Introduction
Curcumin is an important bioactive compound derived
from the rhizome of the plant Curcuma longa Linn, which has
been used in traditional medicine for centuries. This phytochemical
has demonstrated potent antineoplastic activities in various animal
and cellular models of cancer. For example, curcumin exerts
powerful antiproliferative effects in cell lines derived from
colon, and lung cancers (1,2). Similarly, curcumin is able to reduce the
development of breast, lung, and hepatic tumors in various mouse
models (3,4). Importantly, numerous human clinical
trials have also demonstrated the safety and efficacy of curcumin
as an adjuvant for cancer therapy (5,6).
Curcumin features natural anticancer properties and
delivers potent cytostatic and cytotoxic effects against neoplastic
cells. Notably, however, curcumin's cytotoxic effects, including
cell cycle arrest and cell death, are specific to the genetic
background of the tumor-derived cells. For example, curcumin
exposure induced a cell cycle arrest in the G1 phase in prostate
and breast cancer-derived cell lines (7,8); whereas,
it induced a G2/M arrest in glioma and colorectal cancer cells
(9,10). Likewise, curcumin exposure is able to
promote different mechanisms of cell death depending on the tumor
cell type. For example, curcumin treatment of prostate, colon, and
lung cancer cells causes apoptosis (11–13), while
causing mitotic catastrophe in pancreatic and esophageal
tumor-derived cells (14,15). Furthermore, curcumin exposure was
related to autophagy in melanoma and hepatocellular carcinoma cell
lines (16,17).
In hematological malignancies, previous reports have
shown the potential of curcumin to provoke a reduction in cellular
proliferation and an increase of cell death in cell lines derived
from chronic and acute myeloid leukemia (18–22).
However, controversial results have been obtained from these
studies. For example, Sarkar et al (19) and Zhang et al (20) demonstrated that curcumin induced a
cell cycle arrest at the G2/M phase in the acute myeloid
leukemia-derived cell line HL-60; whereas in the same cell line,
Liu et al (21) and Chen et
al (22) reported a
curcumin-related cell cycle arrest in G1 phase. In addition,
previous reports showed that treatment of the chronic myeloid
leukemia-derived cell line K562 with curcumin promoted apoptosis
through the activation of caspases-9 and −3 (23,24).
Another study found that curcumin activates both apoptosis and
autophagy in K562 cells (25).
It is worth noting that a comparative study
investigating the different cytotoxic and cytostatic effects of
curcumin on cell lines derived from chronic or acute myeloid
leukemia cells has not been carried-out. Therefore, in the present
study, we compared the cytostatic and cytotoxic effects of curcumin
on both K562 and HL-60 cell lines that are derived from chronic and
acute myeloid leukemia cells, respectively. Our results illustrate
that curcumin activates different mechanisms for cell cycle arrest
and cell death in each type of leukemic cells. In HL-60 cells,
curcumin caused a cell cycle arrest in G1 and displayed classical
apoptosis, involving activation of caspases-9 and −3. In contrast,
in K562 cells, curcumin induced a G2/M arrest, followed by a cell
death process similar to mitotic catastrophe, with partial
activation of caspases-9 and −3.
Materials and methods
Cell cultures
The cell lines derived from chronic myeloid leukemia
(K562) and from acute promyelocytic leukemia (HL-60) were obtained
from the American Type Culture Collection (ATCC; Manassas, VA,
USA). Cell cultures were grown in RPMI-1640 medium containing 10%
fetal bovine serum (FBS), 50 U/ml penicillin, 50 µg/ml streptomycin
and 1% (v/v) non-essential amino acids (Gibco, Grand Island, NY,
USA). Additionally, medium for HL-60 cells was supplemented with 2
mM GlutaMAX (Gibco, Grand Island, NY, USA). Cell cultures were
maintained in an incubator at 37°C with 95% humidity and 5% of
CO2. Curcumin was purchased from Sigma-Aldrich (St.
Louis, MO, USA) and dissolved in dimethyl sulfoxide (DMSO)
(Sigma-Aldrich) at 30 mM. Stock solution was kept at −20°C and
protected from light until use.
Cellular treatment
To compare the cytostatic and cytotoxic effects of
curcumin on HL-60 and K562 cell lines, 2.5×105 cells/ml
from each cell line were grown for 24 h, after which they were
incubated with 5, 10, 15, 20 or 30 µM of curcumin for 24 h
(dose-response assays) or with 30 µM of curcumin for 6, 12, 18, or
24 h (time-response assays). DMSO 0.1% (v/v) (curcumin vehicle)
treatment for 24 h was used as a control culture in both cell
lines.
Cell viability assays
After incubation with the indicated treatment, cells
were washed once with phosphate-buffered saline (PBS; 1X) and the
number of viable cells was determined with the trypan blue
exclusion test by direct counting of cells on a light microscope
Olympus CKX41 (Olympus, Miami, FL, USA). Results were expressed as
a percentage relative to the respective control culture (which was
set=100% of viability).
