) with the loss
of CDKN2A/2B and 5 (5/15, 33.3%) with the deletion of ≥3
genes.
Of the patients with T-ALL, 13/18 (72.2%) harbored
the following deletions: CDKN2A (9/18, 50%), CDKN2B
(6/18, 33.3%), ETV6 (4/18, 22.2%), RB1 (2/18, 11.1%),
EBF1 (2/18, 11.1%), PAX5 (1/18, 5.6%) and BTG1
(1/18, 5.6%; Fig. 1B). Of the 13
patients with gene deletions, 8 (8/18, 44.4%) exhibited 1 gene
deletion, 4 (4/18, 22.2%) exhibited 2 and 1 (1/18, 5.6%) exhibited
≥3. Three cases (3/18, 16.7%) were identified with the co-deletion
of CDKN2A/2B and other genes. Two (2/18, 11.1%) patients
with ETV6 deletions exhibited concurrent deletions of other
genes. All the T-ALL patients with BTG1 and RB1
deletions also exhibited other deletions, as observed in the
patients with B-ALL. A total of 15 cases of T-ALL displayed
recurrence, including 11 patients with gene deletions, of which 6
(6/9, 66.7%) exhibited CDKN2A/2B deletion.
IKZF1 gene deletion analysis
IKZF1 gene deletion was identified in 28/87
patients (32.2%). The IKZF1 gene deletion is significantly
more common in Ph+ patients compared with Ph−
B-ALL patients (21/29, 72.4% vs. 7/40, 17.5%; P<0.01). The
patients in the HR group exhibited a deletion of the IKZF1
gene more frequently than those in the SR group (27/53, 50.9% vs.
1/16, 6.3%. P=0.001). The deletion of CDKN2A/2B and
IKZF1 together in patients with Ph+ B-ALL was
more frequently observed than in those with Ph− B-ALL
(9/29, 31% vs. 3/40, 7.5%; P=0.021). The frequencies of
CDKN2A/2B and IKZF1 deletions were higher in the HR
group than in the SR group (20.8 vs. 0%; P=0.056; Table I).
 | Table I.Frequencies of gene deletions in
different groups of B-cell acute lymphoblastic leukemia
patients. |
Table I.
Frequencies of gene deletions in
different groups of B-cell acute lymphoblastic leukemia
patients.
|
| Ph Chromosome |
| Risk
stratification |
|
|---|
|
|
|
|
|
|
|---|
| Deleted gene | Ph+
(n=29) | Ph−
(n=40) | P-value | HR (n=53) | SR (n=16) | P-value |
|---|
| IKZF1 | 21 (72.4%) | 7 (17.5%) | 0.00 | 27 (50.9%) | 1 (6.3%) | 0.001 |
| CDKN2A/2B | 10 (34.5%) | 15 (37.5%) | 1 | 21 (39.6%) | 4 (25%) | 0.379 |
| C&I | 8 (27.6%) | 3 (7.5%) | 0.021 | 11 (20.8%) | 0 | 0.056 |
| RB1 | 9 (31%) | 1 (2.5%) | 0.001 | 9 (17%) | 1 (6.3%) | 0.433 |
| ETV6 | 2 (6.9%) | 2 (5%) | 1 | 3 (5.7%) | 1 (6.3%) | 1 |
| PAX5 | 7 (24.1%) | 8 (20%) | 0.771 | 11 (20.8%) | 4 (25%) | 0.736 |
| BTG1 | 5 (17.2%) | 2 (5%) | 0.122 | 6 (11.3%) | 1 (6.3%) | 1 |
| EBF1 | 5 (17.2%) | 1 (2.5%) | 0.122 | 5 (9.4%) | 1 (6.3%) | 1 |
| JAK2 | 2 (6.9%) | 0 | 0.173 | 2 (3.8%) | 0 | 1 |
| CSF2RA | 1 (3.4%) | 0 | 0.42 | 1 (1.9%) | 0 | 1 |
| IL3RA-1 | 2 (6.9%) | 0 | 0.173 | 2 (3.8%) | 0 | 1 |
| P2RY8 | 1 (3.4%) | 0 | 0.42 | 1 (1.9%) | 0 | 1 |
| No deletion | 5 (17.2%) | 19 (47.5%) | 0.011 | 15 (28.3%) | 10 (62.5%) | 0.018 |
| ≥3 deletions | 13 (44.8%) | 5 (12.5%) | 0.004 | 16 (30.2%) | 2
(12.5%) | 0.003 |
A total of 12 cases (12/28, 42.9%) revealed the
deletion of exons 4–7, which were the most common deletions, and
deletions of exons 1–8 were observed in 2 cases (2/28, 7.1%). Only
single cases exhibited a deletion of exons 1 or 6.
