Introduction
Esophageal cancer (EC) is one of the most common
types of malignancy globally, including esophageal squamous cell
carcinoma (ESCC) and esophageal adenocarcinoma. In China, >90%
of ECs are ESCC (1). Currently, the
morbidity of EC is increasing annually and the rate of metastasis
remains high with a poor prognosis (2). The diagnosis of EC usually occurs in the
middle and late stages due to the lack of symptoms in the early
stage, and as a result, the 5-year survival of patients with EC is
18–30% (3). The RAS/rapidly
accelerated fibrosarcoma (RAF)/extracellular signal-regulated
kinase (ERK) signaling pathway is critical for the malignant
biological behavior of ESCC (4). This
pathway can be activated in multiple ways, and has an important
role in the proliferation, metastasis and infiltration of ESCC
(5). Therefore, the identification of
molecules that specifically inhibit activation of the RAS/RAF/ERK
signaling pathway may enable the development of novel treatments
for ESCC.
Raf kinase trapping to Golgi apparatus [also known
as progestin and adipoQ receptor family member 3 (PAQR3)] is a type
III seven-transmembrane protein with an extracellular N-terminus
and a cytosolic C-terminus determined in the membrane of the Golgi
apparatus (6). Binding of PAQR3 to
B-Raf and C-Raf kinases anchors cytosolic RAF to the Golgi
apparatus, and prevents the binding of RAF to RAS (upstream) and
MEK (downstream) (7). Blocking signal
transduction from RAS inhibits the activation of the
Ras/Raf/MEK/ERK signaling pathway, and consequently affects the
proliferation and malignant transformation of cells, as well as the
development and progression of cancer (8,9).
Therefore, PAQR3 is considered a negative regulator
that suppresses RAS/RAF/ERK pathway activation-dependent tumors
(10,11). A study by Wang et al (12) determined that kinases in the
Raf/MEK/ERK signaling pathway were abnormally activated in various
tissues of PAQR3 gene knockout mice. These knockout mice had
significantly increased proliferation of epidermal keratinocytes,
and PAQR3 gene deletion promoted the development and progression of
dimethylbutylamine/12-O-tetradecanoyl-phorbol-12-acetate-induced
skin cancer. Other studies have reported downregulated expression
of PAQR3 in liver (10), gastric
(13,14), colon (15) and breast cancer (11), and determined that this downregulation
was associated with tumor progression, metastasis and prognosis.
However, the role and mechanism underlying the action of PAQR3 in
ESCC has not been reported. Therefore, the purpose of the present
study was to examine the association between PAQR3 and the
prognosis and clinicopathological characteristics of patients with
ESCC, and to increase the understanding of the molecular functions
of PAQR3.
Materials and methods
Ethical statement
All selected patients agreed to participate in the
present study and signed informed consent forms. The present study
was approved by the Ethics Committee of the First Affiliated
Hospital of Xinjiang Medical University (Ürümqi, China), and was
implemented in accordance with ‘The Declaration of Helsinki’.
Tissue specimens
A total of 80 histopathological specimens from
patients surgically diagnosed with ESCC between January and March
2012 were collected from the First Affiliated Hospital and
Affiliated Tumor Hospital of Xinjiang Medical University. All
patients had undergone radical surgery and had not received prior
treatments. The clinicopathological data of these patients,
including age, sex, ethnic group, tumor location, tumor length,
tumor type, degree of differentiation, number of lymph node
metastases and T staging, were obtained from the hospitals. A total
of 40 male and 40 female patients (mean age, 54.7±12.6 years)
underwent tumor resection. Tumor and paracancerous histological
normal tissue (PCHNT) specimens were obtained at the same time
during surgery, and were stored at −80°C. PCHNT is defined as
esophageal tissue that is located ≥5 cm away from the tumor margin
and is confirmed to be normal esophageal tissue by postoperative
histopathology. Patient follow-up was conducted from the day
following surgery, and anastomotic or regional lymph node
recurrence was diagnosed by clinical or pathological examination at
3 months post-operation. Local recurrence within 3 months following
surgery was considered a part of the initial treatment process.
Distant metastasis was defined as distant metastatic disease
diagnosed by clinical examination or imaging. Disease-free survival
(DFS) time was defined as the time from the day following surgery
to the first recurrence or metastasis. Overall survival (OS) time
was defined as the time from day 1 post-operation to last follow-up
or mortality. Complete follow-up data included the time and
location of recurrence or metastasis, and condition and time of
survival. All patients were followed up until March 2017 and the
duration of follow-up was 9–62 months (median, 28 months).
Cell culture
Human ESCC cell lines (ECA-109 and TE-1) were
obtained from the Type Culture Collection of Chinese Academy of
Sciences (Shanghai, China). They were maintained in Dulbecco's
modified Eagle's medium (DMEM) containing 10% fetal bovine serum
(both from Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) at 37°C
and in a 5% CO2 atmosphere.
Plasmid and transfection
Full-length human PAQR3 cDNA was amplified using a
polymerase chain reaction (PCR), as previously described (2). PCR products were purified and inserted
into a pcDNA3.1(+) vector. The PAQR3-expressing plasmid
(pcDNA3.1/PAQR3) or an empty vector were transfected into ECA-109
and TE-1 cells using Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA), according to the
manufacturer's instructions. A total of 24 h after transfection,
cells were examined for gene expression, proliferation and cell
cycle distribution. For stable transfection, 1×105
ECA-109 cells were transfected with the pcDNA3.1/PAQR3 plasmid (0.5
µg) and 24 h later. Subsequent to culturing for another 24 h, the
cells were incubated at 37°C in DMEM with 15% FBS containing 600
µg/ml G418 (Sigma-Aldrich; Merck KGaA) for 3 weeks. The
G418-resistant colonies were pooled together and used for colony
formation and tumorigenic studies.
