Introduction
Lung cancer is the leading cause of
cancer-associated mortalities in China and globally (1–3). A number
of studies have indicated that 85–90% of these cases are a result
of voluntary or involuntary (second-hand) smoking (4–7), with a
notable dose-response association (8). Furthermore, numerous studies have
demonstrated that inhalation of toxicants, including cigarette
smoke, may result in irreversible changes to genetic material,
including DNA mutations and putatively reversible changes to the
epigenetic landscape, and changes in DNA methylation and the
chromatin modification state (9–11).
The tumorigenesis of lung cancer involves genetic
and epigenetic alterations, including DNA methylation and histone
modifications. One of the most common epigenetic regulations is
cytosine methylation, which affects the primary structure of
chromatin and its secondary structure, which is critical to the
regulation of gene expression (12),
as well as the interactions within DNA and histones. Recently,
aberrant methylation of CpG islands in the promoter region of tumor
suppressor genes (TSGs) has been identified as an important
epigenetic mechanism for gene silencing, which may have a limited
effect on tumor proliferation and cellular apoptosis for global
hypomethylation of genomic DNA and the hypermethylation of gene
promoter regions occur simultaneously in a wide variety of
malignancies, including lung cancer (8). The aim of the present study was to
evaluate the growth and apoptotic index of non-small cell lung
cancer (NSCLC) with different smoking statuses.
Histone modification, particularly methylation of
lysine may be associated with the transcriptional status of genes
and chromatin structure (13).
Trimethylated histone H3 at lysine 27, a transcription-suppressive
histone modification, is methylated via the enhancer of zeste
homolog 2 (EZH2). EZH2, the catalytic subunit of polycomb
repressive complex 2 (PRC2), contributes to the maintenance of cell
identity, cell cycle regulation and tumorigenesis (14,15).
Overexpression of the EZH2 gene occurs in a variety of human
malignancies, including breast, prostate, endometrial, gastric,
colon, hepatocellular, bladder and oral cancer (16). Recently, a number of studies have
reported that trimethylation of histone H3 at lysine 27 (H3K27me3)
serves a significant role in the development and/or progression of
various human cancer types, including lung cancer (13,17).
Theoretically, as the methyltransferase for H3K27me3, the
expression of EZH2 should change simultaneously with the expression
of H3K27me3; however, a previous study (17) revealed that the association in the
expression levels did not match this theory. The underlying
mechanism of this paradox remains unknown. It was theorized that
cigarette smoking, which frequently results in epigenetic
alterations, may have a limited effect on this phenomenon. To date,
the expression dynamics of H3K27me3 in lung cancer with different
smoking statuses have not been investigated (13).
In the present study, the major gene involved in
inactivating histone modification was investigated, including the
expression of H3K27me3, and the DNA methylation status at CCGG
sites in smoking and non-smoking patients with NSCLC, along with
the possible clinical significance.
Materials and methods
Patients and tissue preparation
Archived formalin-fixed paraffin-embedded tissue
sections from a series of 42 patients [22 adenocarcinomas (ADC) and
20 squamous cell carcinomas (SCC); 27 males and 15 females; age
range, 39–87 years] who had undergone surgical resection for NSCLC
at the First Department of Surgery, Nagasaki University Hospital
(Nagasaki, Nagasaki) between February 2000 and November 2006 were
selected. None had received chemo- or radiotherapy prior to tissue
collection. The histopathological features of cancerous specimens
were classified in accordance with the 8th American Joint Committee
on Cancer criteria on lung cancer, and the TNM staging system
(18). The study protocol was
approved by the Human Ethics Review Committee of Nagasaki
University School of Medicine and signed informed consent was
obtained from each patient. Serial sections were cut at a thickness
of 4 µm and placed onto 3-aminopropyltriethoxysilane-coated glass
slides. A number of sections were stained with hematoxylin and
eosin (H&E) in a routine manner for histological examination
(19).
Chemicals and biochemicals
Bovine serum albumin (BSA; essentially fatty acid
and globulin-free), Trizma base, 2-mercaptoethanol,
3-aminopropyltriethoxysilane and Brij-35 were obtained from
Sigma-Aldrich (Merck KGaA, Darmstadt, Germany);
3,3′-Diaminobenzidine-4HCl (DAB) was purchased from Chemical Dojin
Co., Ltd. (Kumamoto, Japan). Biotin-16-dUTP, digoxigenin-11-dUTP,
Rhodamine anti-Dig and terminal deoxynucleotidyltransferase (TdT)
were from Roche Diagnostics (Mannheim, Germany). Dideoxy ATP
(ddATP) and dideoxy TTP (ddTTP) were from Jena Bioscience (Jena,
Germany). HpaII and MspI were purchased from Takara
Bio, Inc. (Otsu, Japan). DAPI was obtained from Invitrogen (Thermo
Fisher Scientific, Inc., Waltham, MA, USA). Permount was from
Thermo Fisher Scientific, Inc. All other reagents used in the
present study were obtained from Wako Pure Chemicals Industries,
Inc. (Osaka, Japan) and were of analytical grade.
Immunohistochemistry (IHC)
IHC was performed with the indirect enzyme-labeled
antibody method, as described previously (17,20–22). All
experimental procedures were performed in room temperature unless
otherwise indicated. IHC using anti-H3K27me3 polyclonal (1:200;
cat. no. 9733) antibody, anti-EZH2 monoclonal (1:400; cat. no.
5246) antibody (both from Cell Signaling Technology, Inc., Danvers,
MA, USA) and anti-PCNA monoclonal (PC10; 1:400, cat. no. M0879)
antibody (DakoCytomation; Agilent Technologies, Inc., Santa Clara,
CA, USA) was performed according to the manufacturer's protocol.
For detection, paraffin-embedded sections were deparaffinized with
toluene and rehydrated in graded alcohol. Following autoclaving for
15 min at 120°C in 10 mM citrate buffer (pH 6.0) for antigen
retrieval, endogenous peroxidase was inactivated with 0.3% hydrogen
peroxide in methanol for 15 min. The sections were then
pre-incubated with 500 µg/ml normal goat immunoglobulin G (IgG;
cat. no. I-5256, Sigma-Aldrich; Merck KGaA) dissolved in 1% BSA in
PBS (pH 7.4) for 1 h, reacted with primary antibodies for 16 h,
washed with 0.075% Brij™-35 in PBS, and then incubated
with horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG
(H3K27me3/EZH2, 1:200) or HRP-conjugated goat anti-mouse IgG (PCNA)
in 1% BSA in PBS for 1 h (cat. no. AP307P; EMD Millipore,
Billerica, MA, USA). Following washing with 0.075%
Brij™-35 in PBS, the sites of HRP were visualized with
DAB and H2O2 in the presence of nickel, and
cobalt ions (23). As a negative
control, a number of sections were reacted with normal rabbit IgG
or normal mouse IgG (1:500; cat. nos. X-0903 and X-0931,
respectively, both Dako Cytomation; Agilent Technologies, Inc.)
instead of the specific antibodies.
Quantitative evaluation
Staining results were examined by two pathologists
(D. Hu and W. Jiang) blinded to the clinical information of
patients. Another reading by a third observer was needed to reach a
consensus when there was a significant discrepancy between initial
readings. At least five high-power fields and >2,000 cells were
analyzed in each case with a Zeiss 2021-85 light microscope (Carl
Zeiss AG, Oberkochen, Germany) at ×400 magnification.
Immunostaining results were evaluated via a semi-quantitative
scoring system as previously described (24), and the final score ranged from
0–12.
