Introduction
Chemotherapy remains the best first line therapy for
treatment of aggressive cancer. Whilst it can be effective in the
short term, the high doses required can give rise to cancer cells
that exhibit drug resistance, which is a major problem in current
cancer treatment protocols. Recently, anti-mitotic drugs, including
those targeting aurora kinases, mitotic spindle proteins and
polo-like kinases, have proven disappointing underscoring the
urgent need for the development of novel therapeutic strategies to
overcome drug-resistance (1).
Cluster of differentiation (CD)147 (also known as
EMMPRIN, basigin, M6, and tumor cell-derived collagenase
stimulating factor), a glycoprotein belonging to the immunoglobulin
superfamily, is enriched on the plasma membrane of tumor cells
(2). The expression of CD147 is
closely related to expression of the classical multi-drug
resistance (MDR)-related transporter (MDR1) and its upregulation
leads to a decrease in the chemosensitivity of some
chemotherapeutic agents such as paclitaxel and curcumin. Studies in
a variety of drug-resistant cell lines have shown that CD147
overexpression followed by RNA interference or use of anti-CD147
blocking antibodies can increase the sensitivity of tumor cells to
chemotherapy drugs (3–5). Thus, overexpression of CD147 on MDR cell
lines may play an important role in the resistance to chemotherapy
drugs and CD147 is considered a potential therapeutic target
(6). While antibodies against CD147
have been screened for cancer treatment, cell immunotherapy using
CD147 as a target has yet to be explored. Therefore, in this study
we investigate whether drug resistance can be overcome by targeting
CD147 expressed on drug-resistance cells.
Cell immunotherapy represents a profound shift in
the treatment of cancer and because it is a specifically targeted
therapy it provides the possibility of fewer side effects compared
to chemotherapy (7,8). Moreover, an optimal target can be
identified for treatment of resistant tumor cells and cell
immunotherapy applied for their removal. For example, generation of
CD147-peptide specific reactive CTLs can be achieved using
dendritic cells (DCs) loaded with the CD147 TAA peptide. However,
some clinical trials have indicated that TAA peptide vaccines
designed with tumor-associated antigen (TAA) fail to achieve a
satisfactory effect in vivo. This may be owing to central
and peripheral immune tolerance making activation and expansion of
low affinity T cells difficult in vivo. Therefore,
strategies to modify the CD147 peptide in order to enhance its
binding to MHC and boost affinity of the peptide MHC complex for
the TCR thereby inducing peptide-specific CTL activation and
expansion in vitro are necessary (9).
Based on these findings, we believe CD147 could be a
optimal target of CD8+ cytotoxic T lymphocytes (CTLs).
However, TAA peptide vaccine designed directly with TAA failed to
achieve a satisfactory effect in vivo (10). This may owing to the central and
peripheral tolerance, it also make low affinity T cell difficult to
be activate and expansion. Therefore, strategies should be taken to
modify CD147 epitope peptide enhance its affinity to MHC molecule
in order to boost the affinity of the peptide MHC complex to the
TCR, thus leading peptide specific CTL activation and expansion
in vitro. In our previous study, a mutated survivin epitope,
identified by point mutation, could elicit specific CTL with
crossreactivity against tumor cells expressing a wild-type survivin
peptide in vitro (11,12).
In our previous study, we identified a point
mutation in the survivin epitope that could elicit a specific CTL
response in vitro with cross-reactivity against tumor cells
expressing a wild-type survivin peptide. In this study, we
identified CD147126–134, a low binding score wild-type
peptide, using a computer-based program and then used
point-mutation technology to substitute the L(leu) at position 2 of
the wild-type peptide with K(lys), to generate a peptide capable of
inducing specific CTLs. We found that these CTLs could recognize
and lyse the wild-type CD147126–134 peptide expressed on
the surface of drug-resistant cells.
Materials and methods
Cells and cell culture
The T2 cell line was purchased from ATCC and
maintained in RPMI 1640 with 10% FBS (both Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA), 100 IU/ml penicillin, 100 g/ml
streptomycin (both Sigma-Aldrich, Madrid, Spain). The MCF-7
(HLA-A*0201+, CD147+), SKOV3
(HLA-A*0201+, CD147−), Hela
(HLA-A*0201−, CD147+) was cultured in DMEM
(Life Technologies, New York, NY, USA) containing 10% FBS, 100
IU/ml penicillin, 100 g/ml streptomycin. The SKOV3 cell line was
transfected with expression vector pcdna3.1 containing HLA-A*0201
cDNA. The MCF-7/Adr (HLA-A*0201+, CD147+)
cell line was cultured in DMEM supplemented with 10% FBS with 1
µg/ml Adriamycin (Selleck, Shanghai, China) (13). K562 cell line purchased from ATCC were
used as natural killer cell-sensitive targets. K562 were cultured
in IMDM (Gibco; Thermo Fisher Scientific, Inc.) supplemented
containing 10% FBS, 100 g/ml streptomycin, 100 IU/ml
penicillin.
Peptide epitope prediction and
synthesizing
The sequences of CD147 was obtained from GenBank and
analyzed for HLA-A*0201 binding motifs using BIMAS (http://www-bimas.cit.nih.gov/molbio/hla_bind/) and
SYPEITHI (www.syfpeithi.de) (14). The wild-type peptide,
CD147126–134, and mutated peptide,
CD147126–134L2, were selected for additional evaluation.
