Introduction
The most prevalent primary malignancy in the central
nervous system is the glioma, which constitutes ~77% of all
malignant brain tumors worldwide in 2007 (1,2). At
present, the pathogenesis of gliomas remains unknown. Nomenclature
of gliomas is specific to the type of cell affected, including
astrocytomas, oligodendrogliomas, ependymomas and mixed glioma
(1–3).
The glioma shares histological features with each specific cell
type, but does not necessarily originate from the cell type
depicted in its nomenclature (4). A
variety of grading systems for tumor types are available, but the
World Health Organization (WHO) grading system (2) is the most common for gliomas. This
system categorizes tumor types from grade I to grade IV, with grade
I being the least advanced and having the most positive prognosis
to IV being the most advanced and having the worst prognosis.
The conventional therapeutic regimen for glioma
consists of varying combinations of surgical procedures,
radiotherapy and chemotherapy. However, this approach has been
ineffective for a number of patients, resulting in a rapid
progression of the disease (5). For
refractory forms of the disease, previous studies have focused on
molecular biomarkers to identify novel drug targets, resulting in
the development of novel therapeutic approaches, which demonstrate
encouraging results (6).
The tumor suppressor candidate 3 (TUSC3) gene, also
referred to as N33, M33, MRT7, MRT22, oligosaccharyltransferase
(OST) 3 homolog A (OST3) and D8S1992, is positioned on chromosome
region 8p22 and encodes the 34 kDa TUSC3 protein (7). It was originally discovered by MacGrogan
et al (8) in 1996. The TUSC3
mRNA may be present in large quantities in a variety of cells and
tissues including prostate, colon, lung, liver, ovary, placenta,
testis, brain and adipose tissues (9), yet it is rare in other tissues,
including in bone marrow. TUSC3 shares a substantial amount of
homologous sequences with Ost3p, a subunit of the OST complex, and
serves a function in the N-linked glycosylation reactions of the
protein folding process (10–12). With emerging research conducted over
the past several years, it has been determined that TUSC3 is
associated with autosomal recessive mental retardation (ARMR)
(13–15). Previously, studies had identified the
deletion of TUSC3 in a number of cancer types including oral
squamous cell carcinoma and lung cancer. This resulted in the
identification of TUSC3 as a potential tumor suppressor gene
(16–17). Multiple studies have been conducted to
determine the association between TUSC3 expression and the
development of a tumor (16–20). However, to the best of our knowledge,
there has been no previously published data on the expression of
TUSC3 in gliomas. In the present study, an investigation
surrounding the expression of TUSC3 in brain glioma was conducted
and the association between TUSC3 expression and the
clinicopathological parameters of gliomas was determined. The
results of the present study may help to identify the potential
prognostic and therapeutic targets for brain gliomas.
Materials and methods
Tissue samples
A total of 12 pairs of glioma tissue samples were
surgically removed from patients at the Qianfoshan Hospital
(Shandong, China). A total of 6 male and 6 female patients
participated in the study, and the mean age was 53.1 years (range,
34–73). The tissues were obtained between August 2015 and October
2015. None of the patients had been exposed to any other
therapeutic interventions, including chemotherapy or radiotherapy,
prior to this surgery. Tissue microarray slides were purchased from
Alenabio (Xi'an, China). The slides included 38 cases of
astrocytoma, 14 cases of glioblastoma, 6 cases of
oligodendrogliomas, 1 case of anaplastic ependymoma, 1 case of
medulloblastoma and 13 cases of normal adjacent tissue. Patients
providing adequate histological material and complete clinical data
were eligible to participate in the present study. Patients with a
history of chemotherapy, radiotherapy or other therapeutic
interventions which may affect the results of the present study
were excluded. Table I represents the
clinicopathological characteristics of patients with brain glioma,
who were clinically staged according to the WHO grading system
(2). The WHO grading scale is as
follows: WHO grade I are well-differentiated, WHO grade II are
moderately-differentiated, WHO grade III are poorly differentiated
and WHO grade IV are undifferentiated. WHO grade I–II are referred
to as low-grade gliomas and WHO grade III–IV are referred to as
high-grade gliomas. The clinicopathological information of all
participants are provided in Table I.