RNA extraction and reverse
transcriptase-quantitative PCR (RT-qPCR) analysis of tumor
specimens
Total RNA was extracted using the TRIzol®
(cat. no. 15596018; Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's one-step instructions. cDNA was
synthesized from 3 µg total RNA using the M-MLV reverse
transcriptase: 4 µl 5X buffer, 2 µl 10 mM dNTPs, 1 µl RNA inhibitor
and 1 µl Reverse transcriptase from the QIAGEN74903-RNeasy
Plantmini kit 74903 (cat. no. M1705; Promega Corporation, Madison,
WI, USA) according to the manufacturer's instructions. GAPDH
was used as the reference gene for RT-qPCR. The forward and reverse
primers for PAQR3 were: 5′-TGTCGAAGATGGATGGCATTAGA-3′ and
5′-ACCTGACGCCAGTAGTTATTACA-3′, respectively. The forward and
reverse primers for GAPDH were: 5′-TGTTGCCATCAATGACCCCTT-3′ and
5′-CTCCACGACGTACTCAGCG-3′, respectively. Gene-specific
amplifications of the 20 µl PCR mixture were performed using the
ABI 7500HT real-time PCR system (Thermo Fisher Scientific, Inc.).
The PCR mixture contained the following: 1 µl DNA, 2X
SYBR® Select Master mix (cat. no. AT311; Beijing
TransGen Biotech Co., Ltd., Beijing, China), and 10 µl each of
forward and reverse primers (10 µM). The PCR amplification
conditions were as follows: 95°C for 30 sec, 95°C for 60 sec and
72°C for 60 sec for a total of 45 cycles, followed by 72°C for 7
min. DNA concentration was approximately doubled following each
cycle of denaturation, annealing and elongation. The cycle
quantification (Cq) of each sample was calculated, and the relative
expression of the gene was calculated using the 2−ΔΔCq
(16) method.
Immunohistochemistry
Formalin-fixed paraffin-embedded ESCC tissues (as
aforementioned) were sectioned (4-µm) for immunohistochemical
analysis. For antigen retrieval, the slides were immersed in EDTA
[1 mmol/l, (pH 8.0)] and boiled 60°C for 20 min in a microwave
oven. Following rinsing with PBS 3 times, endogenous peroxidase was
blocked using 0.3% hydrogen peroxide for 15 min at room
temperature. The slides were incubated with the PAQR3 primary
antibody (1:100; cat. no. ab174327; Santa Cruz Biotechnology, Inc.,
Dallas, TX, USA), in a humidified chamber at 4°C overnight.
Following additional washing with PBS three times, the sections
were sequentially incubated with horseradish peroxidase-conjugated
the anti-rabbit secondary antibody PV-6001 (1:50;
Envision™ detection kit; cat. no. GK500705; Gene Tech
Co., Ltd., Hong Kong, China) at 37°C for 30 min, and then washed
three times with PBS. Finally, 3,3′-diaminobenzidine
tetrahydrochloride was used for signal development and then the
sections were lightly counterstained with 20% hematoxylin at room
temperature for 20 min. The paraffin slides were dehydrated (using
95% ethanol every 2 h) and mounted on coverslips. For the negative
controls, PBS was used in place of the primary antibody. A light
microscope was used at a magnification of ×200.
Immunohistochemistry scores of 0–4 were considered to be
negative/low expression and 5–11 was considered to be normal/high
expression. Anti-PAQR3 (ab174327; Abcam, Cambridge, UK).
Western blot analysis
ECA-109 and TE-1 cells were lysed using
radioimmunoprecipitation assay buffer (Beyotime Institute of
Biotechnology, Nantong, China) supplemented with protease
inhibitors (Sigma-Aldrich; Merck KGaA). Proteins were quantified
using a BCA assay. Total protein extract was separated using 5%
sodium dodecyl sulfate-polyacrylamide gel electrophoresis and
transferred to nitrocellulose membranes. The membranes were blocked
with tris-buffered saline with 10% Tween-20 three times at room
temperature for 60 min. Following incubation at 4°C for 24 h with
the primary antibodies [anti-phosphorylated-AKT serine/threonine
kinase 1 (p-Akt Ser473; 1:1,000; cat. no. 9271), anti-Akt (1:1,000;
cat. no. 9272), and anti-β-actin antibody (1:500; cat. no. 8457),
E-cadherin antibody (1:500; cat. no. 15148), (all from Cell
Signaling Technology, Inc., Danvers, MA, USA)], membranes were
probed with horseradish peroxidase-conjugated the anti-rabbit
secondary antibodies (1:3,000; cat. no. 074-1506; Santa Cruz
Biotechnology, Inc.) for 30 min at room temperature. Signals were
visualized using an enhanced chemiluminescent system (Cell
Signaling Technology, Inc.) and quantified using Quantity One
software v.4.6 (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Statistical analysis
All statistical analyses were performed using the
SPSS 17.0 statistical software (SPSS, Inc., Chicago, IL, USA).