A total of 50 cases, including 38 cases of B-ALL and
12 cases of T-ALL, were analyzed by nested RT-PCR for IKZF1
deletion. A total of 32 patients with IKZF1 deletion (64%;
20 patients with IK6 subtype), including 28 patients with B-ALL
(73.7%; 16 patients with Ph+) and 4 patients with T-ALL
(33.3%), were evaluated by PCR. However, MLPA indicated that only
16/38 patients with B-ALL exhibited the deletion of IKZF1 (8
cases of IK6 subtype), while none of the 12 patients with T-ALL
presented with an IKZF1 deletion. Therefore, the sensitivity
of the two methods was different.
Analysis of other gene deletions
RB1 deletions in the Ph+ group of
patients with B-ALL were more frequent than in the Ph−
group (8/29, 31% vs. 1/40, 2.5%; P=0.001). However, single-gene
defects in RB1 were not observed. More than three gene
deletions were commonly observed in the Ph+ group of
patients with B-ALL compared with the Ph− group (13/29,
44.8% vs. 5/40, 12.5%; P=0.004). This phenomenon was more common in
the SR group than in the HR group (16/53, 30.2% vs. 2/16, 12.5%;
P=0.003). Furthermore, no significant differences were observed in
the distribution of other gene deletions across different
groups.
Analysis of gene amplification
Amplification of 12 genes was detected in 15
patients (15/87, 17.2%). The common amplifications were noted for
SHOX-AREA (3/15, 20%), BTG1 (3/15, 20%) and
EBF1 (3/15, 20%) genes. A total of 4/15 patients harbored