Tumor classification of each of the patients was determined by two
pathologists from Shandong Provincial Qianfoshan Hospital
(Shandong, China) through use of the WHO grading system. The
Ethical Committee of Shandong Provincial Qianfoshan Hospital
ethically approved present study, and all patients prior to the
study provided written informed consent.
 | Table I.Baseline clinicopathological
characteristics of patients with glioma and patients providing
control tissues. |
Table I.
Baseline clinicopathological
characteristics of patients with glioma and patients providing
control tissues.
|
|
| TUSC3 expression |
|
|
|---|
|
|
|
|
|
|
|---|
| Characteristics | Number | Positive | Negative | Positive rate
(%) | P-valuea |
|---|
| Control patients |
|
|
|
|
|
| Age
median (range), years | 46
(26–76) |
|
|
|
|
| Gender
(%) |
|
|
|
|
|
|
Male | 6
(46.2) |
|
|
|
|
|
Female | 7
(53.8) |
|
|
|
|
| Patients with
glioma |
|
|
|
|
|
| Age
median (range), years | 43
(14–66) |
|
|
| 0.007 |
|
<43 | 30 (50.0) | 6 | 24 | 20.0 |
|
|
≥43 | 30 (50.0) | 17 | 13 | 56.7 |
|
|
Gender |
|
|
|
| 1.000 |
|
Male | 35 (58.3) | 12 | 23 | 34.3 |
|
|
Female | 25 (41.7) | 8 | 17 | 32.0 |
|
|
Pathological
classification |
|
|
|
|
|
|
Astrocytoma | 38 (63.3) | 18 | 20 | 47.4 |
|
|
Oligodendroglioma | 6
(10.0) | 5 | 1 | 83.3 |
|
|
Glioblastoma | 14 (23.4) | 0 | 14 | 0 |
|
|
Other types | 2 (3.3) | 0 | 2 | 0 |
|
| WHO
gradeb |
|
|
|
|
|
|
I | 1 (1.7) | 1 | 0 | 100 |
|
|
II | 36 (60.0) | 20 | 16 | 55.6 |
|
|
III | 9
(15.0) | 3 | 6 | 33.3 |
|
|
IV | 14 (23.3) | 0 | 14 | 0 |
|
Western blot analysis
Tissue samples were homogenized using Radio
Immunoprecipitation Assay buffer (Beyotime Institute of
Biotechnology, Haimen, China) with proteinase inhibitors, resulting
in cell lysis. The proteins were quantified by BCA protein
quantitative kit (Beyotime Institute of Biotechnology, Haimen,
China) SDS-PAGE (12%) was administered to equal amounts (30 µg)
protein from each lysate and the resulting products were
transferred onto a polyvinylidene membrane. Blocking of
non-specific binding was achieved by placing the membrane in 5%
non-fat dried milk containing 0.1% Tween-20 at room temperature for
3 h. The membrane was incubated at 4°C overnight in the presence of
primary antibodies for human TUSC3 goat polyclonal antibody
(dilution, 1:400; cat no. sc-98191; Santa Cruz Biotechnology, Inc.,
Dallas, TX, USA) and β-actin (dilution, 1:1,000; cat no. sc-98191;
Santa Cruz Biotechnology, Inc., Dallas, TX, USA). Subsequent to 3
washes with PBST buffer (PBS with 0.05% Tween-20) to remove unbound
primary antibodies, the membrane was further incubated for 1 h at
room temperature with horseradish peroxidase (HRP)-conjugated
secondary antibodies (rabbit anti-goat IgG, dilution, 1:5,000; cat
no. TA130031; OriGene Technologies, Inc., Beijing, China).
Following washing, the protein was identified through detection of
immunoreactive bands using Western Lightning Chemiluminescence
Reagent (Millipore; Merck KGaA, Darmstadt, Germany) and the bands
were analyzed using ImageJ (version 1.5; National Institutes of
Health, Bethesda, MD, USA).
Immunohistochemistry (IHC) assay
IHC staining was performed directly on the 5-µm
tissue slides. The first step was to dewax the slides using xylene.