Count data were analyzed using the χ2 test and Fisher's
exact test. Paired-samples Student's t-test was used to compare
mRNA/protein expression of PAQR3 in ESCC tissues with paired
adjacent non-tumor tissue samples. Spearman's correlation analysis
was used to compare the mRNA/protein expression of PAQR3 in ESCC
tissues. Survival analysis was performed using the Kaplan-Meier
estimator method, and compared using the log rank test. Prognostic
analysis was performed using the Cox proportional hazards
regression model. P<0.05 was considered to indicate a
statistically significant difference
Results
PAQR3 mRNA expression is downregulated
in ESCC tissue
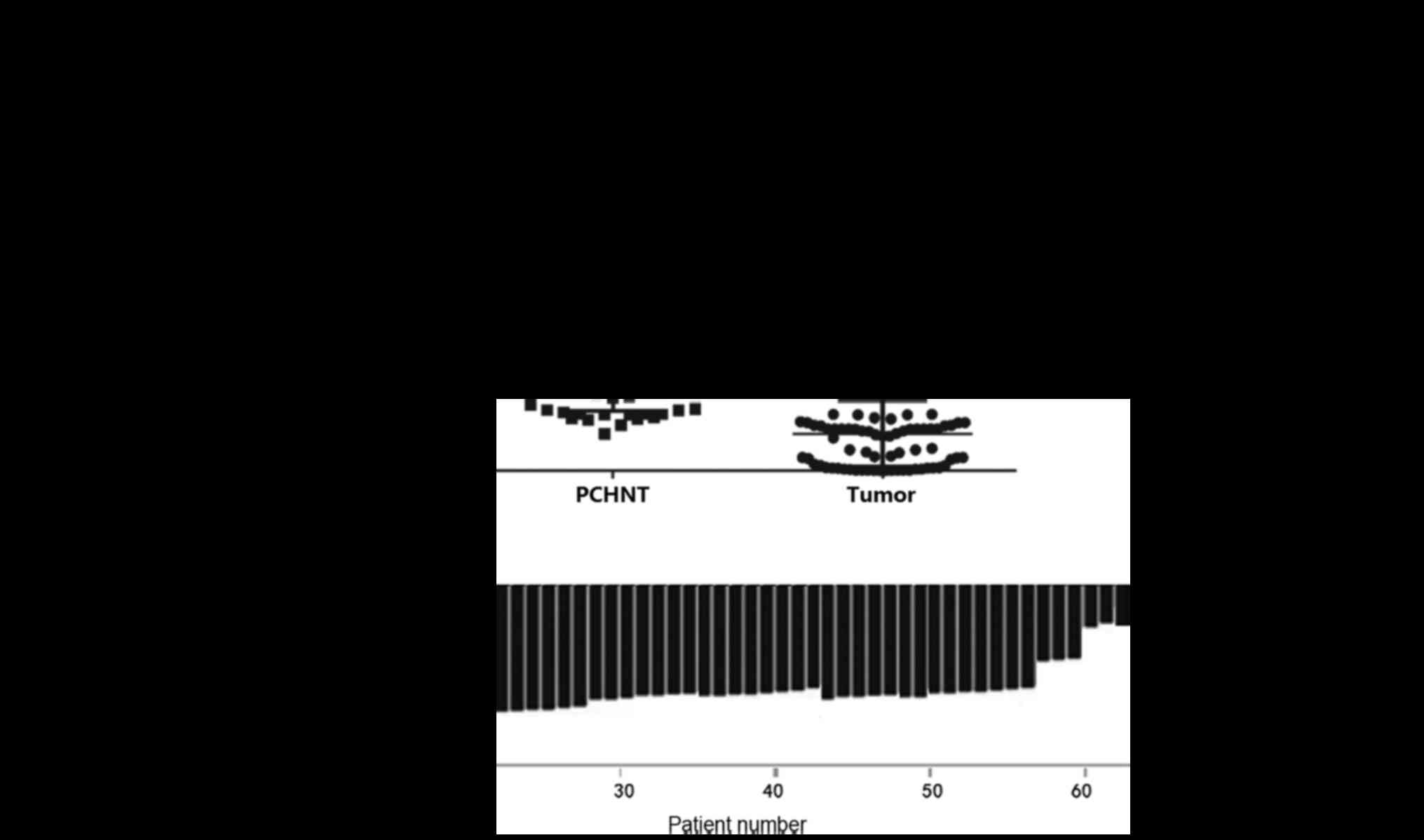
The levels of PAQR3 mRNA expression in the 80 ESCC
and corresponding PCHNT specimens were measured using RT-qPCR. The
mRNA expression of PAQR3 was significantly lower in ESCC tissues
compared with that in the corresponding PCHNT tissues (P<0.0001;
Fig. 1A). A total of 71.25% of
subjects (57/80) had significantly lowered PAQR3 mRNA expression in
their ESCC tissues compared with the PCHNT tissues (Fig. 1B). Low PAQR3 mRNA expression was
defined as ESCC/PCHNT <0.5. All 80 pairs of specimens were
independently measured twice.
PAQR3 protein expression is
downregulated in ESCC tissue
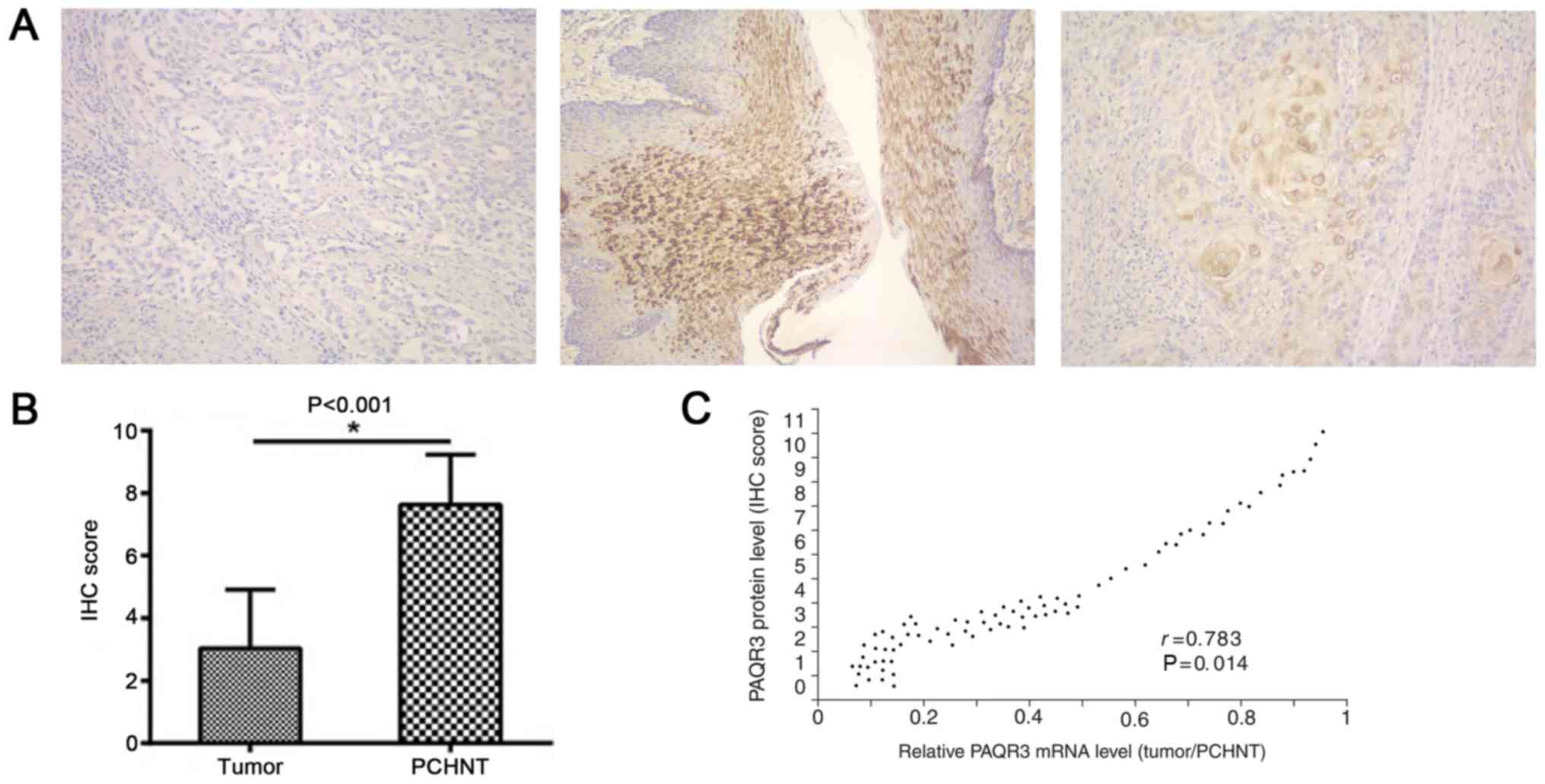
PAQR3 protein was detected by immunohistochemical
staining. The normal and adjacent tissues stained positive, in
contrast with the staining of PAQR3 in cancer tissues, which was
mostly negative (Fig. 2A). PAQR3
protein expression scores were also significantly reduced in EC
tissues compared with adjacent normal tissues (P<0.0001;
Fig. 2B). The protein levels of PAQR3
were significantly correlated with its mRNA levels (r=0.783;
P=0.014; Fig. 2C).
Association between PAQR3 expression
and clinicopathological characteristics of ESCC
The association between PAQR3 expression and