The HIVpol476–484 was used as a positive control for
HLA-A*0201 binding ability. The HIVpol476–484 peptide
was used as an irrelevant peptide to assess cytotoxicity in a
Calcein-AM release assay. All peptides were synthesized by
Chinapeptide (Shanghai, China) and the purity was detected to an
average of approximately 98 percent by analytical mass spectrometry
and high performance liquid chromatography. Peptides were dissolved
at 10 mg/ml in DMSO (Sigma, St Louis, MO, USA) and stored at −70°C
for long-term preservation. All peptides are list in Table I.
 | Table I.Predicted CD147 peptides. |
Table I.
Predicted CD147 peptides.
| Peptide name | Position | Amino acid
sepuence | BIMAS score | SYFPEITHI
score |
|---|
|
CD147126–134 | 126–134 | CKSESVPPV | 0.911 | 17 |
|
CD147126–134La | 126–134 | CLSESVPPV | 655.875 | 27 |
|
HIVpol476–484 | 476–484 | ILKEPVHGV | 39.025 | 30 |
Peptide-binding assay. A peptide-induced
stabilization assay was performed using the T2 cell line expressing
the HLA A*0201 molecule (15).
Briefly, T2 cells (1×106/group) were incubated in the
presence of 20 µg/ml peptide in AIMV medium (Gibco; Thermo Fisher
Scientific, Inc.) supplemented with 5 µg/ml human β2-microglobulin
(Sigma-Aldrich, Spain) at 37°C in 5% CO2 for 18 h. T2
cells were washed twice with PBS to remove unbound peptide and
resuspended in PBS containing 2% FBS. T2 cells loaded with peptide
were incubated with FITC-conjugated HLA-A2 monoclonal antibody
(BB7.2; BioLegend, San Diego, CA, USA). The expression level of
HLA-A*0201 was measured using flow cytometry (Beckman Coulter,
Miami, FL, USA) and the EXPO32 v1.2 software was used to analyze
the results.
Flow cytometric analysis of CD147
expression
Cells (1×106 cells/group) were washed
with PBS two times followed by resuspension in PBS with 2% FBS.
Cells then were then incubated for 30 min at 4°C with
FITC-conjugated monoclonal anti-CD147 antibody (BD Biosciences, San
Diego, CA USA) or FITC-conjugated anti-mouse IgG1 isotype control
antibody (BD Biosciences). After two washes with PBS, cells were
resuspended in PBS to measure expression of CD147 by the flow
cytometry and the EXPO32 v1.2 software was used to analyze the
results.
Induction of peptide-specific
CTLs
All subjects in this study were Han Chinese from
Guangdong province, China, and all gave a written informed consent.
This study was performed with the approval of the Institute
Research Medical Ethics Committee of Guangzhou Pharmaceutical
University. PBMCs used were isolated from buffy coats obtained from
healthy HLA-A*0201 volunteer donors. Adherent monocyte-enriched
PBMCs were maintained in X-VIVO (Lonza, Benicia, CA, USA) in the
presence of 10 ng/ml recombinant human IL-4 and 1,000 U/ml
recombinant human GM-CSF (both from Peprotech, London, UK). Half of
the medium was replaced every 3 days. After 6 days, 10 ng/ml tumor
necrosis factor-α (TNF-α) was added to the culture. On day 10, all
mature DCs were collected, and partly mature DCs
(1×105/group) were loaded with 20 µg/ml peptide at 37°C
in 5% CO2 for 4 h. DCs (1×105/group) loaded
with peptide were cocultured with PBLs (1×106/group)
plated at a 1:10 ratio in 2 ml X–VIVO medium containing 10% FBS in
6-well plates, and 5 ng/ml IL-2, 5 ng/ml IL-15, and 10 ng/ml IL-7
(all from Peprotech) were added after 24 h. Half of the medium was
replaced with media containing fresh cytokines every 3 days. Seven
days later, the CTLs were reticulated with DCs loaded with peptide.
After 3 cycles of reticulation, an ELISPOT (Dakewe, ShenZhen,
China) assay and Calcein-AM release assay for cytotoxicity were
performed.
ELISPOT assay
A human IFN-γ ELISPOT assay kit was used to
determine the function of the CTLs, according to the manufacturer's
instructions. CTLs induced by peptide CD147126–134 and
CTLs induced by CD147126–134L2 were used as the effector
cells. T2 cells loaded with or without peptide were used as target
cells. Effector cells were incubated in duplicate for 18 h at 37 °C
with target in a 96-well ELISPOT plate coated with anti-human IFN-γ
antibody. A positive control (PHA) and a negative control
(HIVpol476–484peptide) were included in all assays.
Biotinylated antibody, streptavidin-enzyme conjugate and the enzyme
substrate nitroblue tetrazolium was added to the plates in order,
followed by a thirty-minute incubation at room temperature. Images
of spots were captured by using a dissection microscope, then
counted using Image Master Total Lab v1.10 software (Amersham
Biosciences, Uppsala, Sweden).
Cytotoxicity calcein-AM release
assay
To measure the cytotoxic response of the CTLs
induced by target cells with different peptides, a calcein AM
(Nippon Chemical Research TongRen Institute, Japan) release-based
cytotoxic assay was performed as described previously. MCF-7,
MCF-7/Adr, Hela, SKOV3, K562 and T2 loaded with or without peptide
were used as target cells. CD147126–134-CTLs and
CD147126–134L2-CTLs were used as the effector cells. An
irrelevant peptide, HIV476–484, was used as a negative
control. T2 cells were loaded with or without peptide for 4 h at
37°C in 5% CO2 and washed thrice. Target cells were
labeled with Calcein-AM for 25 min at 37°C in 5% CO2 and
then calcein-AM-labeled target cells were cocultured with effectors
at different ratios (E:T=10:1, 20:1, 40:1) in 96-well-U-bottomed
plates (Guangzhou Jet Bio-Filtration Co., Ltd., Guangzhou, China).
