Introduction
Oral squamous cell carcinoma, characterized by its
poor overall prognosis, complex pathogenesis and high mortality
rate, is the sixth most common type of malignant cancer globally
(1). Approximately 300,000 oral
squamous cell carcinoma cases are reported annually (1), with the likelihood of severe progression
partnered with a high risk of nodal metastasis and locoregional
invasion (2). In addition, the
combined actions of various factors, including genetic factors and
tumor microenvironment are implicated in the complex pathogenesis
of oral squamous cell carcinoma (3,4). Despite
recent advances in surgery, chemotherapy and radiotherapy, the
5-year survival rate of oral squamous cell carcinoma has remained
at ~50% for the past 10 years (5).
Therefore, to potentially identify targets that may aid therapeutic
intervention, further research is required to investigate the
underlying mechanisms implicated during progression of oral
squamous cell carcinoma.
Vascular cell adhesion molecule 1 (VCAM1) is a
member of the immunoglobulin superfamily that binds integrin
receptors α4β1 and α4β7 (6). The
VCAM1 gene is ~25 kb long and is located in the lp31-32
human chromosomal region (6). The
VCAM1 protein may potentially be released from the cell membrane
and function in a soluble form in response to environmental cues
(7,8).
As an immunoglobulin-like adhesion molecule, VCAM1 serves a
significant role in various pathophysiological tissues (9,10).
Aberrant expression of VCAM1 frequently occurs in various types of
cancer (10). For example, abnormal
expression of VCAM1 is associated with the metastasis of gastric
carcinoma (11,12). Furthermore, VCAM1 is implicated in the
preferential attachment of highly metastatic melanoma cells to
microvesicles within the tumor microenvironment (13). VCAM1 is also key metastasis of breast
cancer to the lung and, thus, may present a potential therapeutic
target (10). Functioning as an
environmental sensor that regulates adult neurogenesis, VCAM1
expression is induced in the neural stem cell niche of the
subventricular zone (14). Therefore,
a potentially useful approach for assessing prognosis in patients
with oral tongue squamous cell carcinoma may be monitoring changes
in VCAM1 expression in lymphatic vessels (15).
RNA interference (RNAi) refers to the gene-silencing
phenomenon induced by small molecules of double-stranded RNA
(dsRNA) (16), in which RNA molecules
inhibit gene expression or translation, by neutralizing targeted
mRNA molecules (17,18). RNAi is controlled by the RNA-induced
silencing complex (RISC), and initiated in the cell cytoplasm by
short double-stranded RNA molecules that interact with the
catalytic RISC component Argonaute (16). The synthetic dsRNA, which is then
introduced into cells, has the ability to induce the suppression of
target genes (16). Recently, RNAi
has become a key technology for identifying the components within
particular cell processes.
VCAM1 expression has been demonstrated to be induced
in oral tongue squamous cell carcinoma (15). In addition, a previous study
demonstrated that overexpression of the VCAM1 gene serves a
potential role in oral squamous cell carcinoma development, which
was closely associated with lymph node metastasis and angiogenesis
(19). However, there remain few
studies on the effect of VCAM1 silencing on the
proliferation of human oral squamous carcinoma (15). Therefore, to provide an experimental
basis for the diagnosis and colorectal cancer treatment, the
present study aimed to investigate the effect of VCAM1
silencing on the proliferation of human oral squamous carcinoma
HN12 cells.
Materials and methods
Construction of VCAM1 silencing HN12
cell lines
HN12 human oral squamous carcinoma cells (Michigan
State University, East Lansing, MI, USA) were cultured in
Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10%
fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc., Waltham,
MA, USA), and incubated at 37°C with 5% CO2. To create
the VCAM1 short hairpin RNA (shRNA)-silenced sub-cell lines,
the following shRNA sequence was designed against the VCAM1
gene:
5′-GGCTGGAGATAGACTTACTTTCAAGAGAAGTAAGTCTATCTCCAGCCTTTTTTACGCGT-3′.