only the gene amplification and the remaining 11 patients displayed
concurrent gene deletions. A single gene amplification was reported
in 12 cases and >2 amplifications were observed in 3 patients.
The gene amplifications were identified in 14/15 cases of B-ALL, 1
case of T-ALL, 1 case in the SR group and 14 cases in the HR group
(6.25 vs. 26.4%). The specific gene amplifications are presented in
Fig. 1C.
Effects of gene deletion on survival
Prognostic significance of IKZF1
deletion
In B-ALL, the 2-year OS and RFS rates in patients
with IKZF1 deletions were slightly worse than in those
without, although no significant difference was observed (OS, 60.8
vs. 51.2%, P=0.247; RFS, 51.3 vs. 35.5%, P=0.169). Furthermore, no
significant differences in the 2-year OS (57.5 vs. 47.6%; P=0.256)
and RFS (55.6 vs. 34.3%; P=0.209) rates were observed between
patients with IKZF1 deletion and those without in the
Ph− B-ALL group. Additionally, no significant
differences in the 2-year OS (50 vs. 52%, P=0.284) and RFS (50 vs.
42.4%, P=0.256) rates were observed between the 21 patients with
IKZF1 deletion and those without in the Ph+ B-ALL
group.
Prognostic analysis of gene
deletions
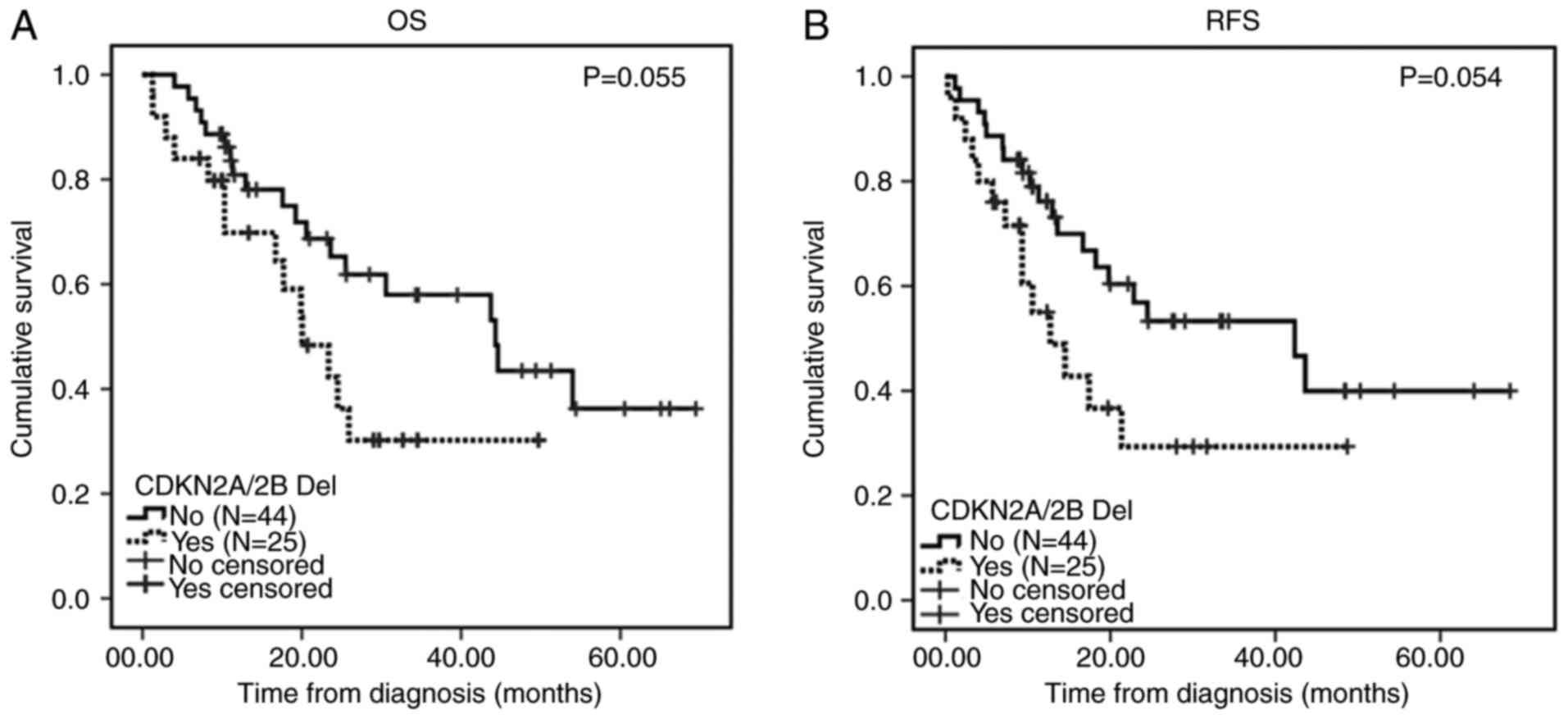
Among the patients with B-ALL, the 2-year OS (61.8
vs. 30.2%; P=0.055) and RFS (53.3 vs. 29.3%; P=0.054) rates in
patients with CDKN2A/2B deletions were slightly worse than
in those without these deletions (Fig.
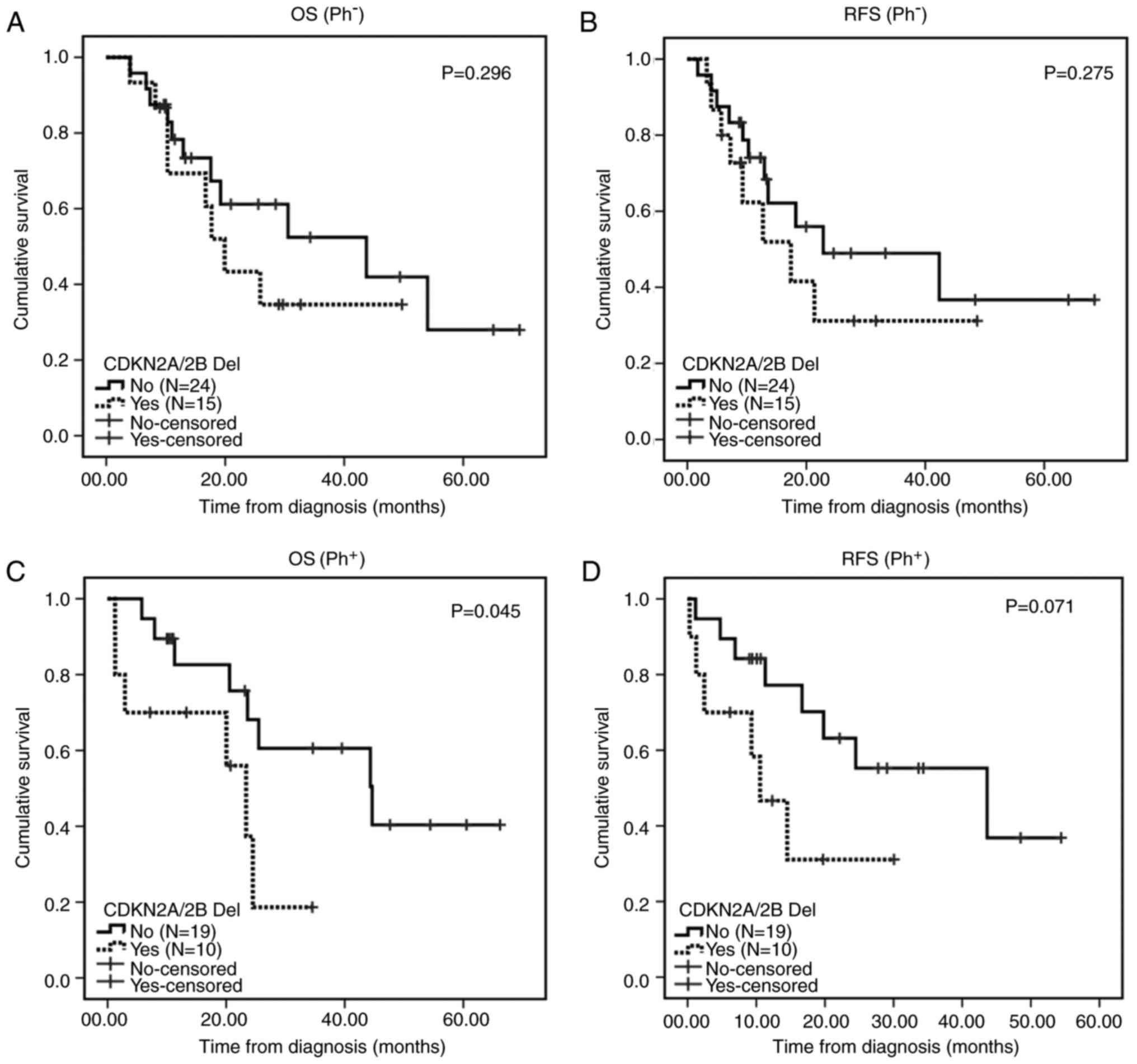