After incubation for 4 h at 37°C in 5% CO2, cell-free
supernatant was analyzed using a Microplate Reader (Thermo Fisher
Scientific, Inc.) with excitation at 485 nm and emission at 535 nm.
In blocking experiments, T2 cells loaded with peptide or tumor cell
lines were preincubated with 10 µg/ml anti-HLA-A2 antibody (BB7.2:
mouse IgG2a) or isotype control antibody (L243: mouse IgG2a) for 1
h. Each assay was performed in triplicate. The percentage of
specific lysis was determined as: (ODexperimental
release-ODspontaneous release)/(ODmaximal
release-ODspontaneous release) ×100. The labeled
targets in the spontaneous release well were incubated with 2%
Triton X-100 and the labeled targets in the maximum release well
were incubated with medium alone.
Statistical analysis
Statistical analysis was performed using GraphPad
Prism 5 software (GraphPad Software, La Jolla, CA, USA). All
results are expressed as the mean ± SEM and statistical analyses
were performed using the Student's t-test. P<0.05 was considered
to indicate a statistically significant difference and ns, no
statistical significance.
Results
Expression of CD147 in drug-resistant
and drug-sensitive cell lines
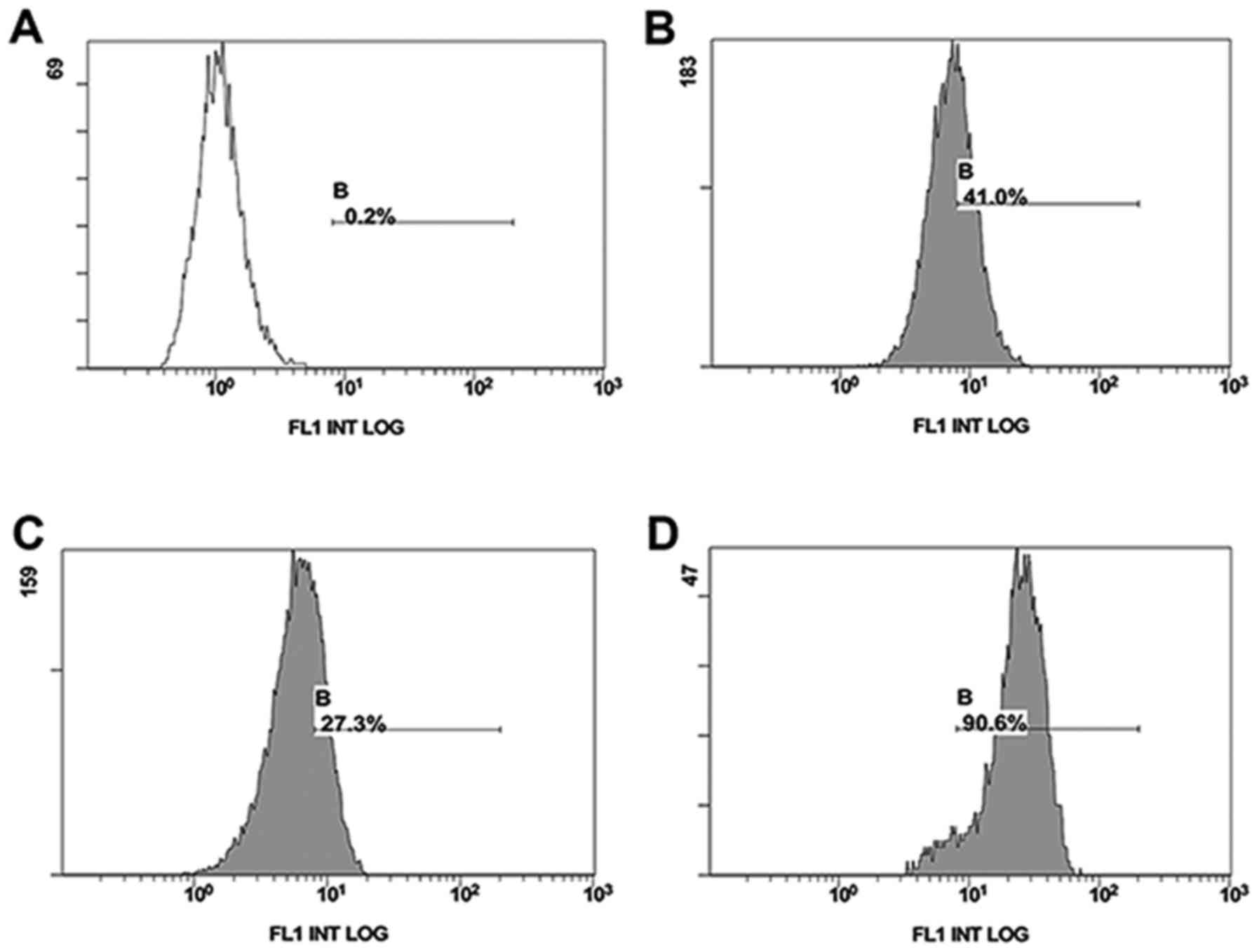
Flow cytometry was used to compare the surface
expression of CD147 on drug-resistant and drug-sensitive cell
lines. drug-resistant cell lines MCF-7/Adr (90.6%) expressed a
higher level than drug-sensitive MCF-7 (27.3%) or Hela
drug-sensitive cell lines (40.0%) (Fig.