The VCAM1 RNAi was purchased from the Daan Gene Co., Ltd.
(Guangzhou, China), and the HN12 cells were transfected with the
plasmids, psPAX2 and pMD2.G using Lipofectamine® 2000
transfection reagent according to the manufacturer's protocol
(Invitrogen; Thermo Fisher Scientific, Inc.). After 48 h of
transfection, HN12 Cells were divided into three groups: The
untreated blank control cell group (CK), the negative control group
transfected with non-homologous vector (NC) and the positive group
transfected with the VCAM1-kncodown shRNA sequence (KD).
Infection efficiency measurement
For cell infection, the HN12 cells
(2.5×105 cells/well) were seeded in 24-well plates and
cultured in an incubator for 12 h. Following this, 1, l.0, 5.0 or
10.0 µl of blank lentiviruses with green fluorescence protein (GFP)
[1×109 transducing units (TU)/ml] were added to the
wells. For each concentration of lentiviruses, three wells were
used. After 12 h, lentiviruses were washed with PBS and the cells
were further cultured in DMEM containing 10% fetal bovine serum in
an incubator at 37°C with 5% CO2. The infection
efficiency was observed after 48 h, and the multiplicity of
infection (MOI) was evaluated using a fluorescence microscope to
analyze the expression level of GFP.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
In 6-well plates, 3×105 HN12 cells were
cultured for 3 days at 37°C with 5% CO2, and cell
culture was continued for an additional 2 days at 37°C with 5%
CO2 when the MOI was >50%. Following digestion and
centrifugation (400 × g) for 5 min at 37°C, total RNA was extracted
using TRIzol® reagent (Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) according to the manufacturer's protocol. RNA was
reverse-transcribed using the FastQuant RT kit (Tiangen Biotech
Co., Ltd., Beijing, China) at 37°C. Next, SYBR Green I-labeled PCR
product (Takara Bio, Inc., Otsu, Japan) was used according to the
manufacturer's protocol for fluorescent qPCR to obtain
quantification cycle (Cq) values. The following thermocycling
conditions were maintained: 95°C for 3 min; 95°C for 10 sec and
60°C for 30 sec for 39 cycles; and melting curve analysis using
increase from 65.0 to 95.0°C in 0.5°C increments for 5 sec.
Finally, the 2−ΔΔCq method (20) was used to analyze differences in
relative gene expression in each sample, using β-actin as the
internal reference gene. Primer sequences are listed in Table I.
 | Table I.Primers for ACTB and
VCAM1. |
Table I.
Primers for ACTB and
VCAM1.
| Gene | Accession
numbera | Primer sequences
(5′-3′) | Product size, bp |
|---|
| ACTB | NM_001101.3 | F:
TGTTACAGGAAGTCCCTTGCCATC | 85 |
|
|
| R:
CTGTGTGGACTTGGGAGAGGAC |
|
| VCAM1 | NM_001078.3 | F:
TTCTGTGCCCACAGTAAGG | 95 |
|
|
| R:
GCAGCTTTGTGGATGGATTC |
|
Western blot analysis
The HN12 cells were collected 7 days the lentivirus
infection and lysed in SDS sample buffer [100 mM Tris-HCl (pH 6.8),
10 mM ethylenediaminetetraacetic acid, 4% SDS, and 10% glycine].
Next, the protein content was evaluated using the Lowry method
(21). To detect target proteins,
equal amounts of protein samples were separated by SDS-PAGE (12%
gel) and transferred to polyvinylidene difluoride membranes. Next,
the membranes were incubated with TBST [25 mM Tris (pH 7.4) 150 mM
NaCl, 0.1% Tween-20] containing 5% non-fat dry milk at room
temperature for 1 h. Following washing with TBST for 3 times, the
membranes were probed with the primary antibody anti-VCAM1 rabbit
mAb (dilution, 1:500; cat. no. ab106777; Abcam, Cambridge, UK) and
anti-β-actin rabbit mAb (dilution, 1:500; cat. no. ab6272; Abcam)
at 4°C overnight, followed by incubation with a goat anti-rabbit
IgG horseradish peroxidase-linked antibody (dilution, 1:1,500; cat.