2). In addition, no significant differences in the 2-year OS
(55.2 vs. 34.7%; P=0.296) and RFS (51.7 vs. 31.2%; P=0.275) rates
were observed between patients with and without the
CDKN2A/2B deletions in the Ph− B-ALL group. The
2-year OS and RFS rates in patients with CDKN2A/2B deletions
were worse than in the other patients (60.6 vs. 18.7%, P=0.045) and
(63.2 vs. 31.1%, P=0.071) in the Ph+ B-ALL group
(Fig. 3). No significant differences
were observed in the 2-year OS (72.2 vs. 50%; P=0.544) and RFS
(70.7 vs. 50%; P=0.726) rates of patients with CDKN2A/2B
deletion in the SR group. The HR group carrying the
CDKN2A/2B deletion exhibited poor 2-year OS (58.4 vs. 24.7%,
P=0.037) and RFS (50.7 vs. 25%, P=0.047) rates compared with the
other patients (Fig. 4).
Prognostic analysis of PAX5
deletion
In the present study, 16 patients carried the
PAX5 gene deletion. Among the patients with B-ALL, no
significant difference in the 2-year OS (76.7 vs. 54%; P=0.432) and
RFS (54.3 vs. 45%; P=0.44) rates was observed between those with
and without PAX5 deletions. The Ph− B-ALL
patients with PAX5 deletions exhibited poor 2-year OS (90.1
vs. 19%; P=0.004) and RFS (83.7 vs. 21.4%; P=0.016) rates compared
with those without this deletion. No significant difference was
observed in the OS (59.1 vs. 53.6%; P=0.749) and RFS (63.7 vs.
57.1%; P=0.785) rates of Ph+ B-ALL patients with
PAX5 deletion with respect to those in the Ph+
B-ALL group (Fig. 5).