1).
Identification of CD147 peptide
candidates
We first screened for a low affinity epitope peptide
from the CD147 protein sequence and position 2 is a hydrophilic
amino acid followed by substitution with a hydrophobic amino acid.
CD147126–134 and CD147126–134L2 peptides were
identified from candidate HLA-A*0201 CD147 epitopes using two
different HLA-peptide-binding prediction programs, BIMAS and
SYFPEITHI. In CD147126–134L2 the Lys(K) at position 2 of
CD147126–134 is substituted with (L)leu. As shown in
Table I, mutated peptide
CD147126–134L2 showed significantly higher binding to
the HLA-A*0201 molecule compared with the wild-type
CD147126–134. Moreover, this binding was even higher
than the positive control peptide, HIVpol476–484, which
was generated from the HIV pol protein and was previously reported
to have high binding affinity for the HLA-A*0201.
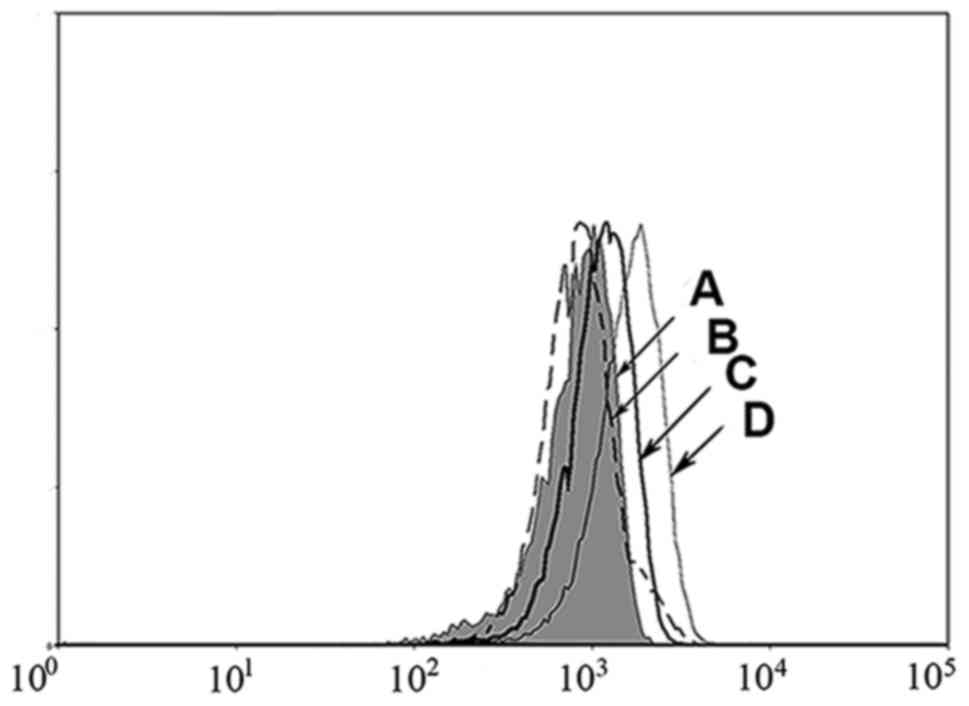
MHC stabilization assay
A T2 cell peptide-binding test was used to evaluate
the binding ability of mutated peptides to HLA-A*0201 molecules.
Because peptide binding to HLA-A2 molecules can increase the
expression of HLA-A*0201 molecules, high affinity peptides can
significantly upregulate HLA-A*0201 compared to low affinity
peptides. As shown in Fig. 2, the
CD147126–134L2 (Fig. 2D)
peptide induced an increase in cell surface HLA-A*0201
stabilization compared to the positive control,
HIVpol476–484 peptide (Fig.
2C). In contrast, the wild-type peptide CD147126–134
(Fig. 2B) showed no increase over
background (T2 cells without peptide) (Fig. 2A). Thus, the high binding score of the
mutated CD147126–134L2 peptide correlates with high
affinity to HLA-A*0201 molecules, as demonstrated by this MHC
stabilization assay. These results suggest that the mutated
CD147126–134L2 peptide may be more immunogenic than the
wild-type CD147126–134 peptide.
CD147 reactive CTLs can lyse
peptide-pulsed T2 target cells
Previous studies have shown that a variety of known
CTL epitopes exhibit high to intermediate affinity binding to HLA
class I molecules and have the capacity to induce peptide-specific
CTL responses. Therefore, to investigate the antigen specificity of
peptide-induced CTLs, we evaluated their ability to secrete IFN-γ
in response to target cells. To this end, T2 cells pulsed with the
mutated CD147126–134L2 or wild-type
CD147126–134 peptide were used as targets in IFN-γ
ELISPOT and cytotoxicity assays.