TdT-mediated dUTP nick end labeling
(TUNEL) staining for apoptotic cancerous cells in NSCLC with
different smoking statuses
To identify nuclei with DNA strand breaks at a
cellular level, TUNEL was performed according to the method by
Gavrieli et al (25). In
brief, paraffin sections (5 µm) were cut onto silane-coated glass
slides, dewaxed with toluene and rehydrated in an ethanol series.
Following washing with PBS, the sections were treated with 5 µg/ml
and of proteinase K (Takara Bio, Inc.) in PBS at 37°C for 15 min.
The sections were then washed once with deionized distilled water
and incubated with TdT buffer (25 mM Tris/HCl buffer, pH 6.6,
containing 0.2 M potassium cacodylate and 0.25 mg/ml BSA) alone at
room temperature for 30 min. Following incubation, the slides were
reacted with 200 U/ml TdT dissolved in TdT buffer, supplemented
with 5 µM biotin-16-dUTP, 20 µM dATP, 1.5 mM CoCl2 and
0.1 mM dithiothreitol, at 37°C for 1 h. The reaction was terminated
by washing with 50 mM Tris/HCl buffer (pH 7.4) for 15 min.
Endogenous peroxidase activity was inhibited by immersing the
slides in 0.3% H2O2 in methanol at room
temperature for 15 min. The signals were detected
immunohistochemically with a HRP-conjugated goat anti-biotin
antibody (1:100, cat. no. AP132B; EMD Millipore), as described
previously (17,22). For statistical analysis, >10,000
cancer cells/patient were counted, and the number of TUNEL-positive
cells was expressed per 1,000 of the total cells (mean ± standard
error of the mean). Data for different groups were compared for
statistical differences using the Student's t-test. P<0.05 was
considered to indicate a statistically significant result.
In situ evaluation of DNA
methylation
To evaluate the DNA methylation level of NSCLC at
the CCGG sites, histoendonuclease-linked detection of methylation
sites of DNA (HELMET) was performed (17,26).
Paraffin sections were dewaxed and digested with 10 µg/ml
proteinase K in PBS at 37°C for 15 min. Then the sections were
incubated with TdT buffer alone at room temperature for 30 min.
Following incubation, the slides were reacted with 800 U/ml TdT
dissolved in TdT buffer containing 20 µM ddATP, 20 µM ddTTP, 1.5 mM
CoCl2 and 0.1 mM dithiothreitol at 37°C for 2 h.
Following washing with PBS, the sections were fixed with
freshly-prepared 4% paraformaldehyde (PFA) in PBS for 5 min at room
temperature and then rinsed with PBS. The non-methylated CCGG sites
were digested at 37°C for 2 h with 100 U/ml HpaII dissolved
in 10 mM Tris/HCl buffer (pH 7.5), containing 10 mM
MgCl2 and 1 mM dithiothreitol. The HpaII-cut
sites were labeled with biotin-16-dUTP via TdT reaction for 90 min
at 37°C. Then, the 3′-OH ends were blocked with a mixture of
dideoxynucleotides by TdT at 37°C as aforementioned, for 2 h.
Following fixation with 4% PFA in PBS, the methylated CCGG sites
were digested at 37°C for 2 h by 100 U/ml MspI dissolved in
Tris/HCl buffer (pH 7.9), containing 10 mM MgCl2, 0.5 mM
dithiothreitol, 66 mM potassium acetate and 0.1% BSA. The
MspI-cut sites were then labeled with digoxigenin-11-dUTP
via TdT reaction for 90 min at 37°C. Finally, the sections were
incubated with a mixture of 500 µg/ml normal goat IgG and normal
sheep IgG (cat. no. B3148; Sigma-Aldrich; Merck KGaA) in 5% BSA/PBS
for 1 h at 37°C, and then visualized by fluorescein
isothiocyanate-labeled goat anti-biotin and rhodamine-labeled sheep
anti-digoxigenin (both 1:100, cat. no. SP-3040; Vector
Laboratories, Ltd., Peterborough, UK; and cat. no. 11082736103 from
Roche Diagnostics) for 1 min. The nuclei were stained with 0.5
µg/ml DAPI for 1 min at room temperature. The stained slides were
analyzed under a laser scanning microscope (×400 magnification, LSM
5 PASCAL; Zeiss GmbH, Jena, Germany).
Statistical analysis
The X-tile software program version 3.6.1 (Yale
University School of Medicine, New Haven, CT, USA), described
previously (27), was used to
determine the threshold value of H3K27me3 for classifying samples
into groups of high and low expression. The SPSS statistical
software package (version 23; IBM Corp., Armonk, NY, USA) was
employed for all analyses. The association between markers, and the
clinicopathological characteristics of the patients, either
separated or combined, including age, sex, tissue type, tumor
differentiation, P-factor, LV-factor, V-factor, smoking status,
Brinkman index, CCGG methylation ratio, relapse, postoperative
metastasis and carcinoembryonic antigen (CEA) level, were evaluated
by the Pearson's χ2 and Fisher's exact tests, or
Spearman's rank correlation as appropriate. P<0.05 was
considered to indicate a statistically significant difference.
Results
Clinicopathological data of
patients
The clinicopathological data of the patients with
cancer are depicted in Table I. The
cohort included 27 males and 15 females, with a mean age of 69
years. By histological classification, 20 cases were SCC and 22
were ADE. In the SCC group, the number of well-, moderately- and
poorly-differentiated cancer types were 3, 9, and 8, respectively.
In the ADE group, the number of cancer types with predominant
growth patterns for lepidic, acinar, papillary and solid with mucin
were 7, 6, 4 and 5, respectively. All cases were TNM stage 1 and
lymph node-negative. Postoperative follow-up data were available in
all cases, and the median follow-up duration in the ADC and SCC
groups were 75.2, and 52.9 months, respectively.
 | Table I.Clinicopathological data of
patients. |
Table I.
Clinicopathological data of
patients.
|
| No. of cases
(%) |
|---|
|
|
|
|---|
| Parameter | ADC | SCC |
|---|
| Median age,
years | 68.50 | 69.75 |
| Age, years |
|
|
|
≤69 | 12 (54.5) | 12 (60) |
|
>69 | 10 (45.5) | 8 (40) |
| Sex |
|
|
|
Male | 10 (45.5) | 17 (85) |
|
Female | 12 (54.5) | 3 (15) |
| Serum CEA,
ng/ml |
|
|
| ≤5 | 22 (100) | 17 (85) |
|
>5 | 0 | 3 (15) |
| P-factor |
|
|
|
Positive | 1 (4.5) | 0 |
|
Negative | 21 (95.5) | 20 (100) |
| LV-factor |
|
|
|
Positive | 18 (81.8) | 13 (65) |
|
Negative | 4 (18.2) | 7 (35) |
| V-factor |
|
|
|
Positive | 8 (36.4) | 10 (50) |
|
Negative | 14 (63.6) | 10 (50) |
| T-stage |
|
|
| 1a | 16 (72.7) | 13 (65) |
| 1b | 5 (22.7) | 7 (35) |
| 2a | 1 (4.6) |
|
| Nodal status |
|
|
| N0 | 22 (100) | 20 (100) |
| Differentiation
(SCC) |
|
|
|
Well |
| 3 (15) |
|
Moderate |
| 9 (45) |
|
Poor |
| 8 (40) |
| Predominant growth
pattern (ADC) |
|
|
|
Lepidic | 7 (31.8) |
|
|
Acinar | 6 (27.3) |
|
|
Papillary | 4 (18.2) |
|
| Solid
with mucin | 5 (22.7) |
|
| Relapse |
|
|
|
Yes | 2 (9.1) | 8 (40) |
| No | 20 (90.9) | 12 (60) |
| Smoking status |
|
|
|
Smoker | 7 (31.8) | 20 (100) |
|
Non-smoker | 15 (68.2) | 0 (0) |
| Postoperative
metastasis |
|
|
|
Yes | 3 (13.6) | 7 (35) |
| No | 19 (86.4) | 13 (65) |
| Median follow-up,
months | 75.2 | 52.9 |
As depicted in Table
I, among the 42 patients with lung cancer, the majority of the
smokers were male (21/27), whereas the majority of females (12/15)
did not smoke. SCC was predominant in smokers (20/20), whereas ADE
was the main histological type in non-smokers (15/22); therefore,
smoking resulted in a ≥2-fold higher risk of SCC than ADE
(20:7).