Cell death assays
Cell death was measured with the
LIVE/DEAD® Fixable Dead Cell Stain Kit (Thermo Fisher
Scientific, Inc., Waltham, MA, USA). This method uses red
fluorescent reactive dye to stain the cells with damaged membranes.
As a positive control of cell death, both cell lines were treated
with 40 mJ/cm2 of UV irradiation for 2 min in a
cross-linker GS Gene linker-UV chamber (Bio-Rad, Hercules, CA, USA)
and recovered 24 h post-irradiation. After the indicated
treatments, cells were pelleted, washed with PBS 1X, and stained
with 1 µl of fluorescent reactive dye from the
LIVE/DEAD® Stain Kit. Cell suspensions were protected
from light and incubated on ice for 30 min. Data was captured using
the FACSCalibur flow cytometer system (Beckman Coulter, San Jose,
CA, USA). At least 20,000 events were recorded for each sample.
Data analysis was performed with the Summit Software (version
5.1.0.5563; Beckman Coulter).
Cell cycle assays
The K562 and HL-60 cells exposed to dose- and
time-response assays, were pelleted, washed with cold-PBS 1X, and
fixed overnight at −20°C with 1 ml of ice-cold 70% ethanol. Pellets
were resuspended in 250 µl of PBS 1X and treated with RNAse A (0.5
mg/ml) for 1 h at 37°C. Then, the cell suspensions were incubated
on ice with 10 µg/ml of propidium iodide (Sigma-Aldrich) for 30 min
in the dark. DNA content was measured using a FACSCalibur flow
cytometer (Beckman Coulter), and cell cycle analysis performed with
ModFit LT™ software (version 4.1; Verity Software House,
Topsham, ME., USA). At least 20,000 events were recorded for each
sample.
Western blot analysis
To obtain total protein extracts, cell cultures
treated with curcumin were collected and lysed with
ProteoJET™ mammalian cell lysis reagent (Fermentas,
Waltham, MA, USA), following the manufacturer's instructions.
Whole-cell extracts (30 µg) were separated by SDS-PAGE, transferred
onto a polyvinylidene difluoride (PVDF) membrane (Perkin-Elmer,
Boston, MA, USA), blocked with 5% non-fat milk in TBS-Tween-20 (10
mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1% Tween-20), and incubated
overnight at 4°C with specific primary antibodies used at a
dilution of 1:1,000. Primary antibodies used were rabbit anti-human
polyclonal caspase-9, rabbit anti-human polyclonal caspase-3 and
rabbit anti-human polyclonal PARP (Cell Signaling Technology, Inc.,
Boston, MA, USA; cat. nos. 9502, 9662 and 9542, respectively).
Additionally, we used a mouse anti-human monoclonal BubR1, goat
anti-human polyclonal p-Chk1 (Ser 345), mouse anti-human monoclonal
Chk1 (Santa Cruz Biotechnology, Santa Cruz, CA, USA; cat. nos.
sc-47744, sc-17922 and sc-8408) and rabbit anti-human monoclonal
Securin (GeneTex, Inc., Irvine, CA, USA; cat. no. GTX62173).
Membranes were washed twice with TBS-Tween-20 and incubated at room
temperature for 90 min with horseradish peroxidase-conjugated goat
anti-mouse or goat anti-rabbit secondary antibodies (Invitrogen,
Camarillo, CA, USA; cat. nos. 62–6520 and 65–6120, respectively) at
a dilution of 1:5,000. Signals were detected using SuperSignal West
Pico Maximum Sensitivity Substrate (Thermo Fisher Scientific,
Rockford, IL, USA). Membranes were reblotted and incubated with
mouse anti-human monoclonal β-actin (1:500) (US Biological,
Swampscott, MA, USA; cat. no. A0760-40) to account for well-to-well
variation in loading.
Caspases activity assay
Activity of caspases-9 and −3 was evaluated in cell
cultures treated with 30 µM of curcumin for 24 h using specific
fluorescence. Briefly, after treatment, cell lines were pelleted,
washed, and lysed in lysis buffer (50 mM HEPES, 5 mM DTT and 1%
Triton X-100) for 30 min. Cellular extracts were clarified by
centrifugation at 16,000 × g for 15 min at 4°C, and 100 µg of
proteins were incubated with 2 ml of caspase specific reaction
buffer and 1 µM of a fluorescent substrate to either caspase-3
(Ac-DEVD-AMC) or caspase-9 (Ac-LEDH-AFC) from Sigma-Aldrich at 32°C
for 24 h, in the presence or absence of 2 µM of specific inhibitors
for caspase-3 (Ac-DEVD-CHO) or caspase-9 (Ac-LEDH-CHO), obtained
from Enzo Life Sciences (Lausen, Switzerland). Caspase activity was
measured as a function of fluorescence signal produced by the
cleavage of synthetic substrate. Activity of caspases was
determined by subtracting the fluorescence value obtained in the
presence of caspase inhibitor from the value of total
fluorescence.