no. 7074 CST Biological Reagents Co., Ltd., Shanghai, China) for 1
h at room temperature. Finally, the blots were detected with an
enhanced chemiluminescence detection kit (Pierce; Thermo Fisher
Scientific, Inc.) following the manufacturer's protocol. β-actin
was used as the reference control. The relative expression level of
VCAM1 was acquired based on the gray values, and analyzed with
Quantity One 1-D analysis software v4.6 (Bio-Rad Laboratories,
Inc., Hercules, CA, USA).
Cell Counting Kit-8 (CCK-8) assay
The lentivirus transfected HN12 cells were seeded
into 96-well plates at a density of 2×103 cells per well
and incubated for 12 h at 37°C. Then cells were treated with 20 ml
Lipofectamine®-small interfering RNA (siRNA) complexes,
which contained 2 nM siRNA. Cells treated with scrambled siRNA were
used as negative controls. Cell proliferation rate was measured 5
days following transfection, by adding 10 ml CCK-8 solution (Takara
Bio, Inc.) to each well, followed by incubation at 37°C for 2 h.
Absorbance was evaluated at 450 nm by spectrophotometry using a
SpectraMax 190 Microplate Reader (Molecular Devices, LLC,
Sunnyvale, CA, USA). In each assay, six parallel wells were
included, and the results were collected to evaluate the mean of
three independent experiments.
Statistical analysis
SPSS 17.0 statistical software (SPSS, Inc., Chicago,
IL, USA) was used for data analysis. Data were expressed as the
mean ± standard deviation. One-way analysis of variance was
performed to analyze the effect of VCAM1 silencing on the
proliferation of HN12 cells. P<0.05 was considered to indicate a
statistically significant difference.
Results
Transfection efficiency of
lentivirus
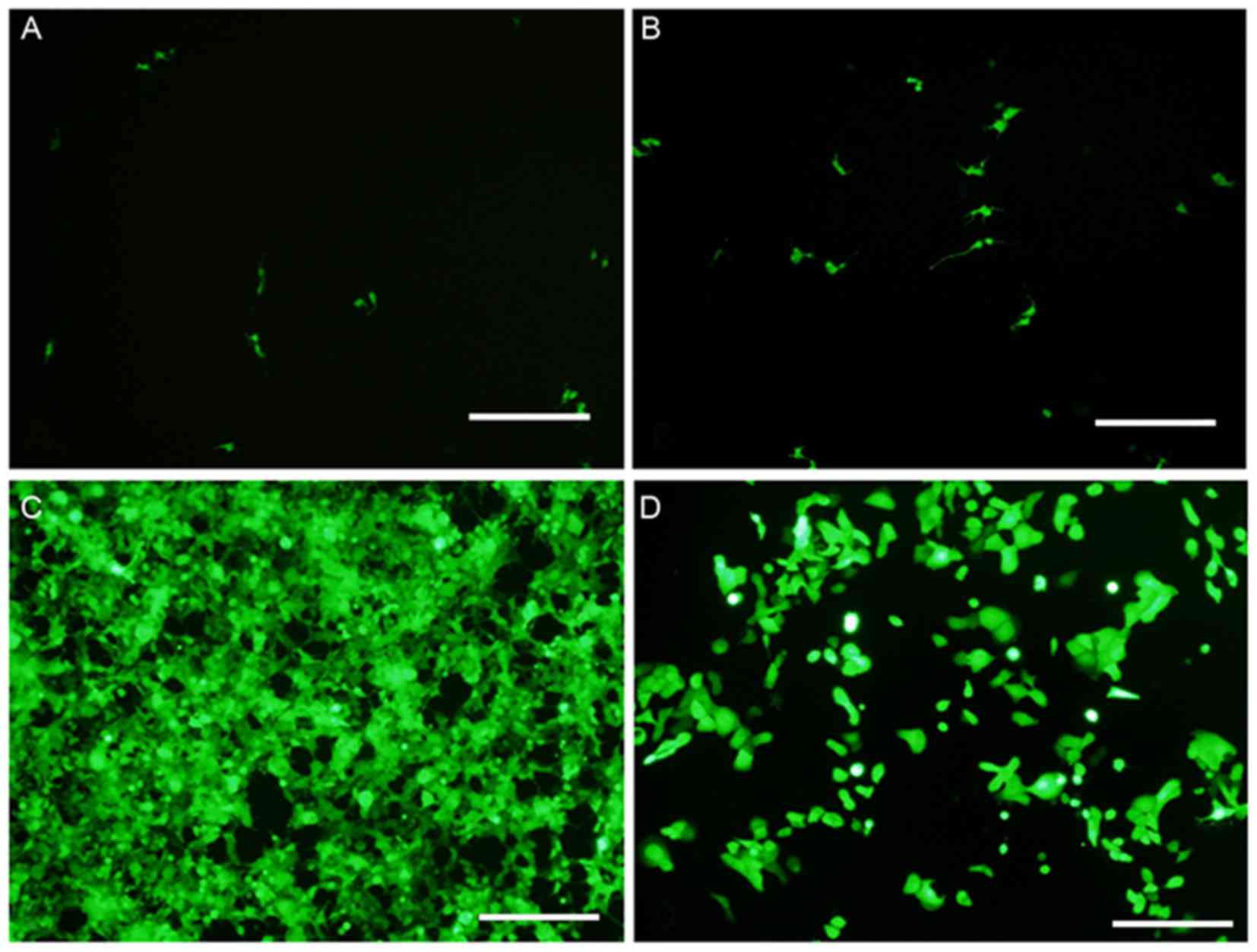
Fluorescence microscopy was used to detect the
expression level of GFP. Following this, the transfection
efficiency of viruses was evaluated by analyzing the MOI of HN12
cells. It was observed that the infection rate was <10% when the
MOI was 1.0 (Fig. 1A). Furthermore,
the infection rate was 10–20% at an MOI of 10.0 (Fig. 1B). At a MOI of 50, the toxic side