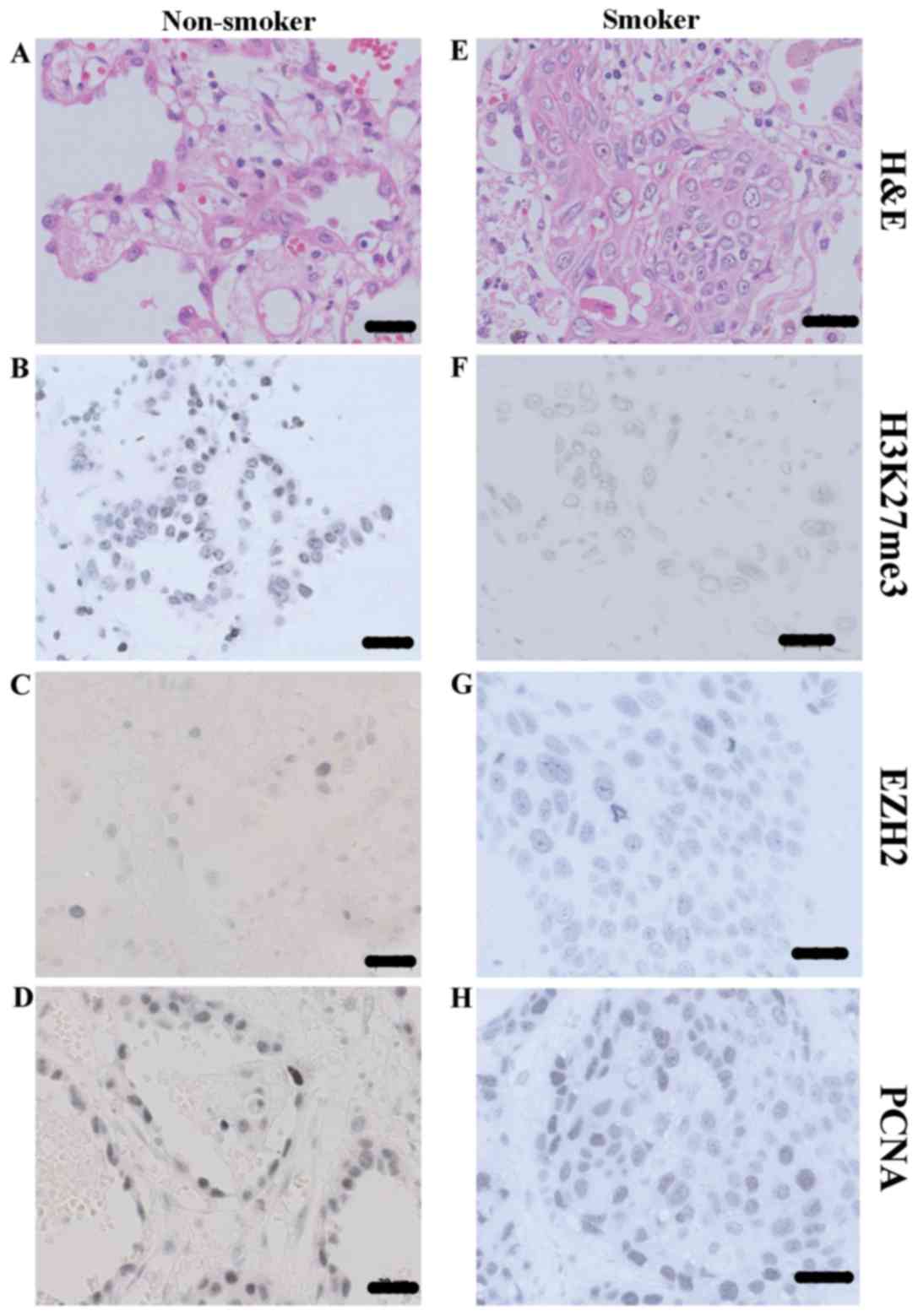
Expression of H3K27me3, EZH2 and PCNA
in NSCLC tissues of smoking and non-smoking patients
The NSCLC tissues from smokers and non-smokers were
stained using various antibodies in order to determine the
expression H3K27me3, EZH2, and PCNA (Fig.
1). H&E staining of NSCLC tissue demonstrated the normal
status of histological status used in the present study (Fig. 1A and E). H3K27me3, EZH2 and PCNA were
all predominantly localized in the nuclei in NSCLC (Fig. 1B-D and F-H). The calculated staining
score of immunopositive cells for H3K27me3 and EZH2 ranged from
0–12 in all tested tissues, whereas PCNA staining was expressed as
a percentage of immunopositive cells. According to the X-tile plots
(data not shown), the samples were categorized into low (IHC score
≤3) and high (IHC score >3) expression subgroups for H3K27me3,
based on a cutoff point determined by X-tile software associated
with survival status. As for EZH2 and PCNA, the average score was
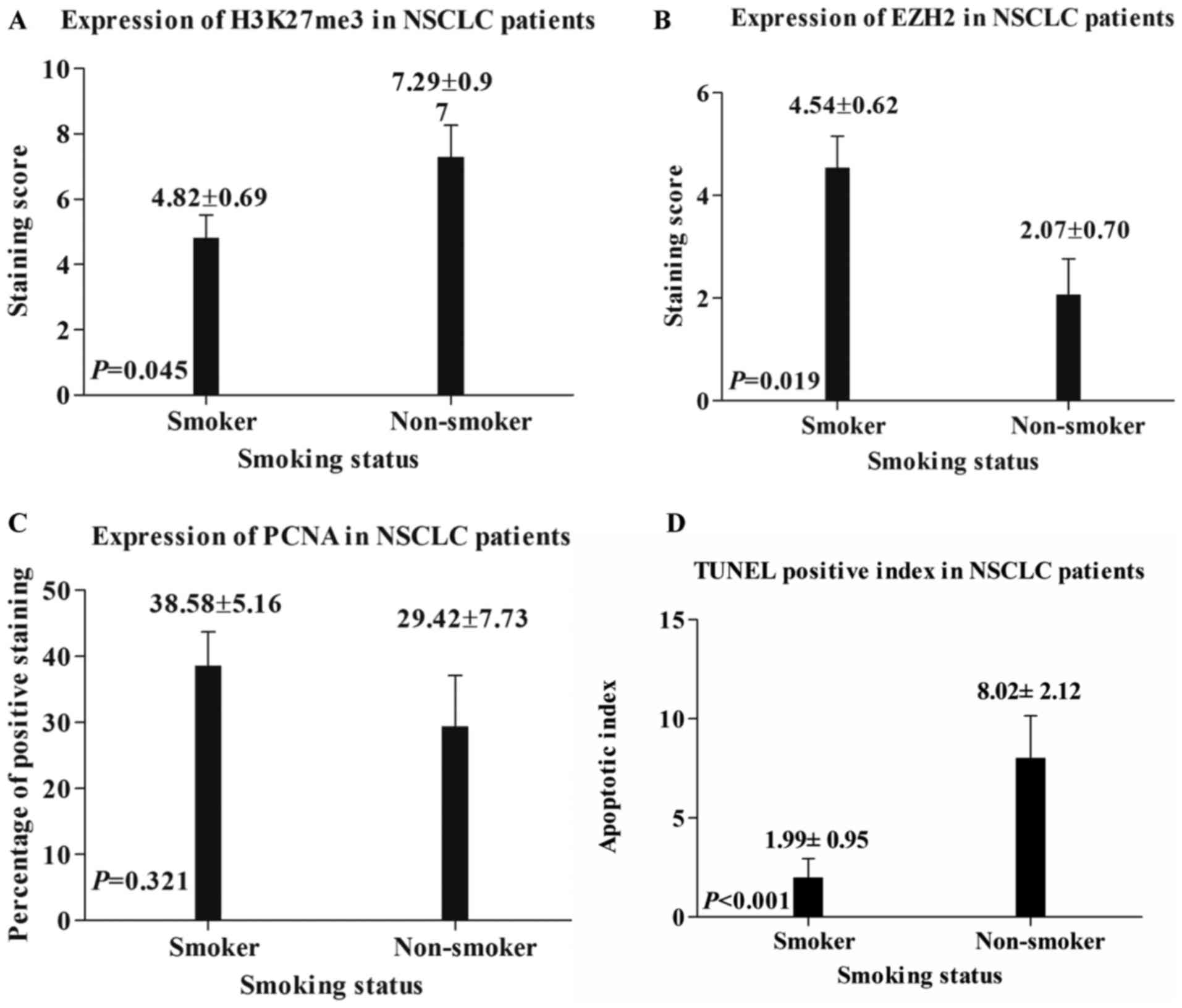
used to evaluate their expression level. As depicted in Figs. 1B, F and 2A, the staining score of H3K27me3 was
significantly higher in non-smoker NSCLC tissue, compared within
those of smokers (7.29±0.97 vs. 4.82±0.69; P=0.045); however, the
staining score of EZH2 was significantly higher in smoker NSCLC
tissue, compared within those of non-smokers (4.54±0.62 vs.