Prognosis of patients with ≥3 gene
deletions
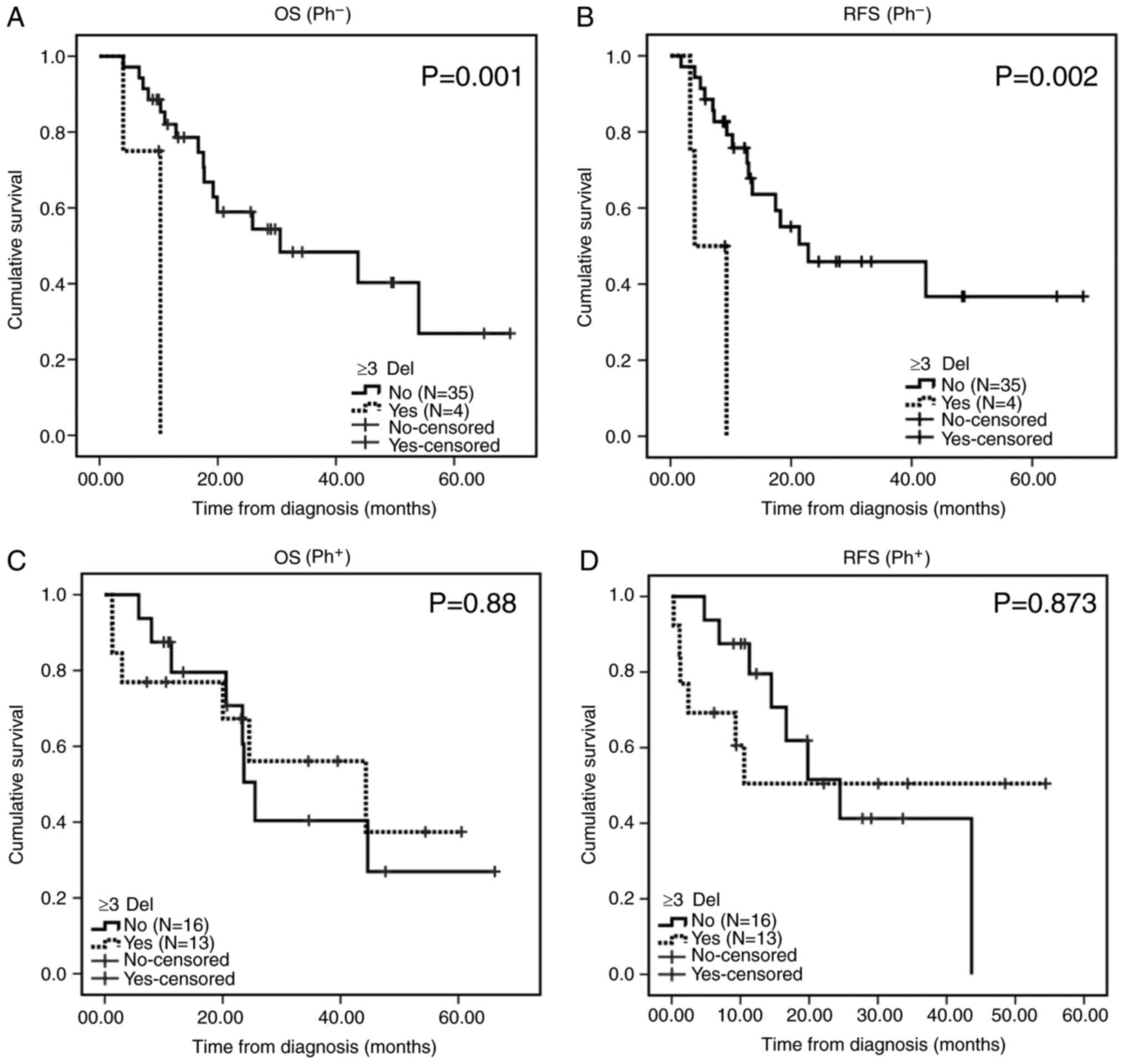
No notable differences were observed in the 2-year
OS (56.7 vs. 49.4%; P=0.738) and RFS (60.1 vs. 49.7%; P=0.455)
rates of B-ALL patients with ≥3 gene deletions (18 cases) compared
with the B-ALL patients who harbored <3 gene deletions. Patients
with ≥3 gene deletions exhibited poor 2-year OS (0 vs. 85.3%;
P=0.001) and RFS (0 vs. 79.2%; P=0.002) rates compared with those
in the Ph− B-ALL group. No marked differences were
observed in the 2-year OS (50.5 vs. 56.1%, P=0.88) and RFS (61.5
vs. 50.5%, P=0.873) rates of patients carrying ≥3 gene deletions
compared with those in the Ph+ group (Fig. 6).
Multifactor analysis of Cox multiple
regression
The OS and RFS rates of different patient groups
were analyzed using Cox regression based upon multiple factors. The
white blood cell count for OS and RFS demonstrated independent
prognostic significance in patients with B-ALL [P=0.056 and hazard
ratio (HR)=1.004, and P=0.011 and HR=1.005, respectively]. In the
Ph− group of patients with B-ALL, the white blood cell
count (OS, P=0.003, HR=1.007; RFS, P=0.001, HR=1.007) and ≥3 gene
deletions were independent prognostic factors (OS, P=0.007,
HR=12.4; RFS, P=0.06, HR=10.301). The CDKN2A/2B gene
deletions (OS, P=0.056, HR=3.0) demonstrated independent prognostic
significance in the Ph+ B-ALL group. The white blood
cell count (RFS, P=0.004, HR=1.005) and CDKN2A/2B gene
deletions (OS, P=0.059, HR=2.322) demonstrated independent
prognostic significance in the HR group. The PAX5 gene
deletion (OS, P=0.049, HR=2.322; RFS, P=0.056, HR=104.7)
demonstrated independent prognostic significance in the SR group of
patients with B-ALL.