clinicopathological characteristics of ESCC was examined using the
χ2 test. Low PAQR3 expression was significantly
associated with ethnic group (P=0.032), tumor length (P=0.019),
lymph node metastasis (P=0.011) and local recurrence (P=0.009).
However, PAQR3 expression was not significantly associated with age
(P=0.369), sex (P=0.446), tumor location (P=1.327), tumor type
(P=2.117), tumor differentiation (P=0.242), T staging (P=0.331) or
distant metastasis (P=0.885; Table
I).
 | Table I.Correlations between the PAQR3
expression level and clinicopathological characteristics of 80
cases of ESCC. |
Table I.
Correlations between the PAQR3
expression level and clinicopathological characteristics of 80
cases of ESCC.
|
|
| PAQR3 mRNA level |
|
|
|---|
|
|
|
|
|
|
|---|
| Clinicopathological
parameters | N | Low | High/normal | χ2 | P-value |
|---|
| All cases | 80 | 57 | 23 |
|
|
| Age, years |
|
|
| 1.807 | 0.369 |
|
<50 | 37 | 24 | 13 |
|
|
| ≥50 | 43 | 33 | 10 |
|
|
| Sex |
|
|
| 0.975 | 0.446 |
|
Female | 40 | 27 | 13 |
|
|
| Male | 40 | 30 | 10 |
|
|
| Nationality |
|
|
| 3.101 | 0.032 |
| Han | 25 | 15 | 10 |
|
|
|
Kazak | 30 | 23 | 7 |
|
|
|
Uygur | 25 | 19 | 6 |
|
|
| Tumor length, cm |
|
|
| 5.782 | 0.019 |
|
<5 | 35 | 19 | 16 |
|
|
| ≥5 | 45 | 38 | 7 |
|
|
| Subsection |
|
|
| 0.291 | 1.327 |
|
Upper | 24 | 17 | 7 |
|
|
|
Middle | 36 | 24 | 12 |
|
|
|
Lower | 20 | 16 | 4 |
|
|
| Gross pathologic
classification |
|
|
| 0.104 | 2.117 |
|
Medullary | 30 | 23 | 7 |
|
|
|
Mushroom | 23 | 16 | 7 |
|
|
|
Ulcer | 17 | 13 | 4 |
|
|
|
Narrowing | 10 | 5 | 5 |
|
|
|
Differentiation |
|
|
| 2.177 | 0.242 |
|
Moderate/high | 50 | 36 | 14 |
|
|
|
Poor | 30 | 21 | 9 |
|
|
| T stage |
|
|
| 1.909 | 0.331 |
|
T1/T2 | 43 | 30 | 13 |
|
|
|
T3/T4 | 37 | 27 | 10 |
|
|
| Lymph node
metastasis |
|
|
| 7.398 | 0.011 |
|
Negative | 38 | 23 | 15 |
|
|
|
Positive | 42 | 34 | 8 |
|
|
| Recurrence |