2.07±0.70; P=0.019; Figs. 1C, G and
2B). No statistical significance was
determined between the PCNA expression in smoker and non-smoker
NSCLC tissues (38.58±5.16 vs. 29.42±7.73; P=0.321; Figs. 1D, H and 2C).
Apoptotic evaluation in patients with
NSCLC with different smoking status
In comparison to smoking patients with NSCLC, a
higher apoptotic index was observed in non-smokers (8.02±2.12 vs.
1.99±0.95; P<0.001; Fig. 2D).
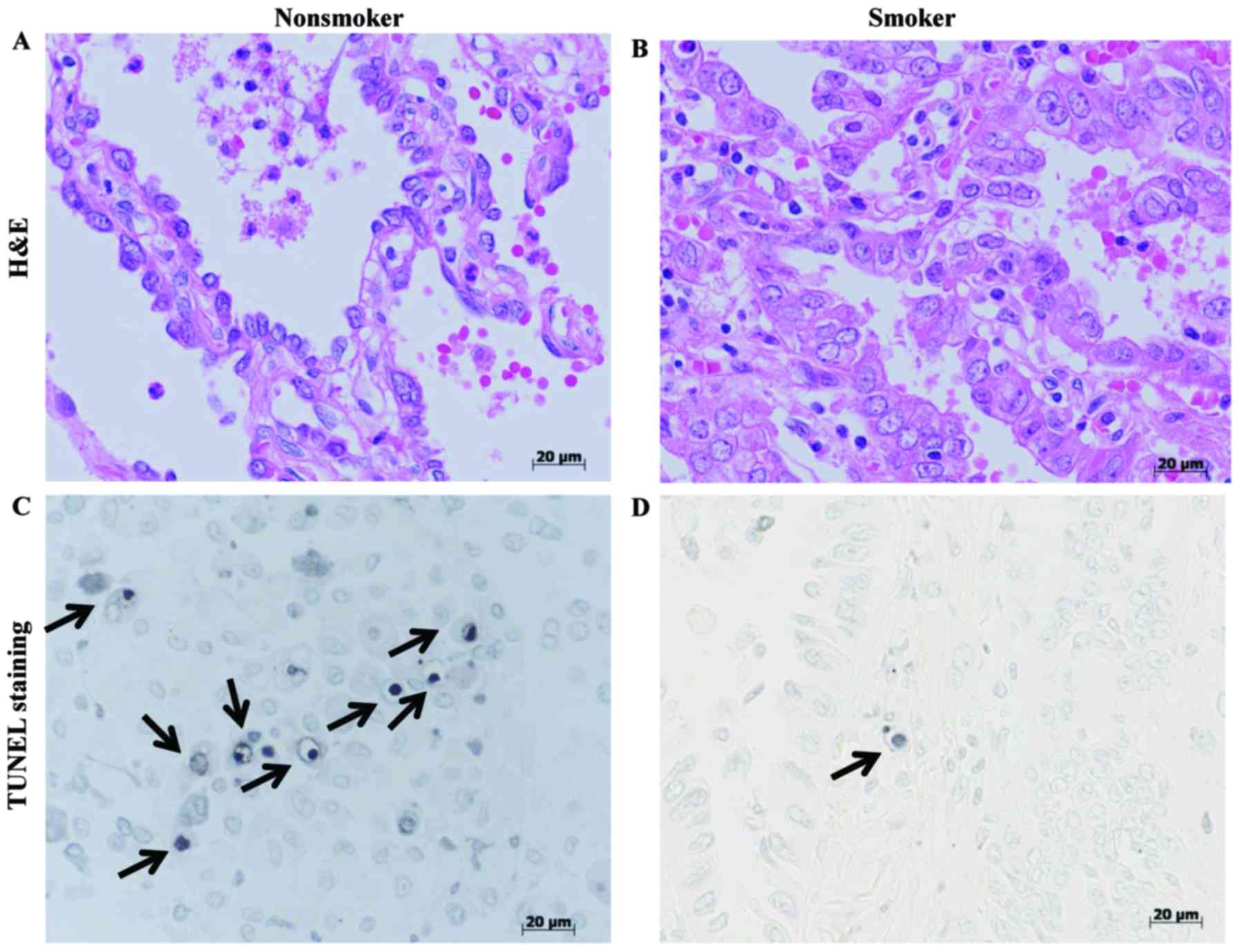
Fig. 3A and B demonstrated the normal
status of tissue used for TUNEL evaluation. In each case, cells
with the morphological characteristics of apoptosis were identified
by: Chromatin condensation; the formation of crescentic caps of
condensed chromatin at the nuclear periphery; and the formation of
visible apoptotic bodies (arrows in Fig.
3C and D).
DNA methylation level at CCGG sites
and its association with clinicopathological parameters
As depicted in Table
II, CCGG sites in non-smokers with NSCLC were generally
hypermethylated, compared with those of smokers (P=0.016).
Furthermore, due to the non-normal distribution of the Brinkman
index and the methylation ratio at CCGG sites, Spearman's rank
correlation analysis (data not shown) was used to determine the
correlation, and a notable statistical significance was determined
(P=0.017).
 | Table II.Significance and correlations of
clinicopathological data in patients with NSCLC with different
smoking status. |
Table II.