Following wax removal, the slides were rehydrated using a series of
decreasing alcohol concentrations (100, 90, 70 and 50% ethanol; 5
min each) followed by incubation for 2 h at 56°C. Then, 3%
H2O2 was administered for ~20 mins at room
temperature to block endogenous peroxidase activity. For antigen
retrieval, sodium citrate (0.01 M, pH 6.0) was used as a buffer.
The slides were incubated at 95°C for 20 min in a household
microwave oven (600 W). Next, 10% normal goat serum (cat no.
ZLI-9022; OriGene Technologies, Inc., Rockville, MD, USA) was added
to the slides to block non-specific binding sites in order to
reduce background staining. The slides were then incubated with the
TUSC3 goat polyclonal antibody (dilution, 1:100; cat no. sc-98191;
Santa Cruz Biotechnology, Inc.) overnight at 4°C.
Phosphate-buffered saline was used to wash the slides 3 times prior
to application of the bio-labeled secondary antibody, Poly-HRP
anti-goat immunoglobulin G secondary antibody (cat no. PV-9003;
OriGene Technologies, Inc.) at a 1:200 dilution for 40 min at 37°C.
The sections were stained with diaminobenzidine at room temperature
for 2 min, followed by a counterstain with hematoxylin at room
temperature for 1 min. The slides were dehydrated using increasing
concentrations of alcohol and mounted using neutral gum.
Photomicrographs were captured under a light microscope
(magnification, ×400; BX-51, Olympus Corporation, Tokyo, Japan).
Aperio Image Scope (version 12.0.1.5027; Aperio Technologies, Inc.,
Vista, CA, USA) was used to score the stained cells using a digital
pathology system.
The cytoplasm and cell membrane were identified as
regions of TUSC3 expression. Two pathologists from the Shandong
Provincial Qianfoshan Hospital independently evaluated the
immunoreactivity of TUSC3 expression, then groups were determined
according to the respective immunoreactive score (IRS). IRS are
calculated by multiplying the staining intensity by the percentage
of positively stained cells. Staining intensity was classified as 0
(negative), 1 (weak), 2 (moderate) or 3 (strong). Percentage of
positively stained cells was scored as 0 (negative), 1 (<25% of
the cells), 2 (25–50% of the cells), 3 (50–75% of the cells) or 4
(>75% of the cells). Through multiplication of these two scores,
a final score was calculated. An IRS value of 6 was determined to
be the cut-off value separating specimens into two groups of
negative or positive expression.
Statistical analysis
Statistical analysis of the results was conducted
using SPSS 13.0 software (SPSS, Inc., Chicago, IL, USA).
Differences in the grayscale value of protein expression were
compared using a paired-t test. The data are presented as the mean
± standard error. Differences in TUSC3 expression between two
groups were compared using a Wilcoxon's rank sum test. The
Spearman's rank correlation coefficient method was used to
determine the strength and direction of the association between two
groups. A Fisher's exact test was performed to compare the positive
rate between two groups. The comparison between clinicopathological
parameters (age, gender, histological classifications and WHO
grading) were performed using a Fisher's exact test. All reported
P-values were two-sided and P<0.05 was considered to indicate a
statistically significant difference. A receiver operating
characteristic curve was used to determine the cut-off IRS value of
6 for TUSC3 expression levels in patients. Relative TUSC3
expression for each band was analyzed using ImageJ (version 1.51;
National Institutes of Health, Bethesda, MD, USA).