|
|
| 9.273 | 0.009 |
| No | 48 | 30 | 18 |
|
|
|
Yes | 32 | 27 | 5 |
|
|
| Distance
metastasis |
|
|
| 0.603 | 0.885 |
| No | 60 | 45 | 15 |
|
|
|
Yes | 20 | 12 | 8 |
|
|
Downregulated PAQR3 expression is
associated with ESCC prognosis
In order to evaluate the feasibility of PAQR3
expression as a prognostic factor for ESCC, multivariate survival
analysis was performed on all parameters using the Cox proportional
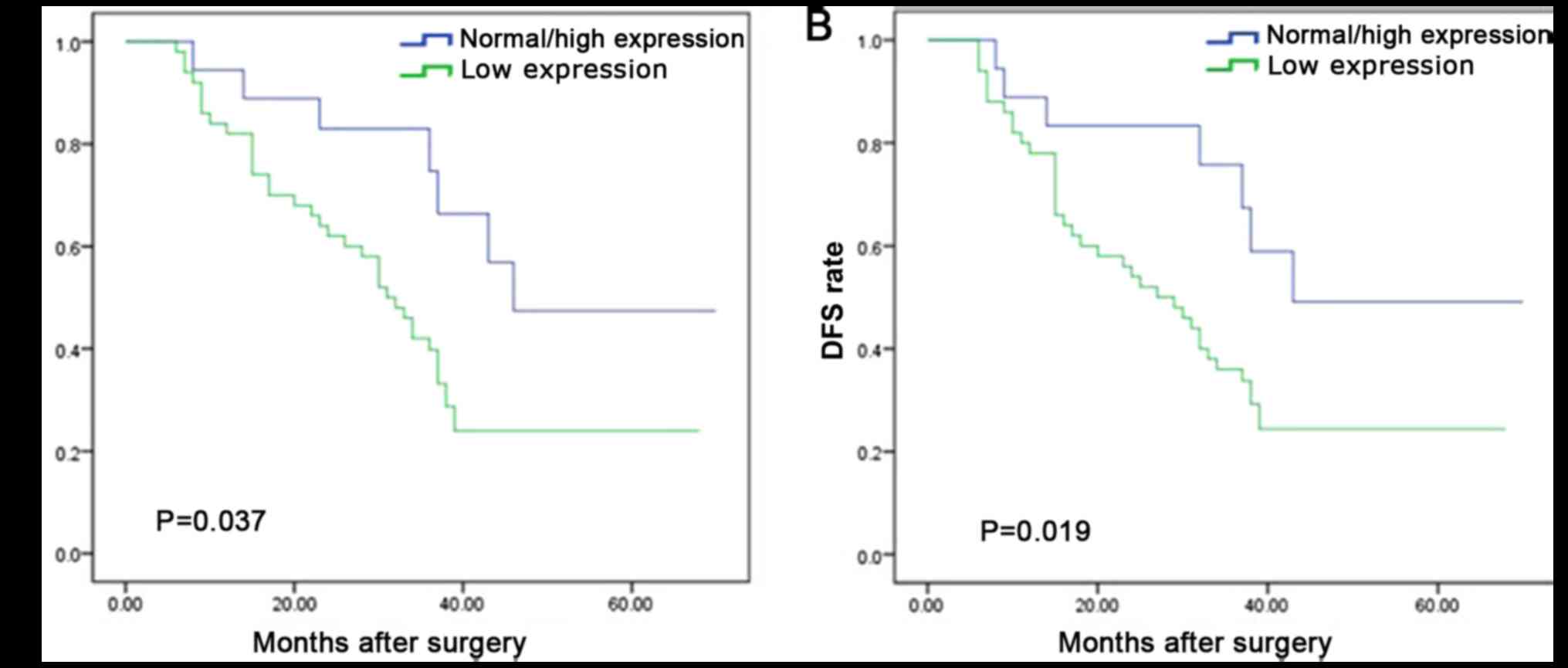
hazards regression model. It was determined that OS time was
significantly dependent on lymph node metastasis (P=0.021), local
recurrence (P=0.036) and PAQR3 expression level (P=0.047; Table II). OS time was significantly reduced
in patients with ESCC and a low PAQR3 expression compared with
those with a high PAQR3 expression (median survival, 30 vs. 46
months; P=0.037; Fig. 3A).
Furthermore, lymph node metastasis (P=0.004), local tumor
recurrence (P=0.029) and PAQR3 expression level (P=0.033) were
independent prognostic factors for the 5-year DFS rate of patients
with ESCC (Table II). DFS time was
significantly reduced in patients with ESCC and a low PAQR3
expression compared with those with high PAQR3 expression (median
survival, 26 vs. 43 months; P=0.019; Fig.
3B). The 1-year, 3-year and 5-year OS rates were as follows:
PAQR3 normal/high group, 93, 68 and 45%; PAQR3 low expression
group, 82, 35 and 25% (χ2=14.497; P<0.05). The 1-, 3-
and 5-years DFS rates were as follows: PAQR3 normal/high group, 88,
74 and 50%; PAQR3 low expression group: 80, 36 and 23%
(χ2=19.038; P<0.05) (data not shown).
 | Table II.Univariate and multivariate analysis
of survival in 80 patients with ESCC according to
clinicopathological characteristics and PAQR3 expression level. |
Table II.
Univariate and multivariate analysis
of survival in 80 patients with ESCC according to
clinicopathological characteristics and PAQR3 expression level.