Significance and correlations of
clinicopathological data in patients with NSCLC with different
smoking status.
|
| Smoking status |
|
|---|
|
|
|
|
|---|
| Variable | Smokers | Non-smokers | P-value |
|---|
| H3K27me3
expressionb,c |
|
| 0.029 |
|
High | 10 | 14 |
|
|
Low | 14 | 4 |
|
| CCGG methylation
ratioa |
|
| 0.016 |
|
High | 7 | 12 |
|
|
Low | 17 | 6 |
|
| EZH2
expressiona,b |
|
| 0.029 |
|
High | 14 | 4 |
|
|
Low | 10 | 14 |
|
| Sexb |
|
| 0.001 |
|
Male | 21 | 6 |
|
|
Female | 3 | 12 |
|
| Histology
typeb |
|
| 0.001 |
|
SCC | 17 | 3 |
|
|
ADC | 7 | 15 |
|
| Trend of
H3K27me3c and EZH2
expressiona,b |
|
| 0.015 |
|
Parallel association | 13 | 1 |
|
|
Diverging association | 15 | 13 |
|
| Trend of
EZH2a and PCNA
expressiona,b |
|
| 0.048 |
|
Parallel association | 12 | 11 |
|
|
Diverging association | 16 | 3 |
|
| Trend of
H3K27me3c and PCNA
expressiona,b |
|
| 0.331 |
|
Parallel association | 13 | 4 |
|
|
Diverging association | 15 | 5 |
|
| CCGG methylation
ratioa and H3K27me3
expressionc |
|
| 0.049 |
|
Parallel association | 17 | 8 |
|
|
Diverging association | 11 | 6 |
|
| CCGG methylation
ratioa and EZH2
expressiona,b |
|
| 0.050 |
|
Parallel association | 11 | 5 |
|
|
Diverging association | 17 | 9 |
|
| CCGG methylation
ratioa and PCNA
expressiona |
|
| 0.382 |
|
Parallel association | 16 | 6 |
|
|
Diverging association | 12 | 8 |
|
| Lymphatic vessel
involvedb |
|
| 0.731 |
|
Yes | 17 | 14 |
|
| No | 7 | 4 |
|
| Pulmonary vein
involved |
|
| 0.280 |
|
Yes | 12 | 6 |
|
| No | 12 | 12 |
|
| Visceral pleura
involvedb |
|
| 0.429 |
|
Yes | 0 | 1 |
|
| No | 24 | 17 |
|
| PCNA
expressiona,b |
|
| 0.375 |
|
High | 22 | 14 |
|
|
Low | 2 | 4 |
|
| Serum CEA
levelb |
|
| 0.247 |
|
High | 3 | 0 |
|
|
Low | 21 | 18 |
|
Correlation between smoking status and
other clinicopathological parameters in patients with NSCLC
As indicated in Table
II, in comparison with non-smokers, smokers exhibited
significantly lower H3K27me3 expression (P=0.029), a lower
methylation ratio at CCGG sites (P=0.016) and higher EZH2
expression (P=0.029). When evaluating the trends in the expression
or alterations to multiples factors, it was identified that a
parallel association between the H3K27me3 and EZH2 expression
levels in the majority of smoking patients with NSCLC, while in
non-smoking patients with NSCLC, the majority of their expression
levels were not significant (P=0.015). There was a diverging
association between PCNA and EZH2 expression levels in the majority
of smoking patients with NSCLC, while in most non-smoking patients
with NSCLC their expression levels identified statistical
significance (P=0.048). Figs. 4 and
5 depict 4 patients with different
smoking statuses. Patients A and B were smokers, whereas patients C
and D were non-smokers. Tissue samples from each patient were
stained with H&E, H3K27me3, EZH2 and PCNA. It was determined
that patients with NSCLC and differing smoking statuses had
distinct immunostaining for H3K27me3, EZH2 and PCNA. IHC staining