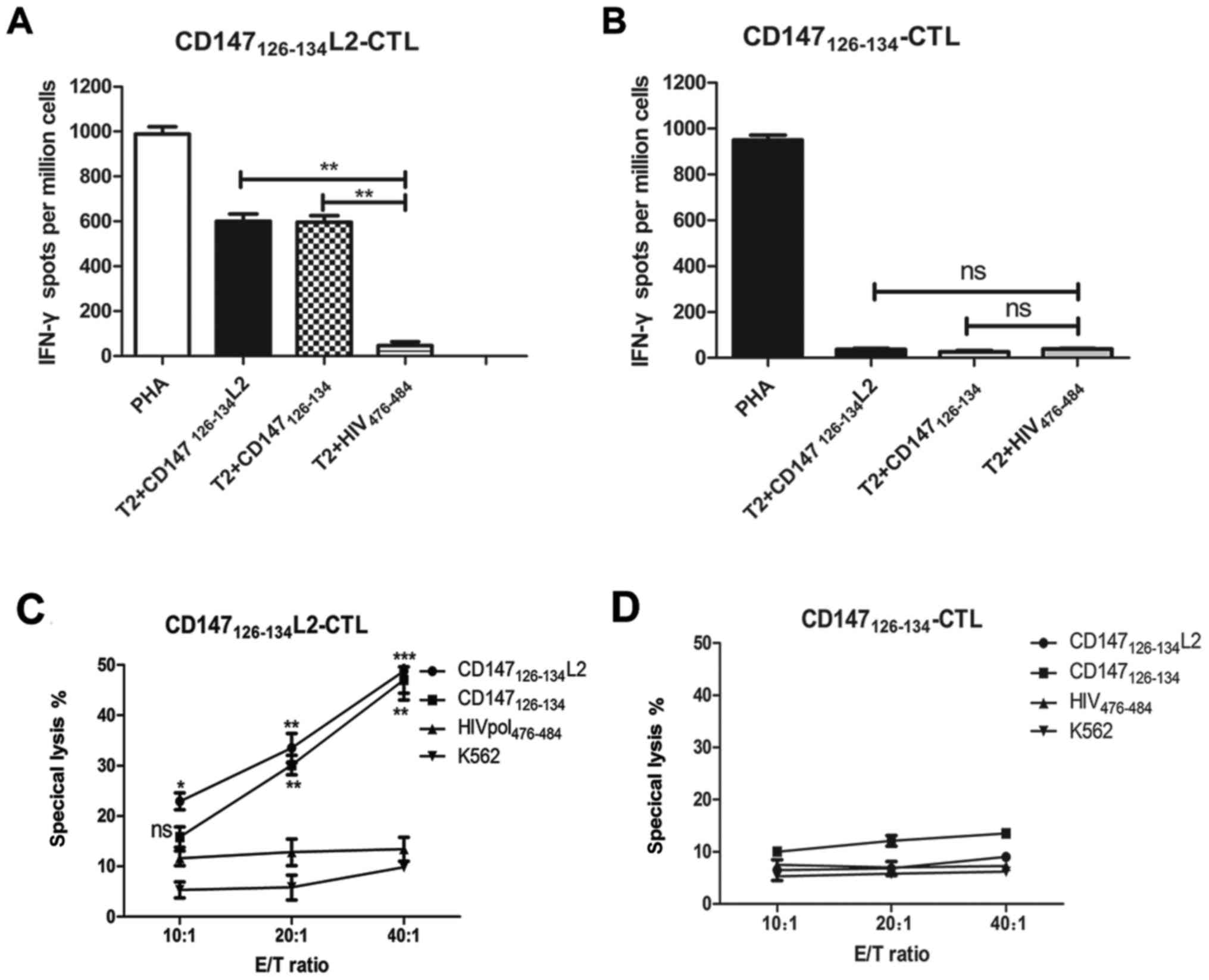
In the IFN-γ ELISPOT assay,
CD147126–134L2 was found to prime significantly more
epitope-specific CTLs than CD147126–134 (Fig. 3). In addition, the frequencies of
IFN-γ producing T cells induced by CD147126–134L2 was
markedly increase compared to the negative control. Importantly,
when T2 cells were loaded with wild-type CD147126–134
peptide, the mutated CD147126–134L2 peptide-induced CTLs
still possessed the capacity for IFN-γ secretion at a level
equivalent to coculturing with T2 cells pulsed with
CD147126–134L2 (Fig. 3A).
In contrast, T2 cells loaded with CD147126–134 elicited
minimal IFN-γ secretion and induced only negligible T-cells
responses (Fig. 3B). Further, T2
cells loaded with wild-type CD147126–134 peptide could
be lysed by the CTLs induced by CD147126–134L2. Also,
CTLs induced by CD147126–134L2 could efficiently lyse
CD147126–134L2 peptide-loaded T2 cells, but did not
irrelevant peptide HIVpol476–484 peptide-loaded T2 cells
at any effector-target ratio (Fig.
3C). In addition, CTLs induced by CD147126–134 only
secrete a small amount of IFN-γ against CD147126–134 or
CD147126–134L2-loaded T2 cells (Fig. 3D). These results demonstrate that the
mutated CD147126–134L2 peptide can elicit CTLs that have
the ability to cross-recognize and specifically lyse T2 cells
loaded with the wild-type CD147126–134 peptide.
CD147 peptide-specific CTLs recognize
CD147 positive MCF-7/ADR cells, but not CD147 negative tumor
cells
We found that CD147126–134L2
peptide-specific CTLs can efficiently recognize wild-type
peptide-pulsed T2 cells and this recognition leads to the
production of IFN-γ. Next, to investigate if these CTLs can lyse
wild-type CD147 peptide naturally presented on tumor cells, we used
the MCF-7 (HLA-A*0201+, CD147low+) and the
MCF-7/Adr (HLA-A*0201+, CD147high+) cell
lines as target cells, and the SKOV3 (HLA-A*0201+,
CD147−) and Hela (HLA-A*0201−,
CD147+) cell lines were included as negative controls.