|
|
| DFS univariate
analysis |
|
| OS univariate
analysis |
|
|
|---|
|
|
|
|
|
|
|
|
|
|---|
| Clinicopathological
parameters | n | χ2 | P-value | Multivariate
analysis HR (95% CI) | P-value | χ2 | P-value | Multivariate
analysis HR (95% CI) | P-value |
|---|
| Age, years |
| 0.385 | 0.535 |
|
| 0.004 | 0.948 |
|
|
|
<50 | 37 |
|
| 1 |
|
|
| 1 |
|
|
≥50 | 43 |
|
| 1.217
(0.453–3.268) | 0.697 |
|
| 1.440
(0.797–2.603) | 0.227 |
| Sex |
| 0.003 | 0.953 |
|
| 0.013 | 0.905 |
|
|
|
Female | 40 |
|
| 1 |
|
|
| 1 |
|
|
Male | 40 |
|
| 1.000
(0.690–1.440) | 0.995 |
|
| 0.522
(0.225–1.209) | 0.128 |
| Nationality |
| 0.026 | 0.873 |
| 0.709 | 1.149 | 0.283 |
| 0.552 |
|
Han | 25 |
|
| 1 |
|
|
| 1 |
|
|
Kazak | 30 |
|
| 1.080
(0.720–1.630) | 0.997 |
|
| 0.330
(0.130–1.330) | 0.458 |
|
Uygur | 25 |
|
| 1.230
(0.480–3.160) | 0.667 |
|
| 0.980
(0.730–1.290) | 0.549 |
| Tumor length,
cm |
| 2.812 | 0.094 |
|
| 1.022 | 0.312 |
|
|
|
<5 | 35 |
|
| 1 |
|
|
| 1 |
|
| ≥5 | 45 |
|
| 1.549
(0.587–4.090) | 0.337 |
|
| 1.530
(0.870–2.693) | 0.140 |
| Subsection |
| 1.813 | 0.178 | 0.864 | 1.251 | 0.262 | 0.807 |
|
|
|
Upper | 24 |
|
| 1 |
|
|
| 1 |
|
|
Middle | 36 |
|
| 0.990
(0.430–2.310) | 0.817 |
|
| 0.920
(0.320–2.590) | 0.869 |
|
Lower | 20 |
|
| 1.000
(0.336–2.780) | 0.986 |
|
| 0.660
(0.310–1.490) | 0.721 |
| Gross pathologic
classification |
| 1.562 | 0.212 |
| 0.580 | 3.042 | 0.083 |
| 0.173 |
|
Medullary | 30 |
|
| 1 |
|
|
| 1 |
|
|
Mushroom | 23 |
|
| 1.540
(0.540–4.440) | 0.419 |
|
| 2.720
(0.780–9.470) | 0.117 |
|
Ulcer | 17 |
|
| 0.660
(0.230–1.930) | 0.451 |
|
| 2.620
(0.560–12.240) | 0.221 |
|
Narrowing | 10 |
|
| 0.840
(0.130–5.550) | 0.856 |
|
| 2.520
(0.770–8.240) | 0.126 |
|
Differentiation |
| 3.801 | 0.051 |
| 0.590 | 2.734 | 0.101 |
| 0.398 |
|
High | 22 |
|
| 1 |
|
|
| 1 |
|
|
Moderate | 28 |
|
| 1.190
(0.540–2.680) | 0.659 |
|
| 3.260
(0.910–11.680) | 0.367 |
|
Poor | 30 |
|
| 1.580
(0.640–3.890) | 0.318 |
|
| 3.220
(0.710–14.750) | 0.452 |
| T-stage |
| 0.004 | 0.948 |
| 0.533 | 1.323 | 0.232 |
| 0.289 |
| T1 | 20 |
|
| 1 |
|
|
| 1 |
|
| T2 | 23 |
|
| 1.390
(0.490–3.910) | 0.525 |
|
| 2.330
(0.740–7.380) | 0.273 |
| T3 | 26 |
|
| 0.550
(0.190–1.540) | 0.255 |
|
| 0.540
(0.120–2.420) | 0.334 |
| T4 | 11 |
|
| 0.490
(0.720–3.350) | 0.469 |
|
| 0.340
(0.080–1.370) | 0.258 |
| Lymph node
metastasis |
| 5.371 | 0.021 |
|
| 4.776 | 0.031 |
|
|
|
Negative | 38 |
|
| 1 |
|
|
| 1 |
|
|
Positive | 42 |
|
| 1.740
(1.200–2.530) | 0.004 |
|
| 2.080
(1.120–3.870) | 0.021 |
| Recurrence |
| 6.363 | 0.012 |
| 5.221 | 0.023 |
|
|
|
| No | 48 |
|
| 1 |
|
|
| 1 |
|
|
Yes | 32 |
|
| 0.636
(1.155–2.609) | 0.029 |
|
| 1.860
(1.920–3.760) | 0.036 |
| Distance
metastasis |
| 4.621 | 0.032 |
|
| 3.135 | 0.075 |
|
|
| No | 60 |
|
| 1 |
|
|
| 1 |
|
|
Yes | 20 |
|
| 1.223
(0.384–3.893) | 0.733 |
|
| 1.070
(0.750–1.540) | 0.698 |
| PAQR3 |
| 4.998 | 0.028 |
|
| 4.094 | 0.043 |
|
|
|
Low | 57 |
|
| 1 |
|
|
| 1 |
|
|
Normal/high | 23 |
|
| 1.760
(1.050–2.960) | 0.033 |
|
| 3.630
(1.020–12.950) | 0.047 |
PAQR3 suppresses
epithelial-mesenchymal transition (EMT) features in human ESCC
cells
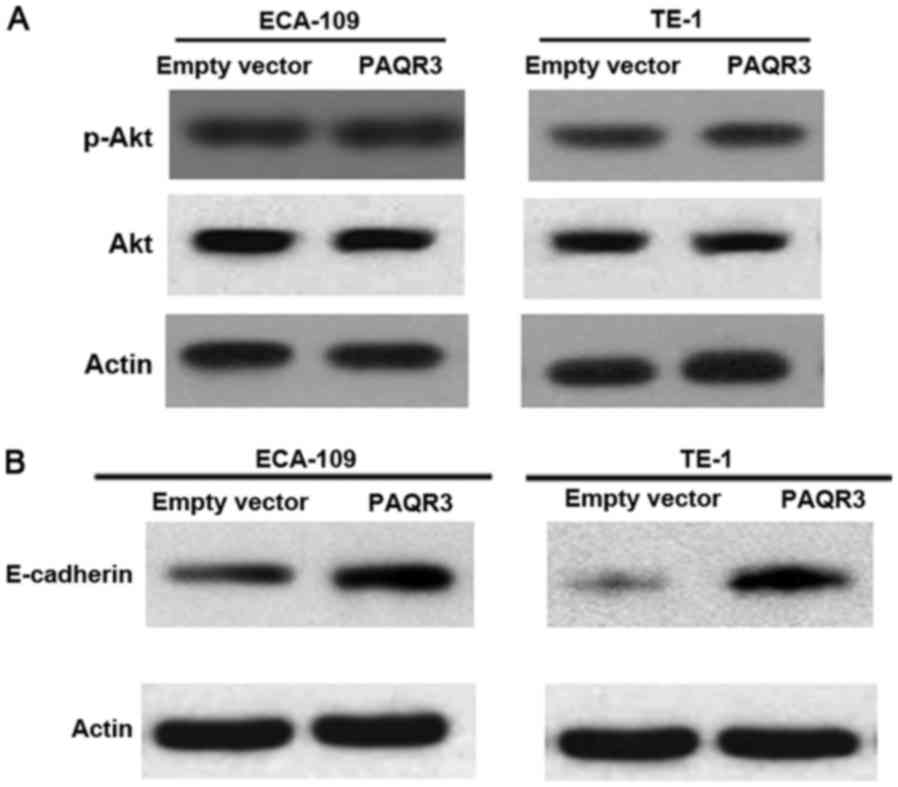
Previous studies have revealed that PAQR3 is able to
inhibit Ras/Raf/MEK/ERK signaling cascades. Whether PI3K/AKT
signaling pathways were also affected by PAQR3 in ESCC cells was
explored (15). Western blot analysis
of p-Akt and Akt proteins in ECA-109 and TE-1 cells transfected
with the PAQR3-expressing plasmid or vector was performed. However,
the activation of Akt was not altered by PAQR3 overexpression
(Fig. 4A).