Due to the limited number of T-ALL patients, and
limited number of patients exhibiting gene amplification, no
significant differences were observed in the survival analysis
between different groups.
Discussion
MLPA was used previously to detect the ALL gene copy
number in children (10), which
revealed that the commonly deleted genes included CDKN2A/B
(41%), PAX5 (35%), ETV6 (26%), RB1 (5.1%),
BTG1 (4.3%) and EBF1 (1.7%). The IKZF1
deletions accounted for 16, and 26% of the patients with
IKZF1 deletions were categorized as the IK6 subtype (4–7
exons deletion). A similar method of detection was observed in
1,644 cases among British children with ALL in 2014 (11). CDKN2A/2B and ETV6 are
the commonly (20–25%) deleted genes in children with ALL; and
IKZF1 and PAX5 gene deletions occurred simultaneously
in 15% of patients. The proportion of other gene deletions was
<10%. In ~43% patients, no gene deletions were detected and
patients with ≥3 types of gene deletions were observed in 10% of
all patients. MLPA facilitated the screening of 204 children with
ALL relapse (12). The common gene
deletions included CDKN2B (37.7%), CDKN2A (37.3%),
IKZF1 (33.3%), PAX5 (26.5%) and ETV6 (25%).
The proportion of IKZF1 gene deletions in these cases of ALL
relapse was ~2-fold that reported previously in children newly
diagnosed with ALL (33 vs. 14–19%). The common gene deletions
identified in 142 cases of adolescent and adult ALL were
CDKN2A/2B (42%), IKZF1 (35%), PAX5 (34%),
RB1 (15%), BTG1 (10%), EBF1 (11%) and
ETV6 (7%) (12). The majority
of the patients with IKZF1 and CDKN2A/2B deletions
also harbored other deletions. The proportion of IKZF1
deletions in Ph+ patients was higher, and the age and
white blood cell count of patients with IKZF1 deletion were
significantly higher than that of those without this deletion.
The results of the present study demonstrated that
58/87 (66.7%) patients with ALL harbored a gene deletion. The genes
that were frequently deleted included IKZF1 (40.6%),
CDKN2A (31.9%), CDKN2B (29%), PAX5 (21.7%),
RB1 (14.5%), BTG1 (10.1%), EBF1 (9.2%) and
ETV6 (7/69, 8%). The 24 patients without gene deletions
comprised 34.8% of the cohort. A total of 25 patients (55.6%)
carried deletions in CDKN2B and/or CDKN2A genes. A
concurrent deletion of IKZF1 and CDKN2A/2B genes was
observed in 11 patients (22.4%). Furthermore, in 16 patients
(35.6%), only 1 gene was deleted, while 11 (24.4%) patients carried
two gene deletions. More than 3 types of gene deletion were
detected in a total of 18 cases (40%). Concurrent mutations,
including 82.1% with a deletion of IKZF1 and other genes,
and 93.3% with a deletion of PAX5 and other genes, were
detected. In addition, IKZF1 gene deletions were more common
in the Ph+ group of patients with B-ALL than in the
Ph− group (72.4% vs. 17.5%), and were more frequent in
the HR group than in the SR group (50.9% vs. 6.3%), which was in
agreement with previous international adolescent and adult ALL
studies (13). A total of 42.9%
(12/28) patients with IKZF1 deletions exhibited the IK6
subtype, a ratio that was higher than that reported in previous
studies regarding pediatric ALL (14).
As demonstrated in Table
II, the gene CNVs differed between adults and children with
ALL. The ratio of ETV6 deletions in children with ALL was
higher, and the ratios of IKZF1, CDKN2A/2B and EBF1
gene deletions were significantly lower than those in the adults.
The prevalence of multiple gene deletions was lower in children
than in adults. In the present study, IKZF1 gene deletions
were predominant, followed by CDKN2A/2B deletions. Ribera et
al (15) reported that German
adolescents and adult patients with ALL exhibited prevalent
deletions of CDKN2A/2B.
 | Table II.Gene copy number variation in ALL
patients across different study groups. |
Table II.