Target cells were seeded and cocultured with the
CD147126–134L2 peptide-specific CTLs at different
effector to target ratios for 4 h at 37°C in 5% CO2. As
shown in Fig. 4A,
CD147126–134L2-specific CTLs can lyse both MCF-7 and
MCF-7/Adr drug-resistance cell lines, but only minimally lysed the
HLA-A*0201-negative (Hela) and CD147-negative (SKOV3) lines at any
effector to target ratio (Fig. 4A).
In contrast, the cytotoxic effect on the MCF-7/Adr
(HLA-A*0201+, CD147high+) cell line was
dramatically increased (approximately 40.6%) at E:T=40:1 (Fig. 4A). In addition, CTLs induced by
wild-type peptide CD147126–134 showed only a very weak
effect on MCF-7/Adr cells (HLA-A*0201+,
CD147high+) (Fig. 4B).
 | Figure 4.Specific lysis of various tumor cell
lines by CTLs generated against DCs pulsed with different peptides.
A cytotoxic calcein-AM release assay was performed to test for the
cytotoxic activity against various tumor cell lines including MCF-7
(HLA-A*0201+, CD147low+), MCF-7/Adr
(HLA-A*0201+, CD147high+), Hela
(HLA-A*0201−, CD147+), SKOV3
(HLA-A*0201+, CD147−). (A) The cytotoxic
activity of the CTLs induced by the CD147126–134 L2
peptide was assessed against different target cells at various E:T
ratios. (B) wild-type peptide CD147126–134 induced CTLs
were cocultured with target cells at different effector to target
ratios for 4 h to test the cytotoxicity. (C) MCF-7/Adr and T2 cells
pulsed with CD147126–134L2 peptide were treated with 10
µg/ml anti-HLA A2 antibody or isotype control antibody for 1 h. A
calcein-AM release assay was performed to demonstrate the cytotoxic
activity of the effector cells generated from the HLA-A*0201 donor
against these target cells 20:1. Data are represented as mean ± SD.
***P<0.001, compared with isotype control group. CTL, cytotoxic
T lymphocytes; CD, cluster of differentiation; HLA, human leukocyte
antigen. |
These results illustrate two points: i) the
wild-type CD147126–134 peptide can be naturally
processed and presented by tumor cells, and ii) CD147 epitopes
processed and presented on tumors can be cross-recognized and lysed
by CD147126–134L2-specific CTLs. Furthermore, these
experiments indicate that the low level of CD147 expression on
drug-free tumor cells is not easily recognized and lysed by
CD147126–134L2 peptide-specific CTLs. Interestingly,
flow cytometry revealed that the CD147 expression level on
MCF-7/Adr cell lines was higher than that of the MCF-7 cell line,
which may explain the higher sensitivity of these cells to
lysis.
Antibody inhibition assay
To confirm whether the reactivity of
CD147126–134L2 peptide-specific CTLs was restricted by
the HLA-A2, an antibody blocking assay was performed and calcein-AM
release used as a readout. For these experiments, the MCF-7/Adr
cell line and peptide-pulsed T2 cells were used as target cells.
The specific lysis of CD147126–134L2-induced CTLs
incubated with T2 cells loaded with wild-type
CD147126–134 peptide or mutated
CD147126–134L2 peptide was blocked by anti-HLA-A2
antibody, but not by the isotype control antibody, as shown in
Fig. 4C. In addition, when
anti-HLA-A2 antibody was added to the cytolytic assay, the specific
lysis of the MCF-7/Adr drug-resistant cell line by CD147-specific
CTLs dropped below 5% (Fig. 4C).
These results indicate that the CD147126–134L2
peptide-induced CTLs recognize and lyse cells expressing the
mutated peptide or the wild-type peptide both in an
antigen-specific and HLA-A*0201-restricted manner.
Discussion
Chemotherapy plays an important role in treatment of
cancer patients; however, the long-term use of chemotherapeutic
drugs can result in MDR and death. Moreover, there has not been
significant progress toward reducing multidrug resistance-induced
morbidity and mortality despite myriad advances in treatment
options (16,17). The targeting of drug-resistance cells
using cell-based immunotherapy is a relatively new strategy that
shows promise towards overcoming multidrug resistance (18).
CD147 is overexpressed in many MDR cell lines, and
the association between its expression and resistance to
chemotherapeutic drugs has been well established. For example,
Toole and Slomiany (19) found that
the interaction of CD147 with CD44 and hyaluronan can co-regulate
MDR to anticancer drugs. Many approaches have been used to deplete
drug-resistance cells such as use of an anti-CD147 antibody to
inhibit tumor cell proliferation in vivo in a mouse model
(20). However, the limitation with
antibody treatments is that often only a small amount of antibody
can penetrate into the tumor tissue, so that antibody therapy in
the body is less effective than in vitro. The overexpression
of CD147 in chemoresistant cells makes this molecule an ideal
target for cell immunotherapy that specifically targets cells that
survive chemotherapy.