The effect of PAQR3 on EMT was also analyzed, a
critical step for tumor migration and metastasis (10). In ECA-109 and TE-1 cells,
overexpression of PAQR3 suppressed EMT features as demonstrated by
the marked increase in epithelial marker, E-cadherin (Fig. 4B).
Discussion
PAQR3 is a member of the PAQR family (17). A search of this gene in the world's
largest cancer microarray database, Oncomine (https://www.oncomine.org/resource/login.html),
demonstrated that PAQR3 is associated with multiple cancer types,
including breast cancer, colon cancer, gastric cancer, leukemia and
lymphoma. In addition, the expression of PAQR3 mRNA is
downregulated in numerous cancer types, including gastric cancer,
but upregulated in a number of cancer types of the circulatory
system, including leukemia and lymphoma (18). However, there are no relevant data on
the expression of PAQR3 in EC. In the present study, the levels of
PAQR3 mRNA expression in 80 ESCC and corresponding PCHNT tissue
specimens were measured. In addition, a complete follow-up database
was constructed to examine the association between PAQR3 and the
prognosis and clinicopathological characteristics of patients with
ESCC, and to increase the understanding of the diagnosis and
prognosis of EC, a highly prevalent disease in Xinjiang, China
(19).
RT-qPCR analysis demonstrated that PAQR3 mRNA
expression was significantly lower in ESCC tissues compared with in
PCHNT tissues, which was consistent with the data in other cancer
types, including breast cancer (11),
colon cancer (15) and gastric cancer
(13), demonstrating that PAQR3 may
be an ESCC suppressor gene. The tumor suppressive function of PAQR3
is primarily achieved by inhibiting activation of the ERK signaling
pathway (9). The broad anticancer
activity of PAQR3 is mediated through the alteration of multiple
different signaling pathways (20).
PAQR3 overexpression decreased the phosphorylation of ERK1/2 in
ESCC cells, without affecting the activation of Akt (21). PAQR3 has demonstrated the ability to
interfere with multiple aspects of tumor biology, including
proliferation, migration, invasion, and EMT (22,23). In
the present study, it was revealed that the overexpression of PAQR3
suppressed EMT features as demonstrated by the marked increase in
the epithelial marker E-cadherin.
In the present study, convincing evidence that PAQR3
is a novel genetic marker associated with the progression,
proliferation, infiltration and prognosis of EC has been indicated.
Analysis of the clinicopathological characteristics of 80 patients
with EC indicated that the level of PAQR3 mRNA expression was
associated with ethnic group, status of lymph node metastasis and
tumor length. In particular, the level of PAQR3 mRNA expression was
significantly higher among Han patients with ESCC than among those
of the Kazakh and Uygur ethnic groups. Although current studies
have indicated improved short-term efficacy of treatment in Kazakh
and Uygur patients with ESCC, their prognosis and long-term
survival were lower compared with those of Han patients. Whether
this characteristic is associated with the difference in PAQR3 mRNA
expression between ethnic groups will require further studies to
confirm. In the present study, it was demonstrated that an
association between patients with low PAQR3 expression, and lymph
node metastasis and poor prognosis, indicating that PAQR3 may be a
potential tumor suppressor. The total of 80 histopathological
specimens was a limiting factor for this study as the number of
samples after grouping was too small, reducing the statistical
reliability. In the future study, the sample size should be
increased to confirm the credibility of the findings of the present
study.
The present study demonstrated that low PAQR3
expression was associated with lymph node metastasis and survival.
Multivariate prognostic analysis of the association between PAQR3
expression and the status of lymph node metastasis indicated
reduced DFS in patients with low PAQR3 expression and lymph node
metastasis. Thus, lymph node metastasis or low PAQR3 expression may
be adverse prognostic factors for EC. The combined evaluation of
PAQR3 expression and lymph node status may be helpful in the
prognosis of patients, and may provide insights into postoperative
adjuvant therapy strategies. Notably, a previous study have
demonstrated that PAQR3 expression may be associated with the
susceptibility of breast cancer to epirubicin, whereby PAQR3
overexpression enhances the susceptibility of breast cancer cells
to epirubicin via caspase-associated signaling pathways and
epirubicin inhibition-induced ERK activation (24). This data indicated that PAQR3 may be a
novel cancer treatment target that can be used to evaluate
radiation therapy and chemotherapy sensitivity. However, further
studies are required to determine the effect of PAQR3 on the
sensitivity of ESCC to treatment.