Results
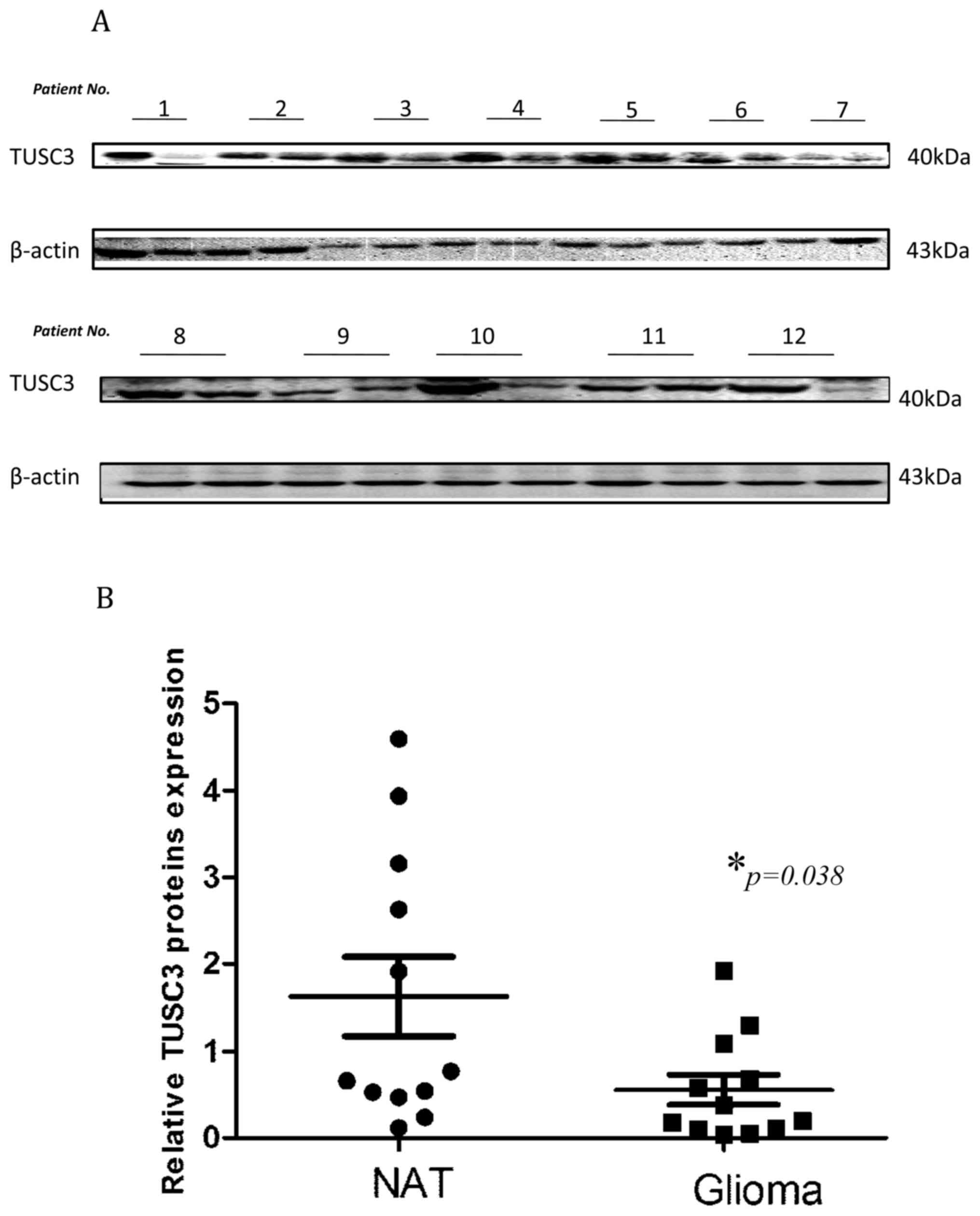
Comparison of TUSC3 protein expression
between tumor and normal adjacent brain tissue in patients with
glioma
To begin with, the TUSC3 expression levels were
determined through western blot analysis in glioma tissues and
normal adjacent tissues, which were surgically collected at
Qianfoshan Hospital. TUSC3 expression was significantly reduced in
glioma tissues when compared with normal adjacent tissue
(1.631±0.4534 vs. 0.5579±0.1723; P=0.038; Fig. 1). These results are indicative of the
pivotal function TUSC3 may serve in glioma development.
Basic clinical feature of patients for
tissue array
Furthermore, the levels of TUSC3 expression were
evaluated using a tissue array. Table
I outlines the basic clinicopathological features of the
patients with glioma and control patients who provided normal
adjacent tissues. The median age of the controls was 46 years
(range, 26–76 years) and that of patients with glioma was 43 years
(range, 14–66 years). Difference in sex and age between normal
controls and patients with glioma were not statistically
significant. However, when the patients with glioma were grouped
according to median age, analyses revealed a significant increase
in positive TUSC3 expression in patients aged≥43 compared with
patients <43 (χ2=8.531, P=0.007; Table I).