T cells recognizing high affinity, immunodominant
epitopes from self-antigens are deleted in the thymus thereby
leading to immune tolerance. T cells that recognize low affinity
epitopes are difficult to be activated. Great effort has been spent
in recent years to design anchor-modified peptides in order to
overcome the failure of activation of T cells that recognize low
affinity epitopes (21). Engels et
al demonstrated that the affinity of peptides and MHC molecules
is particularly critical for peptide cross-presentation and
induction of cytokine production in vivo (22). Thus, peptides that exhibit higher
affinity for MHC molecules may create a peptide-MHC complex which
can interact more efficiently with the peptide-specific TCR
(23). In this study, flow cytometric
analysis revealed that CD147 is overexpressed on drug-resistance
cells, which is consistent with other research. Therefore, we
screened the CD147 protein sequence to identify a low-binding score
peptide using HLA-peptide-binding prediction software and
identified CD147126–134. We then replaced the primary
anchor residue, Lys(K), in position 2 with leu (L), resulting in a
peptide with a very high binding score (CD147126–134L2).
Moreover, the T2 affinity assay clearly showed that
CD147126–134L2 has strong binding capacity compared with
the positive control (HIVpol476–484) and wild-type
CD147126–134 peptide.
In vitro priming and expansion of the CD147
peptide-specific CTLs was clearly shown by IFN-γ Elispot. These
studies also showed that the CD147126–134L2
peptide-specific CTLs secrete markedly more IFN-γ in response to T2
cells loaded with CD147126–134L2 than with
CD147126–134. Moreover, the
CD147126–134L2-stimulated CTLs cocultured with
CD147126–134 loaded T2 cells also showed a similar level
of IFN-γ secretion. Cytotoxicity assays were performed by
coculturing the CD147126–134L2 or
CD147126–134 peptide-primed CTLs with peptide-pulsed T2
target cells. The results showed that CTLs induced by
CD147126–134L2 can not only lyse T2 cells loaded with
CD147126–134L2, but also those loaded with wild-type
CD147126–134 peptide. In contrast, the CTLs induced by
CD147126–134 showed a very weak cytotoxicity to the
CD147126–134 or CD147126–134L2 peptide loaded
T2 cells. Although there is a single amino acid difference between
the mutated and original peptides, CTLs induced by the mutated
peptide can cross-recognize wild-type peptide, as was verified by
our T2 target cell experiment.
Next we used tumor cells as target cells to verify
our hypothesis, and found that HLA-A2 positive MCF-7/Adr cells,
which highly express CD147, can be specifically recognized and
lysed by the CTLs induced by CD147126–134L2. In
contrast, Hela cells (HLAA2−, CD147+) and
SKOV3 cells (HLAA2+, CD147−) could not be
lysed by the CTLs induced by CD147126–134L2, and the
cytotoxic effect was blocked by HLA-A2 antibody. This demonstrates
that the wild-type CD147126–134 peptide is endogenously
processed and presented by MCF-7/Adr cells and that the cytotoxic
effect occurs in an HLA-A2-restricted manner. Thus, high affinity
peptides such as the mutant peptide in this study can bind to MHC
complexes with longer half-lives resulting in more efficient T cell
activation. Once activated, T cells are then able to recognize the
wild-type antigen peptide on the target cell, including antigens on
cancer cells (24).
In conclusion, we identified a novel
HLA-A*0201-restricted peptide (CD147126–134L2) and
showed that specific CTLs can be elicited by priming T cells with
DCs pulsed with this peptide. Moreover, these CTLs are able to
specifically and effectively lyse HLA-A2 positive MCF-7/Adr
drug-resistant cells which highly express CD147. Therefore,
targeting of CTLs against CD147 show promise as an immunotherapy
aimed at eliminating drug-resistant cancer cells.
Acknowledgements
This project was supported by the National Natural
Science Foundation of China (grant nos. 31300737 and 31400149), the
Scientific and Technological Project of Guangdong Province (grant
nos.2014A020212311 and 2016A020215157), and a grant from the
Natural Science Foundation of Guangdong Province (grant nos.
2014A030313586 and 2015A030310310).
References
|
1
|
Amiri-Kordestani L, Basseville A, Kurdziel
K, Fojo AT and Bates SE: Targeting MDR in breast and lung cancer:
Discriminating its potential importance from the failure of drug
resistance reversal studies. Drug Resist Updat. 15:50–61. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Muramatsu T: Basigin (CD147), a
multifunctional transmembrane glycoprotein with various binding
partners. J Biochem. 159:481–490. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Somno A, Anuchapreeda S, Chruewkamlow N,
Pata S, Kasinrerk W and Chiampanichayakul S: Involvement of CD147
on multidrug resistance through the regulation of P-glycoprotein
expression in K562/ADR leukemic cell line. Leuk Res Rep. 6:33–38.
2016.PubMed/NCBI
|
|
4
|
Zhou S, Liao L, Chen C, Zeng W, Liu S, Su
J, Zhao S, Chen M, Kuang Y, Chen X and Li J: CD147 mediates
chemoresistance in breast cancer via ABCG2 by affecting its
cellular localization and dimerization. Cancer Lett. 337:285–292.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pan Y, He B, Song G, Bao Q, Tang Z, Tian F
and Wang S: CD147 silencing via RNA interference reduces tumor cell
invasion, metastasis and increases chemosensitivity in pancreatic
cancer cells. Oncol Rep. 27:2003–2009. 2012.PubMed/NCBI
|
|
6
|
Xu BQ, Fu ZG, Meng Y, Wu XQ, Wu B, Xu L,
Jiang JL, Li L and Chen ZN: Gemcitabine enhances cell invasion via
activating HAb18G/CD147-EGFR-pSTAT3 signaling. Oncotarget.