Gene copy number variation in ALL
patients across different study groups.
|
|
| Gene Del (%) |
|
|---|
|
|
|
|
|
|---|
| Patients | No. | IKZF1 | CDKN2A/2B | PAX5 | RB1 | BTG1 | EBF1 | ETV6 | ≥3 Del | No Del | (Refs.) |
|---|
| UK, Pediatric
ALL | 1,644 | 27 | 11 | 17 | 2 | 6 | 1 | 23 | 10 | 43 | (11) |
| Sweden, Pediatric
ALL | 116 | 41 | 16 | 3.5 | 5.1 | 4.1 | 7.1 | 26 | – | – | (10) |
| Germany, Relapse
Pediatric ALL | 204 | 37.7 | 33.3 | 26.5 | 6.4 | 9.3 | 37.7 | 25 | – | – | (12) |
| Germany, Young and
Adult B-ALL | 142 | 42 | 35 | 34 | 15 | 10 | 11 | 7 | 48.6 | 18 | (13) |
| Adult ALL | 87 | 35.6 | 32.2 | 18.4 | 13.8 | 9.2 | 9.2 | 8 | 21.8 | 33.3 | The present
study |
| Adult B-ALL | 69 | 31.9 | 40.6% | 21.7 | 14.5 | 10.1 | 9.2 | 8 | 40 | 34.8 |
|
Ofverholm et al (10) revealed that deletion of the
IKZF1 gene in childhood ALL was associated with poor OS and
event-free survival (EFS) while no significant differences in OS
and EFS were observed in children with CDKN2A/2B deficiency.
Moorman et al (11) combined
the incidence of CNA with risk stratification and revealed that the
deletion ratios of CDKN2A/2B, PAX5 and IKZF1 in
patients with a poor OS and EFS accounted for 70, 45 and 45%,
respectively. The proportion of patients with a better prognosis
was 1, 5, and 0%, respectively. Ribera et al (15) reported that the 5-year cumulative
incidence rate (CIR) was higher and that the OS was poorer in
patients with IKZF1 deletion than in those without the
deletion. Additionally, CDKN2A/2B deficiency in patients
with B-ALL was associated with a poor OS. The OS of patients with
B-ALL, particularly those with Ph− ALL carrying ≥3 gene
deletions was poor with an increased CIR. Adult Ph+ ALL
patients carrying CDKN2A/2B deletions also exhibited a poor
disease-free survival (DFS) (16).
The present study evaluated the 2-year OS and RFS in different
groups of patients. The deletions of CDKN2A/2B in B-ALL
patients were associated with a poorer prognosis compared with that
of other patients without CDKN2A/2B deletions. Patients with
CDKN2A/2B deletions and those with concurrent IKZF1
and CDKN2A/2B deletions exhibited a poorer prognosis than
the patients in the Ph+ ALL group. Compared with the
patients without PAX5 deletions, those with PAX5
deletions exhibited a poorer prognosis in the SR group of patients
with B-ALL and those with CDKN2A/2B deletion exhibited a
poorer prognosis in the HR group of B-ALL patients than patients
who did not harbor CDKN2A/2B deletion. Patients with PAX5
deletions and ≥3 gene deletions exhibited a poorer prognosis than
the patients in the Ph− group. As mentioned earlier,
patients with IKZF1 gene deletions did not exhibit a poor
prognosis in the present study, which may be attributed to the
small sample size and short follow-up duration. In addition, it was
revealed that MLPA was less sensitive than PCR in analyzing the
IKZF1 gene deletions, which may result in an increased
number of false negative cases and may influence the prognostic
significance of such observations (8).
To conclude, 66.7% of adult patients with ALL in a
Chinese population exhibited variations in gene copy number. The
types and proportions of gene variation were consistent with the
results reported in the literature for adult ALL and it was
concluded that certain gene copy number variations may be used to
predict the prognosis of ALL.
Acknowledgements
The authors would like to thank the Department of
Leukemia, Institute of Hematology and Blood Diseases Hospital,
Chinese Academy of Medical Sciences and Peking Union Medical
College for providing patient samples and the State Key Laboratory
of Experimental Hematology for technical assistance. The present
study was supported by the National Science and Technology Pillar
Program (grant no. 2014BAI09B12), the Tianjin Major Research
Program of Application Foundation and Advanced Technology (grant
no. 15JCZDJC36400) and the Science and Technology Project of
Tianjin (grant no. 15ZXLCSY00010).
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Mullighan CG, Goorha S, Radtke I, Miller
CB, Coustan-Smith E, Dalton JD, Girtman K, Mathew S, Ma J, Pounds
SB, et al: Genome-wide analysis of genetic alterations in acute
lymphoblastic leukaemia. Nature. 446:758–764. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kuiper RP, Schoenmakers EF, van
Reijmersdal SV, Hehir-Kwa JY, van Kessel AG, van Leeuwen FN and
Hoogerbrugge PM: High-resolution genomic profiling of childhood ALL
reveals novel recurrent genetic lesions affecting pathways involved
in lymphocyte differentiation and cell cycle progression. Leukemia.