Expression of TUSC3 in normal adjacent
tissues and tumor tissues
The positive rates of TUSC3 expression in normal
adjacent tissue and glioma tissues were 76.9% (10/13) and 40.0%
(24/60), respectively. The difference in TUSC3 expression levels
between the two groups was revealed to be statistically significant
(χ2=5.854, P=0.029; Table
II). The results displayed in Fig.
1 demonstrating lower TUSC3 expression levels in glioma tissues
support this data.
 | Table II.Comparison of TUSC3 expression
between normal tissue and glioma tissue. |
Table II.
Comparison of TUSC3 expression
between normal tissue and glioma tissue.
|
|
| TUSC3
expression |
|
|
|---|
|
|
|
|
|
|
|---|
|
Characteristics | Number | Positive | Negative | Positive rate
(%) |
P-valuea |
|---|
| Normal adjacent
tissue | 13 | 10 | 3 | 76.9 | 0.029 |
| Glioma tissue | 60 | 24 | 36 | 40.0 |
|
Association between TUSC3 expression
and tumor pathological classification
When the patients were grouped according to tumor
pathological classification, which consisted of astrocytoma,
oligodendroglioma, glioblastoma and other types (including one
medulloblastoma and one anaplastic ependymoma), there was a
statistically significant difference in TUSC3 expression amongst
the different pathological groups (sum rank, P<0.0001; Fig. 2). Furthermore, the positive rates of
TUSC3 expression in the different pathological groups were compared
using Fisher's exact test. Results revealed that the highest TUSC3
expressionwas in the oligodendroglioma group (83.3% positive rate),
with little to no expression in glioblastoma or other types
(Table I).
Expression of TUSC3 in association
with the WHO grade of patients with glioma
The pathology of the tumor was evaluated, resulting
in the categorization of the glioma tissues into grades. According
to the WHO grade system, all of the glioma tissues were separated
into grade I–IV (Fig. 3). As
demonstrated in Fig. 3, TUSC3
expression decreased as the WHO grade increased. As presented in
Fig. 4, comparisons between the
groups revealed that the TUSC3 positive rate was significantly
different amongst different WHO grades (sum rank, P<0.0001;
Fig. 4A). To further confirm the
results above, a numerical value of correlation between TUSC3
expression and the WHO grade was determined using the Spearman
correlation method. TUSC3 expression demonstrated a negative
correlation with WHO grade (P<0.0001; rs=−0.3451; Fig. 4B). Thus, patients with higher grade
tumors had lower TUSC3 expression levels.
Discussion
Clinical treatment results have demonstrated that
gliomas are the main cause of mortality in patients with brain
tumors, particularly in child patients with high-grade glioma
(21). This reveals the urgency to
identify unique indicators for diagnosis and prognosis, and to
develop novel therapeutic approaches for gliomas.
The TUSC3 gene is 224,265 bp in length, with 11
exons spanning the full length of the genomic DNA on chromosome
8p22 (7). TUSC3is considered an
ortholog of the yeast Ost3p, initially identified as a 34 kD
subunit in the yeast OST complex (22,23). Human
TUSC3 protein sequences share ~20% identities of the protein domain
organization with the yeast subunit OST3. Additionally, the two
contain an active binding domain at the amino terminal, central
acidic and zinc-binding domains, and a transmembrane domain at the
carboxyl terminal (11). The
transmembrane segments and N-terminal signal peptide are conserved
(24).
TUSC3 serves notable functions in maintaining normal
cell function. In 2008, Garshasbi et al (14) revealed TUSC3 homozygous deletion in 7
patients with non-syndromic ARMR in 4 sibships. Other studies
additionally demonstrated that TUSC3 mutations or deletions are
involved in the onset of ARMR (15,25).