effects of the virus were negligible and cell density was not
significantly altered (Fig. 1C). At
this MOI, the transfection efficiency was ~95% (Fig. 1C). However, a cytotoxic effect was
observed with a MOI of 100, and the infection rate was >95%
(Fig. 1D). Therefore, an optimum MOI
of 50 was selected for use in the present study. Compared with the
NC group, the knockdown efficiency of the VCAM1 gene was ~85%, and
the expression level of VCAM1 protein was reduced by ~74% in the KD
group.
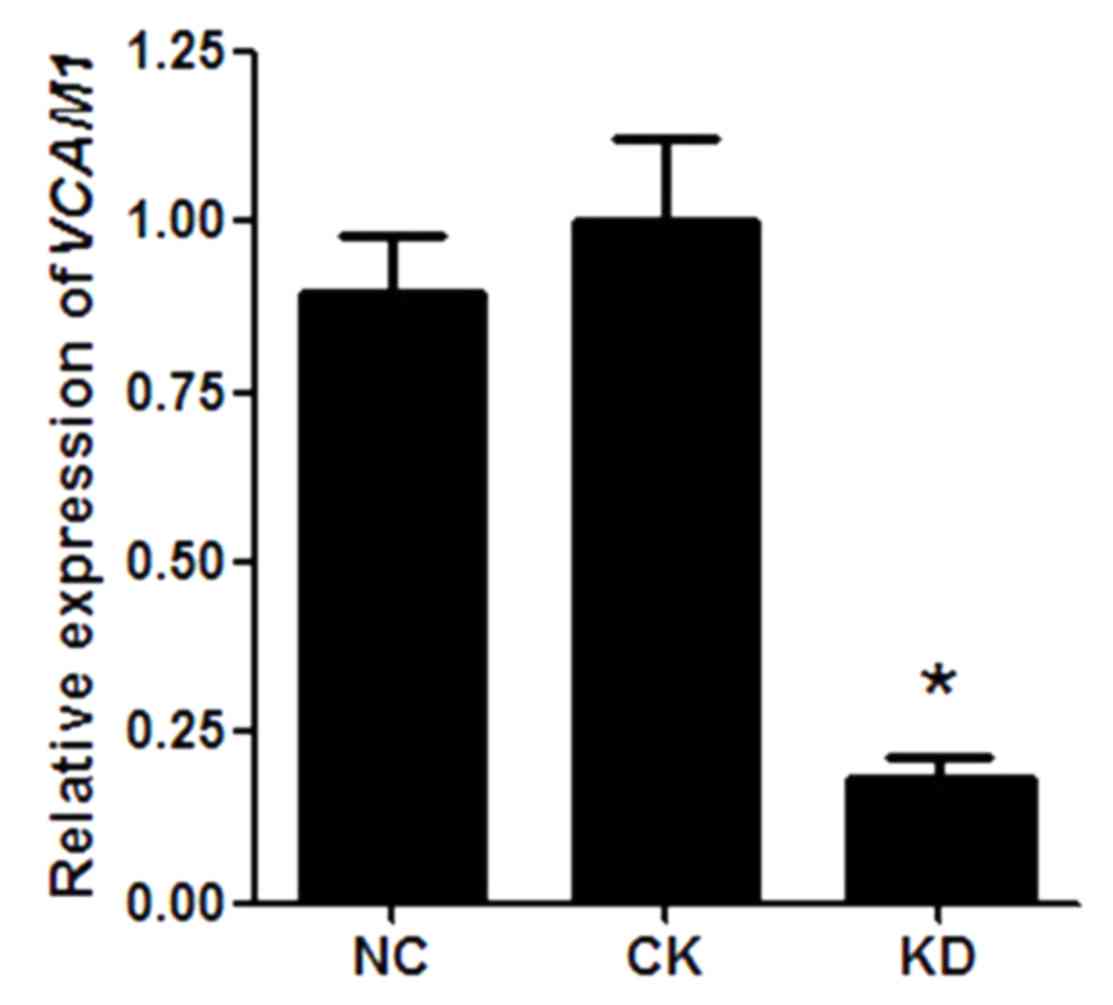
VCAM1 gene expression in HN12
cells
As the lentiviral vector system was efficiently
transduced into HN12 cells, the level of VCAM1 gene
expression was further analyzed. RT-qPCR results indicated that
there was no statistical difference in VCAM1 gene expression
in HN12 cells between the NC group and the CK group (P>0.05;
Fig. 2). However, compared with the
NC group, VCAM1 gene expression was significantly reduced in
the KD group (P<0.05), and the knockdown efficiency of VCAM1 was
~85% (Fig. 2).