of H3K27me3, EZH2 and PCNA for patients A and B were all high
(Fig. 4A, panels 2–4), and all low
(Fig. 4B, panels 2–4), respectively.
While IHC staining of H3K27me3, EZH2 and PCNA for patients C and D
were high, low and low (Fig. 5A,
panels 2–4), and low, high and high (Fig.
5B, panels 2–4), respectively. In addition, the association
between the CCGG methylation ratio and the immunohistochemical
expression of H3K27me3 was parallel in the majority of smoking
patients with NSCLC, while in the majority of non-smoking patients
with NSCLC there was a diverging association (P=0.049). There was
an association between the CCGG methylation ratio and the
immunohistochemical expression of EZH2 (P=0.050), between the CCGG
methylation ratio and the immunohistochemical expression of PCNA
(P=0.382) and between H3K27me3 and PCNA expression (P=0.331), in
patients with NSCLC with different smoking statuses; however, the
associations were identified as having a low significance (Table II). In addition, no statistical
significance was demonstrated between smoking status and the
involvement of the lymphatic vessels (P=0.731), pulmonary vein
(P=0.280), visceral pleura (P=0.429), PCNA expression (P=0.375) and
serum CEA level (P=0.247).
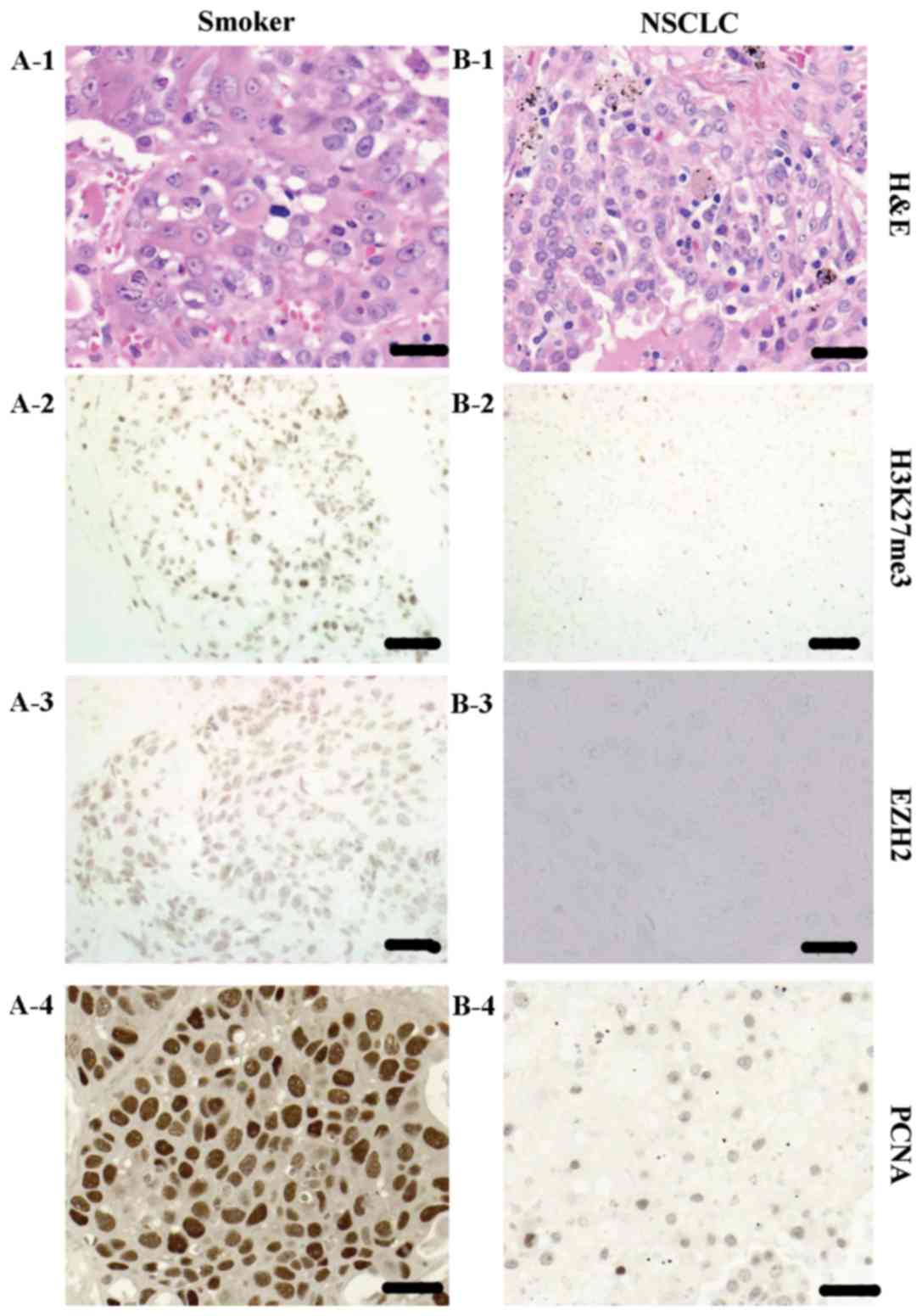
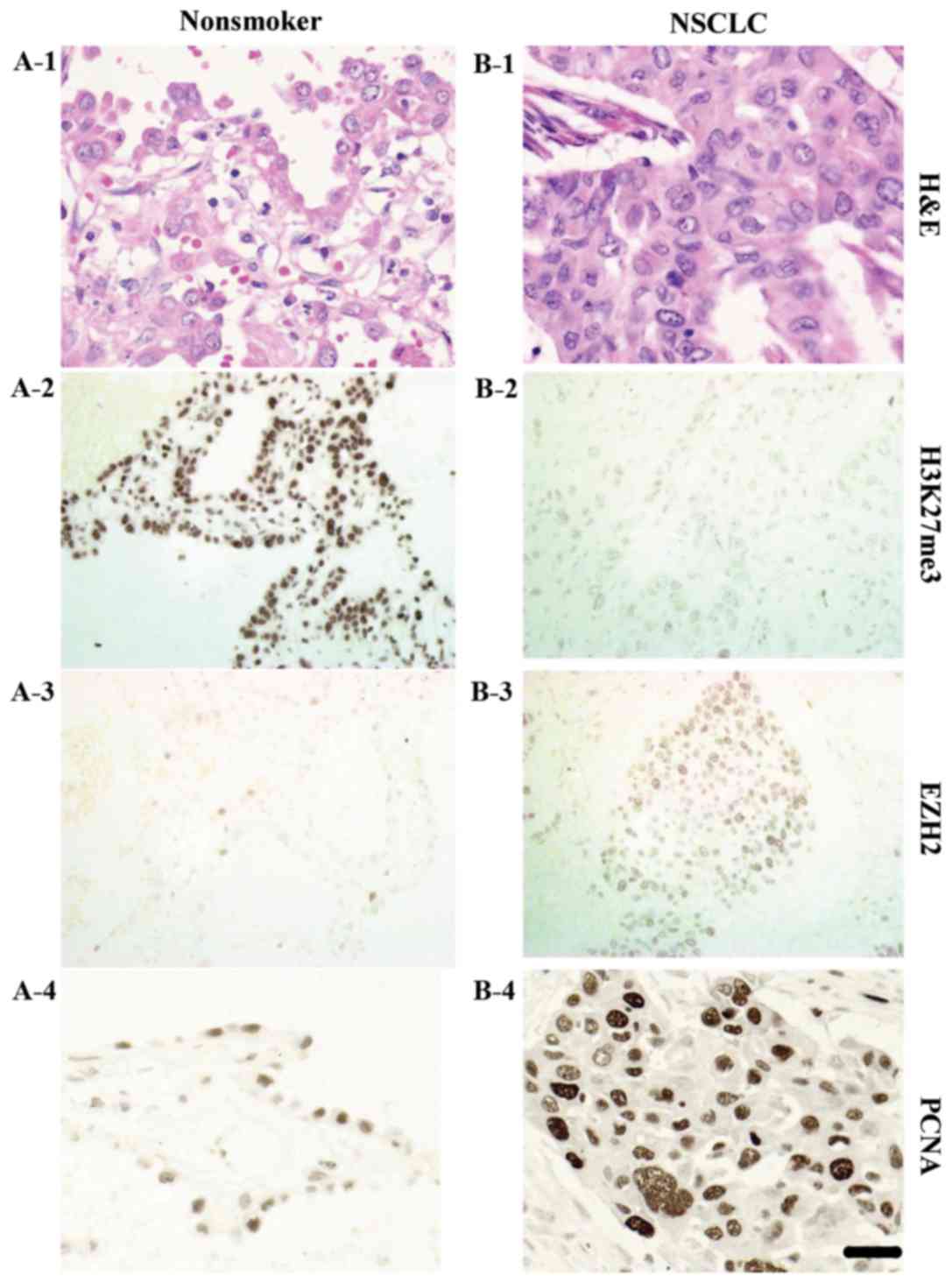 | Figure 5.Non-small cell lung cancer tissues of
smoking patients stained with (A-1 and B-1), H3K27me3, EZH2(c)
H&E; (A-2 and B-2) H3K27me3; (A-3 and B-3) EZH2; and (A-4 and
B-4) PCNA. Patient C exhibited strong (A-2) H3K27me3 expression,
weak (A-3) EZH2 expression and weak (A-4) PCNA expression. Patient
D exhibited weak (B-2) H3K27me3 expression, strong (B-3) EZH2
expressionand strong (B-4) PCNA expression. Scale bar, 20 µm.
NSCLC, non-small cell lung cancer; H3K27me3, trimethylation of
histone H3 at lysine 27; EZH2, enhancer of zeste homolog 2; PCNA,
proliferating cellular nuclear antigen; H&E, hematoxylin and
eosin. |
Discussion
In the present study, epigenetic characteristics,
including histone modification and DNA methylation, in NSCLC were
investigated. Additionally, tumor proliferation, as well as its
apoptotic index in smoking and non-smoking patients with NSCLC were
evaluated. This demonstrated that a lower expression level of
H3K27me3 and a lower level of DNA methylation at CCGG sites were
predominant in smoking patients with NSCLC. In addition, a higher
expression level of EZH2 and a lower apoptotic index had been
observed in smoking patients with NSCLC, in comparison with those
who never smoked; however, no significant difference had been
observed in PCNA expression between two groups.
A previous study demonstrated that patients with
NSCLC and higher expression of H3K27me3 had an increased overall
survival time; therefore, H3K27me3 was demonstrated to be an
independent prognostic risk factor (17). The epigenetic changes concerning the
effect that tobacco smoking may have on epigenetic modifications,
as well as its influence on cellular growth and apoptosis was
examined in the present study.