Accumulated data has established that TUSC3 and cancer are notably
associated (16–20). It has been previously demonstrated
that TUSC3 is deleted or diminished in a number of cancer types,
including prostate, ovarian, gastric, pancreatic, esophageal and
lung cancer (16–20). Ribeiro et al (17) revealed that the loss of the TUSC3 gene
has a potential association with the malignancy of oral squamous
cell carcinoma. It was reported that TUSC3 loss is associated with
a high mortality rate in larynx and pharynx squamous cell carcinoma
(16). Previous observations had
demonstrated lower expression levels of TUSC3 in patients who were
lymph node metastasis positive (LNM+) compared with patients who
were lymph node metastasis negative (LNM-) (18). It was additionally discovered that
decreased TUSC3 levels predict a poor prognosis of patients with
esophageal squamous cell carcinoma (20). However, to the best of our knowledge,
there is no known data on the significance of TUSC3 expression in
patients with brain glioma, and the present study is the first that
analyzes the association between the TUSC3 expression levels and
the clinical characteristics of gliomas.
Through the use of western blot and tissue
microarray assays, it was identified that TUSC3 expression was
significantly lower in glioma tissues compared with normal adjacent
tissues. Furthermore, TUSC3 expression demonstrated a significant
negative correlation with WHO grade in patients with glioma
(P<0.0001; rs=−0.3451). Due to the fact that glioma WHO grade is
an indicator for tumor malignancy and prognosis, the results of the
present study indicated an association between low TUSC3 expression
with a poorly-differentiated tumor grade and higher malignancy. The
present results have the potential to make a notable contribution
to the field of medicine. By combining the present research and
further research in the future, TUSC3 may become a novel grading
tool that assists in evaluating tumor malignancies and consequently
a therapeutic regimen may be used for patients with glioma.
However, when the patients were grouped according to
median age, analyses revealed a significant increase in TUSC3
positive expression in the patients who were age 43 or above
compared with the patients who were younger than age 43
(χ2=8.531, P=0.007; Table
I). These results need to be verified using a larger sample.
Furthermore, due to the screening rules for patient inclusion,
there was a relatively small number of samples included in the
present study. As a result, the TUSC3 expression in I–IV WHO grades
of all histological types were not analyzed separately. Larger
sample sizes will be used in further research.
With the expanding research in this area, the
mechanisms for the involvement of the TUSC3 gene in cancer
development have been illustrated. Vaòhara et al (26) and Horak et al (27) demonstrated that the loss of the
TUSC3gene affects N-glycosylation events in the development of
ovarian cancer and prostate cancer. However, the precise molecular
mechanism concerning the involvement of TUSC3 in the development of
cancer remains unclear. As mentioned previously, a number of
studies have demonstrated that TUSC3 is an integral endoplasmic
reticulum protein involved in N-glycosylation (17,26). In
addition, one previous study indicated that N-glycosylation
abnormalities affect the phosphoinositide 3-kinase-protein kinase B
pathway, thus affecting the growth of tumor cells (28). However, further studies need to be
performed regarding the mechanisms of TUSC3 involvement in glioma
development.
Prior to the present study, limited mechanisms for
TUSC3 gene dysfunction in tumors had been identified. Detected in
multiple cancer types, aberrant TUSC3 reductions were identified
due to a mutation resulting in its deletion (17,29) as
well as its promoter CpG island methylation (27,30).
Further research exploring the causes of TUSC3 dysfunction in
gliomas, including genetic and epigenetic alterations, will aid
understanding of the mechanism of TUSC3-associated
tumorigenesis.
Due to its functional significance in protein
maturation and the development of multiple tumor types, it has been
proposed that TUSC3 has the potential to be a useful biomarker and
a therapeutic target in the diagnosis and therapy of ovarian
cancer, prostate cancer and oral squamous cell carcinoma,
particularly for refractory gliomas. There are various potentials
for the therapeutic targeting of the TUSC3 protein in a tumor. Drug
discoveries to reinstate TUSC3 function in diseases will focus on
avenues including recombinant Ad-TUSC3 gene therapy, targeting the
TUSC3 interactive proteins, or targeting destabilized oncogenic
TUSC3 mutants through the design of mutant-specific TUSC3 rescue
drugs. Although substantial knowledge was gained from the present
study, numerous structural aspects of TUSC3 function have remained
elusive, including details of TUSC3 interactive proteins and the
specifics of TUSC3 mutations in diseases. To be a promising
therapy, TUSC3 targeting therapy requires further exploration.