7:62177–62193. 2016.PubMed/NCBI
|
|
7
|
Kono K, Iinuma H, Akutsu Y, Tanaka H,
Hayashi N, Uchikado Y, Noguchi T, Fujii H, Okinaka K, Fukushima R,
et al: Multicenter, phase II clinical trial of cancer vaccination
for advanced esophageal cancer with three peptides derived from
novel cancer-testis antigens. J Transl Med. 10:1412012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Suzuki H, Fukuhara M, Yamaura T, Mutoh S,
Okabe N, Yaginuma H, Hasegawa T, Yonechi A, Osugi J, Hoshino M, et
al: Multiple therapeutic peptide vaccines consisting of combined
novel cancer testis antigens and anti-angiogenic peptides for
patients with non-small cell lung cancer. J Transl Med. 11:972013.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tsang KY, Fantini M, Fernando RI, Palena
C, David JM, Hodge JW, Gabitzsch ES, Jones FR and Schlom J:
Identification and characterization of enhancer agonist human
cytotoxic T-cell epitopes of the human papillomavirus type 16
(HPV16) E6/E7. Vaccine. 35:2605–2611. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Colella TA, Bullock TN, Russell LB,
Mullins DW, Overwijk WW, Luckey CJ, Pierce RA, Restifo NP and
Engelhard VH: Self-tolerance to the murine homologue of a
tyrosinase-derived melanoma antigen: Implications for tumor
immunotherapy. J Exp Med. 191:1221–1232. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shen H, Shao HW, Chen XH, Wu FL, Wang H,
Huang ZL, Shen J, Wang T, Zhang WF and Huang SL: Identification of
a novel HLA-A2-restricted mutated Survivin epitope and induction of
specific anti-HCC CTLs that could effectively cross-recognize
wild-type Survivin antigen. Cancer Immunol Immunother. 62:393–403.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shao H, Lin Y, Wang T, Ou Y, Shen H, Tao
C, Wu F, Zhang W, Bo H, Wang H and Huang S: Identification of
peptide-specific TCR genes by in vitro peptide stimulation and CDR3
length polymorphism analysis. Cancer Lett. 363:83–91. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang W, Zou L, Zhou D, Zhou Z, Tang F, Xu
Z and Liu X: Overexpression of ubiquitin carboxyl terminal
hydrolase-L1 enhances multidrug resistance and invasion/metastasis
in breast cancer by activating the MAPK/Erk signaling pathway. Mol
Carcinog. 55:1329–1342. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Rammensee HG, Bachmann J, Emmerich NP,
Bachor OA and Stevanović S: SYFPEITHI: Database for MHC ligands and
peptide motifs. Immunogenetics. 50:213–219. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nijman HW, Houbiers JG, Vierboom MP, van
der Burg SH, Drijfhout JW, D'Amaro J, Kenemans P, Melief CJ and
Kast WM: Identification of peptide sequences that potentially
trigger HLA-A2.1-restricted cytotoxic T lymphocytes. Eur J Immunol.
23:1215–1219. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wijdeven RH, Pang B, Assaraf YG and
Neefjes J: Old drugs, novel ways out: Drug resistance toward
cytotoxic chemotherapeutics. Drug Resist Updat. 28:65–81. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chang A: Chemotherapy, chemoresistance and
the changing treatment landscape for NSCLC. Lung Cancer. 71:3–10.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Curiel TJ: Immunotherapy: A useful
strategy to help combat multidrug resistance. Drug Resist Updat.
15:106–113. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Toole BP and Slomiany MG: Hyaluronan, CD44
and Emmprin: Partners in cancer cell chemoresistance. Drug Resist
Updat. 11:110–121. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Walter M, Simanovich E, Brod V, Lahat N,
Bitterman H and Rahat MA: An epitope-specific novel anti-EMMPRIN
polyclonal antibody inhibits tumor progression. Oncoimmunology.
5:e10780562015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tourdot S, Scardino A, Saloustrou E, Gross
DA, Pascolo S, Cordopatis P, Lemonnier FA and Kosmatopoulos K: A
general strategy to enhance immunogenicity of low-affinity
HLA-A2.1-associated peptides: Implication in the identification of
cryptic tumor epitopes. Eur J Immunol. 30:3411–3421. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Engels B, Engelhard VH, Sidney J, Sette A,
Binder DC, Liu RB, Kranz DM, Meredith SC, Rowley DA and Schreiber
H: Relapse or eradication of cancer is predicted by peptide-major
histocompatibility complex affinity. Cancer Cell. 23:516–526. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Valmori D, Fonteneau JF, Lizana CM,
Gervois N, Liénard D, Rimoldi D, Jongeneel V, Jotereau F, Cerottini
JC and Romero P: enhanced generation of specific tumor-reactive ctl
in vitro by selected melan-a/mart-1 immunodominant peptide
analogues. J Immunol. 160:1750–1758. 1998.PubMed/NCBI
|
|
24
|
Slansky JE, Rattis FM, Boyd LF, Fahmy T,
Jaffee EM, Schneck JP, Margulies DH and Pardoll DM: Enhanced
antigen-specific antitumor immunity with altered peptide ligands
that stabilize the MHC-peptide-TCR complex. Immunity. 13:529–538.
2000. View Article : Google Scholar : PubMed/NCBI
|