Environmental factors, particularly cigarette
smoking, have been implicated in the development and progression of
lung cancer by eliciting DNA methylation changes (8). The main epigenetic change in mammals is
the aberrant methylation of CpG islands in the gene promoter
region. As previously identified, global hypomethylationis an early
indicator for colon, lung and breast cancer, and chronic
lymphocytic leukemia, and lung cancer as well (11,28). On
the contrary, Digel and Lübbert (29)
reported that gene hypermethylation was an early event in the
process of lung cancer tumorigenesis. These paradoxical results may
be better interpreted as different degrees of methylation in
various genes, inside or outside the CpG islands. However, in the
present study, the DNA methylation levels at CCGG sites were
evaluated through the HELMET method, which demonstrated that
hypomethylation at CCGG sites was more common in smoking patients
with NSCLC (P=0.016), compared with in non-smokers. In addition, a
decrease in methylation levels at CCGG sites was determined to
closely correlate with a greater Brinkman index (P=0.017), which
was an indicator for heavy smoking. Patients with NSCLC and
different smoking statuses exhibited varying epigenetic markers,
which was revealed via immunohistochemical detection of several
major markers, including H3K27me3, EZH2 and PCNA. In addition to a
lower methylation level at the CCGG sites, the majority of smoking
patients with NSCLC had lower H3K27me3 expression levels and higher
expression levels of EZH2. Due to the methyltransferase of histone
H3 lysine 27, the expression of EZH2 was theorized to be associated
with the expression of H3K27me3; however, using a mirror section
technique, a diverging association was identified between the EZH2
and H3K27me3 expression levels in the previous investigation
(17). It has been repeatedly
reported that mutation in the EZH2 gene is a common phenomenon in a
variety of cancer types (30,31), including breast and lung cancer. Due
to the alteration of methylation status at CCGG sites predominantly
identified in smokers, it was hypothesized that low H3K27me3
expression may result from a mutated EZH2, which had lost its
catalyzing function in the methylation of H3K27 (32,33). To
further investigate the underlying mechanisms, it was theorized
that the alteration of DNA methylation levels, resulting from
tobacco smoking, would cause modifications in the epigenetic
changes. The expression pattern of H3K27me3 and EZH2 in patients
with NSCLC with different smoking statuses were further analyzed,
and it was determined that there was a parallel association between
H3K27me3 and EZH2 expression levels in the majority of smoking
patients with NSCLC, whether it was an increase or a decrease;
however, in non-smoking patients with NSCLC, the majority of the
expression levels had no significant association (P=0.015),
indicating that NSCLC tissues from of non-smoking patients with
high H3K27me3 expression would present with low EZH2 expression,
and vice versa. These results could assist in deciphering
the phenomenon observed in a previous study (17).
Alterations in the status of DNA methylation, as
well as histone modifications, frequently lead to changes in a
number of specific genes, including oncogenes and TSGs, resulting
in proliferation or apoptosis. This was assessed by means of the
immunohistochemical expression of PCNA as well as the TUNEL
apoptotic index, in the present study. This study has demonstrated
a significantly greater apoptotic index in non-smoking patients
with NSCLC tissues, compared with smokers, while no statistical
difference was observed between the PCNA expression levels of the
two groups. This demonstrated that smoking could facilitate the
majority of cancerous cells to obtain cellular immortality,
although their proliferative ability may not have differed
notably.
Changes in the levels of histone modification, as
well as DNA methylation, directly influence disease prognosis.
Site-specific histone modifications at H3K27me3 influence the
clinical outcome of patients with early-stage NSCLC. Based on
published reports (34–36), the novel cigarette smoking-induced
site-specific histone and DNA methylation markers identified in the
present study will have a greater translational impact on
understanding the pathogenetic process involved in smoking-mediated
lung disease, including lung cancer and other cancer types. The
result demonstrated a coexistence of the alteration of DNA
methylation and H3K27me3 expression levels in patients with NSCLC,
which is consistent with the study by Takeshima et al
(37), which is identified
specifically in cancer cells and may be used as a target for
epigenetic therapy. Analysis of DNA methylation and H3K27me3
expression levels in human colon, breast and prostate cancer cell
lines revealed that ~1/4 of DNA methylated genes underwent DNA
methylation and H3K27me3 dual modification in cancer cells, while
there was a presence of ~1/10 in normal cells.
A number of limitations in the present study should
be mentioned. The sample size was limited, which may have led to
bias. Further studies with larger sample sizes are required in
order to validate the present results, and to identify DNA
methylation patterns in specific genes and their associations with
histone modifications.
Conclusively, these results indicated that cigarette
smoking in patients with NSCLC may lead to lower levels of DNA
methylation at CCGG sites, lower H3K27me3 expression levels and a
lower cellular apoptotic index. In addition, patients with NSCLC
with different smoking statuses exhibited distinct changes in
epigenetic markers, which would facilitate the design of epigenetic
drugs targeting specific biomarkers for the immortality of
cancerous cells.
Acknowledgements
The abstract of this study was presented at the ASCO
Annual Meeting 2–6 June 2017 in Chicago, IL, USA and published as
abstract no. e23179 in J Clin Oncol 35: 2017.
Funding
This study was supported in part by a Grant-in-Aid
for Youth Research Project from Health Administration of Fujian
Province (grant no. 2013-1-10 to X. CHEN), the Fujian Provincial
Foundation of Natural Science (grant no. 2016J01515 to X. CHEN),
Fujian Medical University Startup Program (grant no. 2016QH040 to
Y. DENG) and Fujian Medical University Miaopu Program (grant no.
2015MP032 to Y. DENG).
Availability of data and materials
The analyzed data sets generated during the study
are available from the corresponding author, on reasonable
request.
Author contributions
Conception and design, KSZ, YJD, TK, XHC;
administrative support, XWZ, GXW, KSZ; provision of study materials
or patients, KM, TN, TK, XHC; collection and assembly of data, KSZ,
YJD, DH, CH, WHJ, GL, YBC, GBW, XHC; data analysis and
interpretation, KSZ, YJD, GXW, XWZ, XHC; manuscript writing, all
authors; final approval of manuscript, all authors.
Ethics approval and consent to
participate
The protocol of this study was approved by the Human
Ethics Review Committee of Nagasaki University School of Medicine,
and signed informed consent was obtained from all patients.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
NSCLC
|
non-small cell lung cancer
|
|
ADC
|
adenocarcinoma
|
|
SCC
|
squamous cell carcinoma
|
|
H3K27me3
|
trimethylation of histone H3 at lysine
27
|
|
EZH2
|
enhancer of zeste homolog 2
|
|
PRC2
|
polycomb repressive complex 2
|
|
PCNA
|
proliferating cellular nuclear
antigen
|
|
TUNEL
|
terminal
deoxynucleotidyl-transferase-mediated dUTP-biotin nick end
labeling
|
|
HELMET
|
histoendonuclease-linked detection of
methylation sites of DNA
|
References
|
1
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Torre LA, Siegel RL and Jemal A: Lung
cancer statistics. Adv Exp Med Biol. 893:1–19. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hussain SP and Harris CC: Molecular
epidemiology of human cancer. Recent Results Cancer Res. 154:22–36.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Piyathilake CJ, Frost AR, Bell WC,
Oelschlager D, Weiss H, Johanning GL, Niveleau A, Heimburger DC and
Grizzle WE: Altered global methylation of DNA: An epigenetic
difference in susceptibility for lung cancer is associated with its
progression. Hum Pathol. 32:856–862. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Soria JC, Rodriguez M, Liu DD, Lee JJ,
Hong WK and Mao L: Aberrant promoter methylation of multiple genes
in bronchial brush samples from former cigarette smokers. Cancer
Res. 62:351–355. 2002.PubMed/NCBI
|
|
7
|
Wistuba II, Mao L and Gazdar AF: Smoking
molecular damage in bronchial epithelium. Oncogene. 21:7298–7306.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jin Y, Xu P, Liu X, Zhang C, Tan C, Chen
C, Sun X and Xu Y: Cigarette smoking, BPDE-DNA adducts, and
aberrant promoter methylations of tumor suppressor genes (TSGs) in
NSCLC from Chinese population. Cancer Invest. 34:173–180. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tessema M, Yingling CM, Liu Y, Tellez CS,
Van Neste L, Baylin SS and Belinsky SA: Genome-wide unmasking of
epigenetically silenced genes in lung adenocarcinoma from smokers
and never smokers. Carcinogenesis. 35:1248–1257. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sundar IK, Nevid MZ, Friedman AE and
Rahman I: Cigarette smoke induces distinct histone modifications in
lung cells: Implications for the pathogenesis of COPD and lung
cancer. J Proteome Res. 13:982–996. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Søes S, Daugaard IL, Sørensen BS, Carus A,
Mattheisen M, Alsner J, Overgaard J, Hager H, Hansen LL and
Kristensen LS: Hypomethylation and increased expression of the
putative oncogene ELMO3 are associated with lung cancer development
and metastases formation. Oncoscience. 1:367–374. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bjaanaes MM, Fleischer T, Halvorsen AR,
Daunay A, Busato F, Solberg S, Jørgensen L, Kure E, Edvardsen H,
Børresen-Dale AL, et al: Genome-wide DNA methylation analyses in
lung adenocarcinomas: Association with EGFR, KRAS and TP53 mutation
status, gene expression and prognosis. Mol Oncol. 10:330–343. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wiles ET and Selker EU: H3K27 methylation:
A promiscuous repressive chromatin mark. Curr Opin Genet Dev.