Acknowledgements
The present study was supported by the Medical and
Health Science and Technology Development Plan of Shandong Province
(grant nos. 2015WS0213 and 2017GSF18195), the Natural Science
Foundation of Shandong Province (grant nos. ZR2011HQ010,
ZR2015HM077, ZR2016HQ50 and ZR2015CL030), the Chinese Postdoctoral
Science Foundation (grant no. 2016M602154) and the National Natural
Science Foundation of China (grant nos. 30901712 and 81301868).
References
|
1
|
Walsh KM, Ohgaki H and Wrensch MR:
Epidemiology. Handb Clin Neurol. 134:3–18. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Louis DN, Ohgaki H, Wiestler OD and
Cavenee WK: WHO Classification of Tumours of the Central Nervous
System. IARC Press; Lyon: 2007
|
|
3
|
Schwartzbaum JA, Fisher JL, Aldape KD and
Wrensch M: Epidemiology and molecular pathology of glioma. Nat Clin
Pract Neurol. 2:494–503. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Vigneswaran K, Neill S and Hadjipanayis
CG: Beyond the World Health Organization grading of infiltrating
gliomas: Advances in the molecular genetics of glioma
classification. Ann Transl Med. 3:952015.PubMed/NCBI
|
|
5
|
Wen PY and Kesari S: Malignant Gliomas in
Adults. N Engl J Med. 359:492–507. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Reifenberger G, Wirsching HG,
Knobbe-Thomsen CB and Weller M: Advances in the molecular genetics
of gliomas-implications for classification and therapy. Nat Rev
Clin Oncol. 14:434–452. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yu X, Zhai C, Fan Y, Zhang J, Liang N, Liu
F, Cao L, Wang J and Du J: TUSC3: A novel tumour suppressor gene
and its functional implications. J Cell Mol Med. 21:1711–1718.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
MacGrogan D, Levy A, Bova GS, Isaacs WB
and Bookstein R: Structure and methylation-associated silencing of
a gene within a homozygously deleted region of humanchromosome band
8p22. Genomics. 35:55–65. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhou H and Clapham DE: Mammalian MagT1 and
TUSC3 are required for cellular magnesium uptake and vertebrate
embryonic development. Proc Natl Acad Sci USA. 106:pp. 15750–15755.
2009; View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Molinari F, Foulquier F, Tarpey PS,
Morelle W, Boissel S, Teague J, Edkins S, Futreal PA, Stratton MR,
Turner G, et al: Oligosaccharyltransferase subunit mutations in
non-syndromic mental retardation. Am J Hum Genet. 82:1150–1157.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mohorko E, Owen RL, Malojčić G, Brozzo MS,
Aebi M and Glockshuber R: Structural Basis of substrate Specificity
of human oligosaccharyl transferase subunit N33/TUSC3 and its role
in regulating protein N-Glycosylation. Structure. 22:590–601. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jamaluddin MF, Bailey UM, Tan NY, Stark AP
and Schulz BL: Polypeptide binding specificities of Saccharomyces
cerevisiae oligosaccharyl-transferase accessory proteins Ost3p and
Ost6p. Protein Sci. 20:849–855. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang MJ, Xing LX, Cui M, Yang X, Shi JG,
Li J, Zhang KJ, Zheng ZJ, Zhang FC, Li JL and Gao XC: Association
of TUSC3 gene polymorphisms with non-syndromic mental retardation
based on nuclear families in the Qinba mountain area of China.
Genet Mol Res. 14:5022–5030. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Garshasbi M, Hadavi V, Habibi H, Kahrizi
K, Kariminejad R, Behjati F, Tzschach A, Najmabadi H, Ropers HH and
Kuss AW: A defect in the TUSC3 gene is associated with autosomal
recessive mental retardation. Am J Hum Genet. 82:1158–1164. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Garshasbi M, Kahrizi K, Hosseini M, Nouri
Vahid L, Falah M, Hemmati S, Hu H, Tzschach A, Ropers HH, Najmabadi
H and Kuss AW: A novel nonsense mutation in TUSC3 is responsible
for non-syndromic autosomal recessive mental retardationin a
consanguineous Iranian family. Am J Med Genet A. 155A:1–1980.