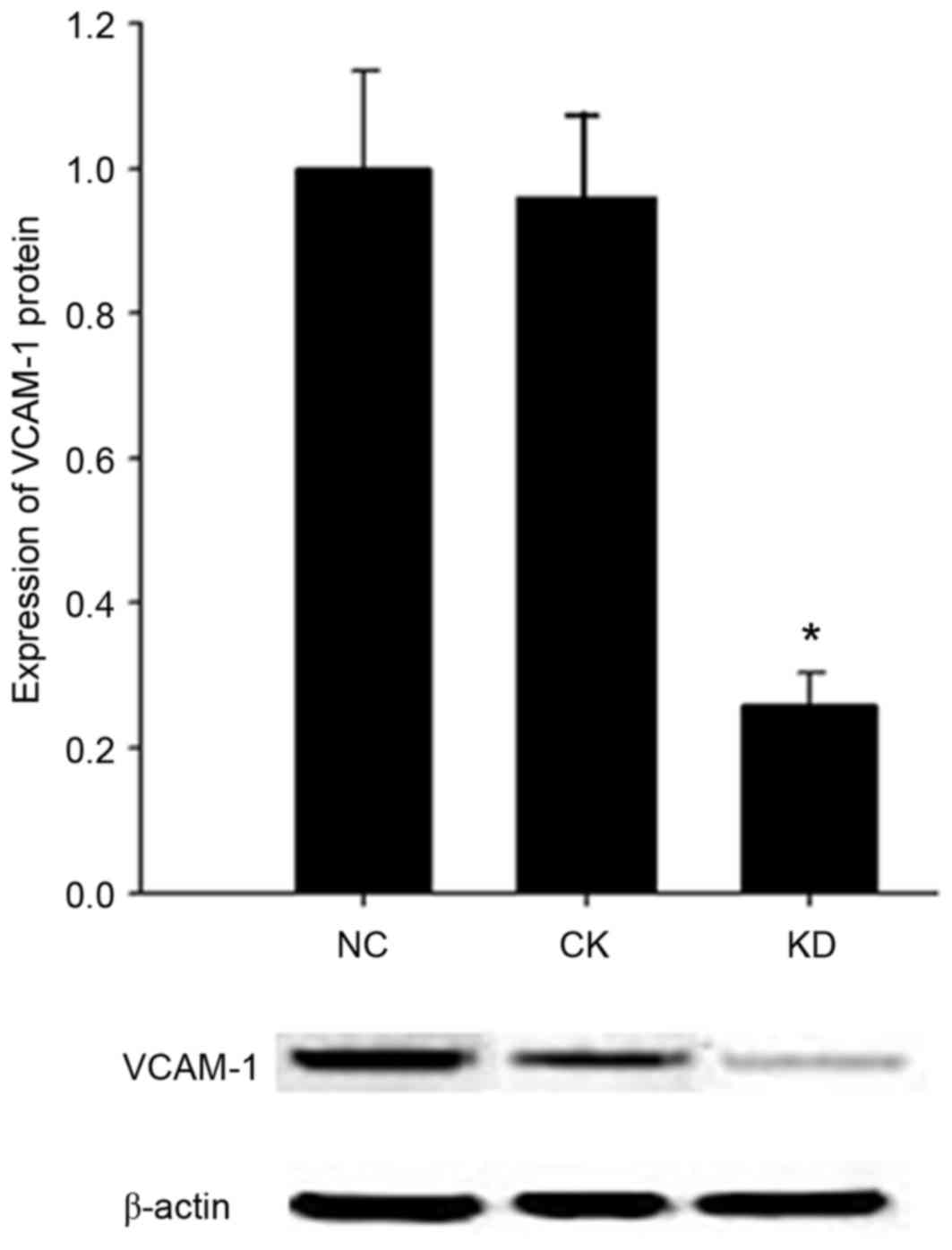
VCAM1 protein expression in HN12
cells
Western blot analysis was applied to detect the
expression level of VCAM1 protein in the HN12 cells infected by
lentiviral RNAi vectors. No significant difference in VCAM1 protein
expression level was observed between the CK group and the NC group
(P>0.05) (Fig. 3). Compared with
the NC group, the expression level of VCAM1 was
significantly decreased, by ~74%, in the KD cell group (P<0.05;
Fig. 3).
Cell growth inhibition rate as
evaluated by a CCK-8 assay
To investigate the effect of VCAM1 silencing on cell
proliferation, the HN12 cell growth inhibition rate was assayed
using the CCK-8 method. Compared with the NC group, cell growth was
significantly inhibited in the KD group (P<0.05), with a growth
inhibition rate of ~33.97% (Fig. 4).
However, no significant inhibitory effect on cell growth was
observed in the CK group (P>0.05; Fig.
4).
Discussion
In the present study, RNAi was used to investigate
the effect of VCAM1 silencing on the proliferation of human
oral squamous carcinoma HN12 cells. The results of RT-qPCR and
western blotting demonstrated that RNAi using a VCAM1-targeted
lentiviral vector significantly inhibited the expression of VCAM1.