21:1258–1266. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Harvey RC, Mullighan CG, Wang X, Dobbin
KK, Davidson GS, Bedrick EJ, Chen IM, Atlas SR, Kang H, Ar K, et
al: Identification of novel cluster groups in pediatric high-risk
B-precursor acute lymphoblastic leukemia with gene expression
profiling: Correlation with genome-wide DNA copy number
alterations, clinical characteristics, and outcome. Blood.
116:4874–4884. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Roberts KG and Mullighan CG: Genomics in
acute lymphoblastic leukaemia: Insights and treatment implications.
Nat Rev Clin Oncol. 12:344–357. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Schwab CJ, Jones LR, Morrison H, Ryan SL,
Yigittop H, Schouten JP and Harrison CJ: Evaluation of multiplex
ligation-dependent probe amplification as a method for the
detection of copy number abnormalities in B-cell precursor acute
lymphoblastic leukemia. Genes Chromosomes Cancer. 49:1104–1113.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Schouten JP, McElgunn CJ, Waaijer R,
Zwijnenburg D, Diepvens F and Pals G: Relative quantification of 40
nucleic acid sequences by multiplex ligation-dependent probe
amplification. Nucleic Acids Res. 30:e572002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhao X, Wie H, Lin D, Wang Y, Zhou C, Liu
B, Li W, Liu K, Wang H, Li C, et al: Optimal treatment of adult Ph
negative acute lymphoblastic leukemia. Zhonghua Xue Ye Xue Za Zhi.
35:873–879. 2014.(In Chinese). PubMed/NCBI
|
|
8
|
Fang Q, Zhao X, Li Q, Li Y, Liu K, Tang K,
Wang Y, Liu B, Wang M, Xing H, et al: IKZF1 alterations and
expressions of CRLF2 predict prognosis in Chinese adult patients
with B-cell precursor acute lymphoblastic leukemia. Leuk Lymphoma.
58:127–137. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gökbuget N and Hoelzer D: Treatment of
adult acute lymphoblastic leukemia. Semin Hematol. 46:64–75. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ofverholm I, Tran AN, Heyman M,
Zachariadis V, Nordenskjöld M, Nordgren A and Barbany G: Impact of
IKZF1 deletions and PAX5 amplifications in pediatric B-cell
precursor ALL treated according to NOPHO protocols. Leukemia.
27:1936–1939. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Moorman AV, Enshaei A, Schwab C, Wade R,
Chilton L, Elliott A, Richardson S, Hancock J, Kinsey SE, Mitchell
CD, et al: A novel integrated cytogenetic and genomic
classification refines risk stratification in pediatric acute
lymphoblastic leukemia. Blood. 124:1434–1444. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Krentz S, Hof J, Mendioroz A, Vaggopoulou
R, Dörge P, Lottaz C, Engelmann JC, Groeneveld TW, Körner G, Seeger
K, et al: Prognostic value of genetic alterations in children with
first bone marrow relapse of childhood B-cell precursor acute
lymphoblastic leukemia. Leukemia. 27:295–304. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yuan T, Zhao XL, Zhang LX, Li QH, Tian Z,
Tang KJ, Wang Y, Lin D, Li W, Liu BC, et al: Expression and
clinical significance of IKZF1 gene IK6 isoforms in adult acute
lymphoblastic leukemia. Zhongguo Shi Yan Xue Ye Xue Za Zhi.
21:539–543. 2013.(In Chinese). PubMed/NCBI
|
|
14
|
Yamashita Y, Shimada A, Yamada T, Yamaji
K, Hori T, Tsurusawa M, Watanabe A, Kikuta A, Asami K, Saito AM and
Horibe K: IKZF1 and CRLF2 gene alterations correlate with poor
prognosis in Japanese BCR-ABL1-negative high-risk B-cell precursor
acute lymphoblastic leukemia. Pediatr Blood Cancer. 60:1587–1592.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ribera J, Morgades M, Zamora L, Montesinos
P, Gómez-Seguí I, Pratcorona M, Sarrà J, Guàrdia R, Nomdedeu J,
Tormo M, et al: Prognostic significance of copy number alterations
in adolescent and adult patients with precursor B acute
lymphoblastic leukemia enrolled in PETHEMA protocols. Cancer.
121:3809–3817. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Iacobucci I, Ferrari A, Lonetti A,
Papayannidis C, Paoloni F, Trino S, Storlazzi CT, Ottaviani E,
Cattina F, Impera L, et al: CDKN2A/B alterations impair prognosis
in adult BCR-ABL1-positive acute lymphoblastic leukemia patients.
Clin Cancer Res. 17:7413–7423. 2011. View Article : Google Scholar : PubMed/NCBI
|