43:31–37. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yu Q, Liu Y, Zheng X, Zhu Q, Shen Z, Wang
H, He H, Lin N, Jiang H, Yu L and Zeng S: Histone H3 lysine 4
Trimethylation, lysine 27 trimethylation, and lysine 27 acetylation
contribute to the transcriptional repression of solute carrier
family 47 member 2 in renal cell carcinoma. Drug Metab Dispos.
45:109–117. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yang X, Li F, Konze KD, Meslamani J, Ma A,
Brown PJ, Zhou MM, Arrowsmith CH, Kaniskan HÜ, Vedadi M and Jin J:
Structure-activity relationship studies for enhancer of zeste
homologue 2 (EZH2) and enhancer of zeste homologue 1 (EZH1)
inhibitors. J Med Chem. 59:7617–7633. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu L, Xu Z, Zhong L, Wang H, Jiang S,
Long Q, Xu J and Guo J: Enhancer of zeste homolog 2 (EZH2) promotes
tumour cell migration and invasion via epigenetic repression of
E-cadherin in renal cell carcinoma. BJU Int. 117:351–362. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen X, Song N, Matsumoto K, Nanashima A,
Nagayasu T, Hayashi T, Ying M, Endo D, Wu Z and Koji T: High
expression of trimethylated histone H3 at lysine 27 predicts better
prognosis in non-small cell lung cancer. Int J Oncol. 43:1467–1480.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen VW, Ruiz BA, Hsieh MC, Wu XC, Ries LA
and Lewis DR: Analysis of stage and clinical/prognostic factors for
lung cancer from SEER registries: AJCC staging and collaborative
stage data collection system. Cancer. 120 Suppl 23:S3781–S3792.
2014. View Article : Google Scholar
|
|
19
|
Taira K, Hiroyasu S, Shiraishi M, Muto Y
and Koji T: Role of the Fas system in liver regeneration after a
partial hepatectomy in rats. Eur Surg Res. 33:334–341. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fujiwara K, Fujimoto N, Tabata M, Nishii
K, Matsuo K, Hotta K, Kozuki T, Aoe M, Kiura K, Ueoka H and
Tanimoto M: Identification of epigenetic aberrant promoter
methylation in serum DNA is useful for early detection of lung
cancer. Clin Cancer Res. 11:1219–1225. 2005.PubMed/NCBI
|
|
21
|
Shirendeb U, Hishikawa Y, Moriyama S, Win
N, Thu MM, Mar KS, Khatanbaatar G, Masuzaki H and Koji T: Human
papillomavirus infection and its possible correlation with p63
expression in cervical cancer in Japan, Mongolia, and Myanmar. Acta
Histochem Cytochem. 42:181–190. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Song N, Liu J, An S, Nishino T, Hishikawa
Y and Koji T: Immunohistochemical analysis of histone H3
modifications in germ cells during mouse spermatogenesis. Acta
Histochem Cytochem. 44:183–190. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Adams JC: Heavy metal intensification of
DAB-based HRP reaction product. J Histochem Cytochem. 29:7751981.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ellinger J, Kahl P, von der Gathen J,
Heukamp LC, Gütgemann I, Walter B, Hofstädter F, Bastian PJ, von
Ruecker A, Müller SC and Rogenhofer S: Global histone H3K27
methylation levels are different in localized and metastatic
prostate cancer. Cancer Invest. 30:92–97. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gavrieli Y, Sherman Y and Ben-Sasson SA:
Identification of programmed cell death in situ via specific
labeling of nuclear DNA fragmentation. J Cell Biol. 119:493–501.
1992. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Koji T, Kondo S, Hishikawa Y, An S and
Sato Y: In situ detection of methylated DNA by histo
endonuclease-linked detection of methylated DNA sites: A new
principle of analysis of DNA methylation. Histochem Cell Biol.
130:917–925. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Camp RL, Dolled-Filhart M and Rimm DL:
X-tile: A new bio-informatics tool for biomarker assessment and
outcome-based cut-point optimization. Clin Cancer Res.
10:7252–7259. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ross JP, Rand KN and Molloy PL:
Hypomethylation of repeated DNA sequences in cancer. Epigenomics.
2:245–269. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Digel W and Lübbert M: DNA methylation
disturbances as novel therapeutic target in lung cancer:
Preclinical and clinical results. Crit Rev Oncol Hematol. 55:1–11.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Usemann J, Ernst T, Schäfer V, Lehmberg K
and Seeger K: EZH2 mutation in an adolescent with Weaver syndrome
developing acute myeloid leukemia and secondary
hemophagocyticlymphohistiocytosis. Am J Med Genet A. 170A:1–1277.
2016.
|
|
31
|
Tiffen JC, Gunatilake D, Gallagher SJ,
Gowrishankar K, Heinemann A, Cullinane C, Dutton-Regester K, Pupo
GM, Strbenac D, Yang JY, et al: Targeting activating mutations of
EZH2 leads to potent cell growth inhibition in human melanoma by
derepression of tumor suppressor genes. Oncotarget. 6:27023–27036.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bosselut R: Pleiotropic functions of
H3K27Me3 demethylases in immune cell differentiation. Trends
Immunol. 37:102–113. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhou Z, Gao J, Popovic R, Wolniak K,
Parimi V, Winter JN, Licht JD and Chen YH: Strong expression of
EZH2 and accumulation of trimethylated H3K27 in diffuse large
B-cell lymphoma independent of cell of origin and EZH2 codon 641
mutation. Leuk Lymphoma. 56:2895–2901. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Vick AD and Burris HH: Epigenetics and
health disparities. Curr Epidemiol Rep. 4:31–37. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Valeri L, Reese SL, Zhao S, Page CM,
Nystad W, Coull BA and London SJ: Misclassified exposure in
epigenetic mediation analyses. Does DNA methylation mediate effects
of smoking on birthweight? Epigenomics. 9:253–265. 2017.PubMed/NCBI
|
|
36
|
Tehranifar P, Wu HC, McDonald JA, Jasmine
F, Santella RM, Gurvich I, Flom JD and Terry MB: Maternal cigarette
smoking during pregnancy and offspring DNA methylation in midlife.
Epigenetics. May 11–2017.(Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Takeshima H, Wakabayashi M, Hattori N,
Yamashita S and Ushijima T: Identification of coexistence of DNA
methylation and H3K27me3 specifically in cancer cells as a
promising target for epigenetic therapy. Carcinogenesis.
36:192–201. 2015. View Article : Google Scholar : PubMed/NCBI
|