In addition, cell growth was significantly inhibited in the KD
group cells, with a cell growth inhibition rate of ~33.97%. A
previous study indicated that VCAM1 serves a significant role as a
molecular switch in the various signal transduction pathways of
tumor cells (6). VCAM1 not only
induces the recruitment of tumor angiogenesis factors, but also
mediates the adhesion of endothelial cell and matrix by sensing the
surrounding environment, leading to further tumor invasion and
metastasis (6). It has been observed
that VCAM1 expression is induced in the vascular endothelium of
oral squamous cell carcinoma (21).
In addition, the abnormal expression of VCAM1 is associated with
the metastasis of gastric carcinoma (11,12). In
addition, the current study indicated that VCAM1-targeted
RNAi is able to effectively inhibit the gene and protein expression
of VCAM1 as well as the proliferation of oral squamous carcinoma
cells, which may provide an experimental basis for the diagnosis
and treatment of colorectal cancer.
Nevertheless, the occurrence and development of oral
squamous cell carcinoma is associated with the aberrant expression
of multiple genes, as well as a variety of in vitro and
in vivo factors (3). Previous
studies have indicated that succinobucol-loaded nanoparticles
exhibit therapeutic efficacy in the metastasis of breast cancer to
the lung by inhibiting VCAM1 expression (22). Furthermore, anti-VCAM1 treatment is
able to significantly decrease cancer-endothelial adhesion and
block fusion (21). Until recently,
studies on gene therapy of oral squamous carcinoma primarily
focused on single-target gene treatments (4). Despite this progress, treatments
targeting multiple genes have not been investigated in vitro
and in vivo. The results of the present study potentially
provide a theoretical basis for future animal studies and
multi-gene targeting therapies in oral squamous carcinoma.
In conclusion, VCAM1-targeted RNAi
effectively inhibits the gene and protein expression of VCAM1, as
well as the proliferation of oral squamous carcinoma cells. These
results may provide an experimental reference for the diagnosis and
treatment of colorectal cancer.
Acknowledgements
The present study was supported by the Shandong
Natural Science Foundation of China (grant no. ZR2013HL008) and the
Science and Technology Project of Shandong Education Department
(grant no. J12LK08).
Glossary
Abbreviations
Abbreviations:
|
VACM1
|
vascular cell adhesion molecule 1
|
|
RISC
|
RNA-induced silencing complex
|
|
NC
|
negative control
|
|
MOI
|
multiplicity of infection
|
|
GFP
|
green fluorescence protein
|
|
SD
|
standard deviation
|
References
|
1
|
Chen YJ, Lin SC, Kao T, Chang CS, Hong PS,
Shieh TM and Chang KW: Genome-wide profiling of oral squamous cell
carcinoma. J Pathol. 204:326–332. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Woolgar JA, Rogers S, West CR, Errington
RD, Brown JS and Vaughan ED: Survival and patterns of recurrence in
200 oral cancer patients treated by radical surgery and neck
dissection. Oral Oncol. 35:257–265. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Aktas E, Uzman M, Yildirim O, Sahin B,
Buyukcam F, Aktas B, Yilmaz B, Yildirim AM, Basyigit S, Yeniova O,
et al: Assessment of hepatic steatosis on contrast enhanced
computed tomography in patients with colorectal cancer. Int J Clin
Exp Med. 7:4342–4346. 2014.PubMed/NCBI
|
|
4
|
Wen F, He S, Sun C, Li T and Wu S: PIK3CA
and PIK3CB expression and relationship with multidrug resistance in
colorectal carcinoma. Int J Clin Exp Pathol. 7:8295–8303.