In conclusion, the present study demonstrated that
PAQR3 downregulation is associated with the progression of tumor
recurrence and increased lymph node metastasis in EC, and is
significantly associated with reduced survival of patients with EC.
This data demonstrated that PAQR3 is a tumor suppressor gene
associated with the survival and prognosis of esophageal
malignancies. PARQR3 may be a potential valuable prognostic factor
for ESCC, and a novel predictor of radiation and chemotherapy
sensitivity. To the best of our knowledge, the present study is the
first to examine the association between PAQR3 and ESCC. However,
further in vitro and in vivo studies are required to
elucidate the molecular mechanism underlying PAQR3 in the
regulation of ESCC.
Acknowledgements
The present study was supported by the State Key Lab
Incubation Base of Xinjiang Major Diseases Research (grant no.
SKLIB-XJMDR-2016-5).
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kamangar F, Dores GM and Anderson WF:
Patterns of cancer incidence, mortality, and prevalence across five
continents: Defining priorities to reduce cancer disparities in
different geographic regions of the world. J Clin Oncol.
24:2137–2150. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Samatar AA and Poulikakos PI: Targeting
RAS-ERK signalling in cancer: Promises and challenges. Nat Rev Drug
Discov. 13:928–942. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tasioudi KE, Saetta AA, Sakellariou S,
Levidou G, Michalopoulos NV, Theodorou D, Patsouris E and
Korkolopoulou P: pERK activation in esophageal carcinomas:
Clinicopathological associations. Pathol Res Pract. 208:398–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Feng L, Xie X, Ding Q, Luo X, He J, Fan F,
Liu W, Wang Z and Chen Y: Spatial regulation of Raf kinase
signaling by PAQR3. Proc Natl Acad Sci USA. 104:pp. 14348–14353.
2007; View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yu X, Li Z, Chan MT and Wu WK: PAQR3: A
novel tumor suppressor gene. Am J Cancer Res. 5:2562–2568.
2015.PubMed/NCBI
|
|
8
|
Fan F, Feng L, He J, Wang X, Jiang X,
Zhang Y, Wang Z and Chen Y: PAQR3 sequesters B-Raf to the Golgi
apparatus and inhibits the proliferation and tumorigenicity of
human malignant melanoma cells. Carcinogenesis. 29:1157–1163. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xie X, Zhang Y, Jiang Y, Liu W, Ma H, Wang
Z and Chen Y: Suppressive function of PAQR3 on chemical
carcinogen-induced skin carcinogenesis in mouse. Carcinogenesis.
29:1632–1638. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wu HG, Zhang WJ, Ding Q, Peng G, Zou ZW,
Liu T, Cao RB, Fei SJ, Li PC, Yang KY, et al: Identification of
PAQR3 as a new candidate tumor suppressor in hepatocellular
carcinoma. Oncol Rep. 32:2687–2695. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li Z, Ling ZQ, Guo W, Lu XX, Pan Y, Wang Z
and Chen Y: PAQR3 expression is downregulated in human breast
cancers and correlated with HER2 expression. Oncotarget.
6:12357–12368. 2015.PubMed/NCBI
|
|
12
|
Wang L, Wang X, Li Z, Xia T, Zhu L, Liu B,
Zhang Y, Xiao F, Pan Y, Liu Y, et al: PAQR3 has modulatory roles in
obesity, energy metabolism, and leptin signaling. Endocrinology.
154:4525–4535. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Qiao S, Guo W, Liao L, Wang L, Wang Z,
Zhang R, Xu D, Zhang Y, Pan Y, Wang Z and Chen Y: DDB2 is involved
in ubiquitination and degradation of PAQR3 and regulates
tumorigenesis of gastric cancer cells. Biochem J. 469:469–480.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ling ZQ, Guo W, Lu XX, Zhu X, Hong LL,
Wang Z, Wang Z and Chen Y: A Golgi-specific protein PAQR3 is
closely associated with the progression, metastasis and prognosis
of human gastric cancers. Ann Oncol. 25:1363–1372. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang X, Li X, Fan F, Jiao S, Wang L, Zhu
L, Pan Y, Wu G, Ling ZQ, Fang J and Chen Y: PAQR3 plays a
suppressive role in the tumorigenesis of colorectal cancers.
Carcinogenesis. 33:2228–2235. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Guo W, You X, Xu D, Zhang Y, Wang Z, Man
K, Wang Z and Chen Y: PAQR3 enhances Twist1 degradation to suppress
epithelial-mesenchymal transition and metastasis of gastric cancer
cells. Carcinogenesis. 37:397–407. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Oncomine database. https://www.oncomine.org/resource/login.html
|
|
19
|
Zhang Y: Xinjiang esophageal cancer
distribution. Xinjiang Medical College. 11:139–144. 1988.
|
|
20
|
Wang L, Pan Y, Huang M, You X, Guo F and
Chen Y: PAQR3 augments amino acid deprivation-induced autophagy by
inhibiting mTORC1 signaling. Cell Signal. 33:98–106. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bai G, Chu J, Eli M, Bao Y and Wen H:
PAQR3 overexpression suppresses the aggressive phenotype of
esophageal squamous cell carcinoma cells through the ERK pathway.
Biomed Pharmacother. 94:1–819. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wu Q, Zhuang K and Li H: PAQR3 plays a
suppressive role in laryngeal squamous cell carcinoma. Tumour Biol.
37:561–565. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Huang W, Guo W, You X, Pan Y, Dong Z, Jia
G, Yang C and Chen Y: PAQR3 suppresses the proliferation, migration
and tumorigenicity of human prostate cancer cells. Oncotarget.
8:53948–53958. 2016.PubMed/NCBI
|
|
24
|
Huang J, Xiang T, Luo X, Huang J, Xiao Y,
Yang B, Yin X, Li H, Xia Li X, Peng W, et al: RKTG, a RAS/RAF/ERK
signaling antagonist regulated by HER2, suppresses malignant
phenotypes and enhances chemosensitivity in breast cancer cells. J
Third Military Med Univ. 8:1658–1662. 2013.
|