2011.PubMed/NCBI
|
|
16
|
Guervós MA, Marcos CA, Hermsen M, Nuño AS,
Suárez C and Llorente JL: Deletions of N33, STK11 and TP53 are
involved in the development of lymph node metastasis in larynx and
pharynx carcinomas. Cell Oncol. 29:327–334. 2007.PubMed/NCBI
|
|
17
|
Ribeiro IP, Marques F, Caramelo F, Pereira
J, Patrício M, Prazeres H, Ferrão J, Julião MJ, Castelo-Branco M,
de Melo JB, et al: Genetic gains and losses in oral squamous cell
carcinoma: Impact on clinical management. Cell Oncol (Dordr).
37:29–39. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yu X, Zhang K, Liu F, Zhang J, Zhai C, Cao
L, Song X, Wang Y, Li B, Sun H and Du J: Tumor suppressor candidate
3 as a novel predictor for lymph node metastasis in lung cancer
patients. Oncol Lett. 12:5099–5105. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bashyam MD, Bair R, Kim YH, Wang P,
Hernandez-Boussard T, Karikari CA, Tibshirani R, Maitra A and
Pollack JR: Array-based comparative genomic Hybridization
Identifies localized DNA amplifications and homozygous deletions in
pancreatic cancer. Neoplasia. 7:556–562. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yu X, Zhang J, Zhong H, Liu F, Liang N,
Wang Y, Meng X and Du J: Decreased tumor suppressor candidate 3
predicts poor prognosis of patients with esophageal squamous cell
carcinoma. Int J Med Sci. 13:963–969. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Rizzo D, Ruggiero A, Martini M, Rizzo V,
Maurizi P and Riccardi R: Molecular biology in pediatric high-grade
glioma: Impact on prognosis and treatment. Biomed Res Int.
2015:2151352015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Das RC and Heath EC:
Dolichyldiphosphoryloligosaccharide-protein
oligosaccharyltransferase; solubilization, purification, and
properties. Proc Natl Acad Sci USA. 77:pp. 3811–3815. 1980;
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kelleher DJ, Karaoglu D, Mandon EC and
Gilmore R: Oligosaccharyl transferase isoforms that contain
different catalytic STT3 subunits have distinct enzymatic
properties. Mol Cell. 12:101–111. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mohorko E, Glockshuber R and Aebi M:
Oligosaccharyl-transferase: The central enzyme of N-linked protein
glycosylation. J Inherit Metab Dis. 34:869–878. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Khan MA, Rafiq MA, Noor A, Ali N, Ali G,
Vincent JB and Ansar M: A novel deletion mutation in the TUSC3 gene
in a consanguineous Pakistani family with autosomal recessive
nonsyndromic intellectual disability. BMC Med Genet. 12:562011.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Vaòhara P, Horak P, Pils D, Anees M, Petz
M, Gregor W, Zeillinger R and Krainer M: Loss of the oligosaccharyl
transferase subunit TUSC3 promotes proliferation and migration of
ovarian cancercells. Int J Oncol. 42:1383–1289. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Horak P, Tomasich E, Vaňhara P,
Kratochvílová K, Anees M, Marhold M, Lemberger CE, Gerschpacher M,
Horvat R, Sibilia M, et al: TUSC3 loss alters the ER stress
response and accelerates prostate cancer growth in vivo. Sci Rep.
4:37392014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Fang M, Shen Z, Huang S, Zhao L, Chen S,
Mak TW and Wang X: The ER UDPase ENTPD5 promotes protein
N-glycosylation, the Warburg effect, and proliferation in the PTEN
pathway. Cell. 143:711–724. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Birnbaum DJ, Adélaïde J, Mamessier E,
Finetti P, Lagarde A, Monges G, Viret F, Gonçalvès A, Turrini O,
Delpero JR, et al: Genome profiling of pancreatic adenocarcinoma.
Genes Chromosomes Cancer. 50:456–465. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Pils D, Horak P, Vanhara P, Anees M, Petz
M, Alfanz A, Gugerell A, Wittinger M, Gleiss A, Auner V, et al:
Methylation status of TUSC3 is a prognostic factor in ovarian
cancer. Cancer. 119:946–954. 2013. View Article : Google Scholar : PubMed/NCBI
|