2014.PubMed/NCBI
|
|
5
|
Siegel R, Ward E, Brawley O and Jemal A:
Cancer statistics, 2011: The impact of eliminating socioeconomic
and racial disparities on premature cancer deaths. CA Cancer J
Clin. 61:212–236. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Francavilla C, Maddaluno L and Cavallaro
U: The functional role of cell adhesion molecules in tumor
angiogenesis. Semin Cancer Biol. 19:298–309. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Garton KJ, Gough PJ, Philalay J, Wille PT,
Blobel CP, Whitehead RH, Dempsey PJ and Raines EW: Stimulated
shedding of vascular cell adhesion molecule 1 (VCAM-1) is mediated
by tumor necrosis factor-alpha-converting enzyme (ADAM 17). J Biol
Chem. 278:37459–37464. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rose DM, Cardarelli PM, Cobb RR and
Ginsberg MH: Soluble VCAM-1 binding to alpha4 integrins is
cell-type specific and activation dependent and is disrupted during
apoptosis in T cells. Blood. 95:602–609. 2000.PubMed/NCBI
|
|
9
|
Wu TC: The role of vascular cell adhesion
molecule-1 in tumor immune evasion. Cancer Res. 67:6003–6006. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen Q and Massagué J: Molecular pathways:
VCAM1 as a potential therapeutic target in metastasis. Clin Cancer
Res. 18:5520–5525. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Semba S, Kodama Y, Ohnuma K, Mizuuchi E,
Masuda R, Yashiro M, Hirakawa K and Yokozaki H: Direct
cancer-stromal interaction increases fibroblast proliferation and
enhances invasive properties of scirrhous-type gastric carcinoma
cells. Br J Cancer. 101:1365–1373. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ding YB, Chen GY, Xia JG, Zang XW, Yang HY
and Yang L: Association of VCAM-1 overexpression with oncogenesis,
tumor angiogenesis and metastasis of gastric carcinoma. World J
Gastroenterol. 9:1409–1414. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhao XP, Wang M, Song Y, Song K, Yan TL,
Wang L, Liu K and Shang ZJ: Membrane microvesicles as mediators for
melanoma-fibroblasts communication: Roles of the VCAM-1/VLA-4 axis
and the ERK1/2 signal pathway. Cancer Lett. 360:125–133. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kokovay E, Wang Y, Kusek G, Wurster R,
Lederman P, Lowry N, Shen Q and Temple S: VCAM1 is essential to
maintain the structure of the SVZ niche and acts as an
environmental sensor to regulate SVZ lineage progression. Cell Stem
Cell. 11:220–230. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yan J, Jiang Y, Ye M, Liu W and Feng L:
The clinical value of lymphatic vessel density, intercellular
adhesion molecule 1 and vascular cell adhesion molecule 1
expression in patients with oral tongue squamous cell carcinoma. J
Cancer Res Ther. 10 Suppl:C125–C130. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hannon GJ: RNA interference. Nature.
418:244–251. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Warnock JN, Merten OW and Al-Rubeai M:
Cell culture processes for the production of viral vectors for gene
therapy purposes. Cytotechnology. 50:141–162. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Aoki T, Shimizu S, Urano E, Futahashi Y,
Hamatake M, Tamamura H, Terashima K, Murakami T, Yamamoto N and
Komano J: Improvement of lentiviral vector-mediated gene
transduction by genetic engineering of the structural protein Pr55
Gag. Gene Ther. 17:1124–1133. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sun LG, Song YF, Liu L, Yang YC, Wang F
and Wang LF: Correlations between the expression of vascular cell
adhesion molecule-1 gene and clinical pathological characteristics
and angiogenesis in oral squamous cell carcinoma. Shanghai Kou
Qiang Yi Xue. 17:569–573. 2008.(In Chinese). PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lee N, Shin S, Chung HJ, Kim DK, Lim JM,
Park H and Oh HJ: Improved quantification of protein in vaccines
containing aluminum hydroxide by simple modification of the Lowry
method. Vaccine. 33:5031–5034. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Song K, Zhu F, Zhang HZ and Shang ZJ:
Tumor necrosis factor-α enhanced fusions between oral squamous cell
carcinoma cells and endothelial cells via VCAM-1/VLA-4 pathway. Exp
Cell Res. 318:1707–1715. 2012. View Article : Google Scholar : PubMed/NCBI
|