Statistical analysis
Results are expressed as means ± standard deviation
(SD) of at least three independent experiments. Differences between
control and curcumin treatments were analyzed by one-way analysis
of variance followed by Tukey's multiple comparison test.
Differences were considered significant at P<0.05. Statistical
analysis was carried out using the GraphPad Prism 5 Software
(version 5.01; Hearne Scientific, Pty Ltd., Melbourne,
Australia).
Results
K562 cell line exhibits a higher
susceptibility than HL-60 to curcumin effects
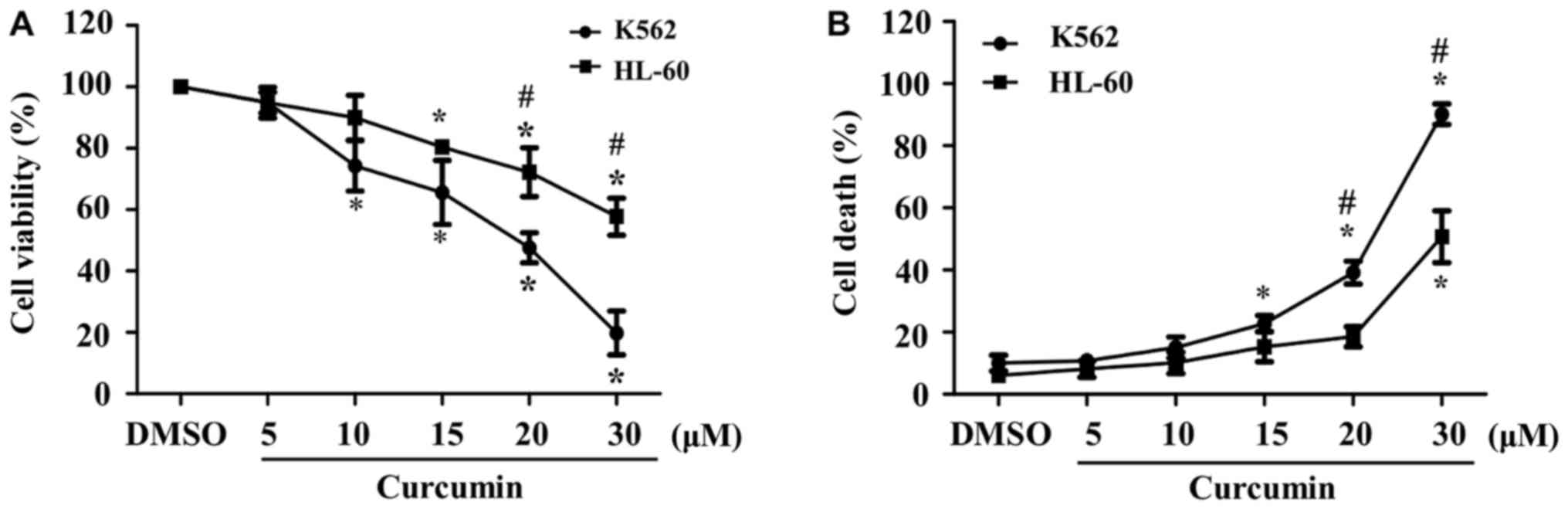
To determine the effect of curcumin on cell
viability in acute (HL-60) and chronic (K562) myeloid
leukemia-derived cell lines, both were treated for 24 h with
increasing doses (5 to 30 µM) of the phytochemical. Although we
observed a dose-dependent reduction in cellular viability in both
cell lines, the percentage of viable K562 cells decreased to
<80% at doses as low as 10 µM, whereas in HL-60 cells, a similar
reduction of the percentage of viability was observed until 20 µM.
Moreover, cellular viability at the highest dose of curcumin was
significantly lower in K562 than in HL-60 cells (20 vs. 60%,
respectively) (Fig. 1A). These data
indicate that the K562 cell line is more sensitive to curcumin than
HL-60 cells.
In addition, we evaluated cytotoxic properties of
curcumin in K562 and HL-60 cells exposed to different
concentrations of the phytochemical for 24 h. As expected, we
observed a dose-dependent increase in the percentage of cell death
in both cell cultures, which started at lower doses of the
phytochemical in K562, in comparison with HL-60 cells (15 vs. 30
µM, respectively). Similar to cell viability, the percentage of
cell death at the highest dose of curcumin was significantly higher
in K562 than in HL-60 cells (90 vs. 50%, respectively) (Fig. 1B).
Curcumin produces different effects on
cell cycle progression in K562 and HL-60 cell lines
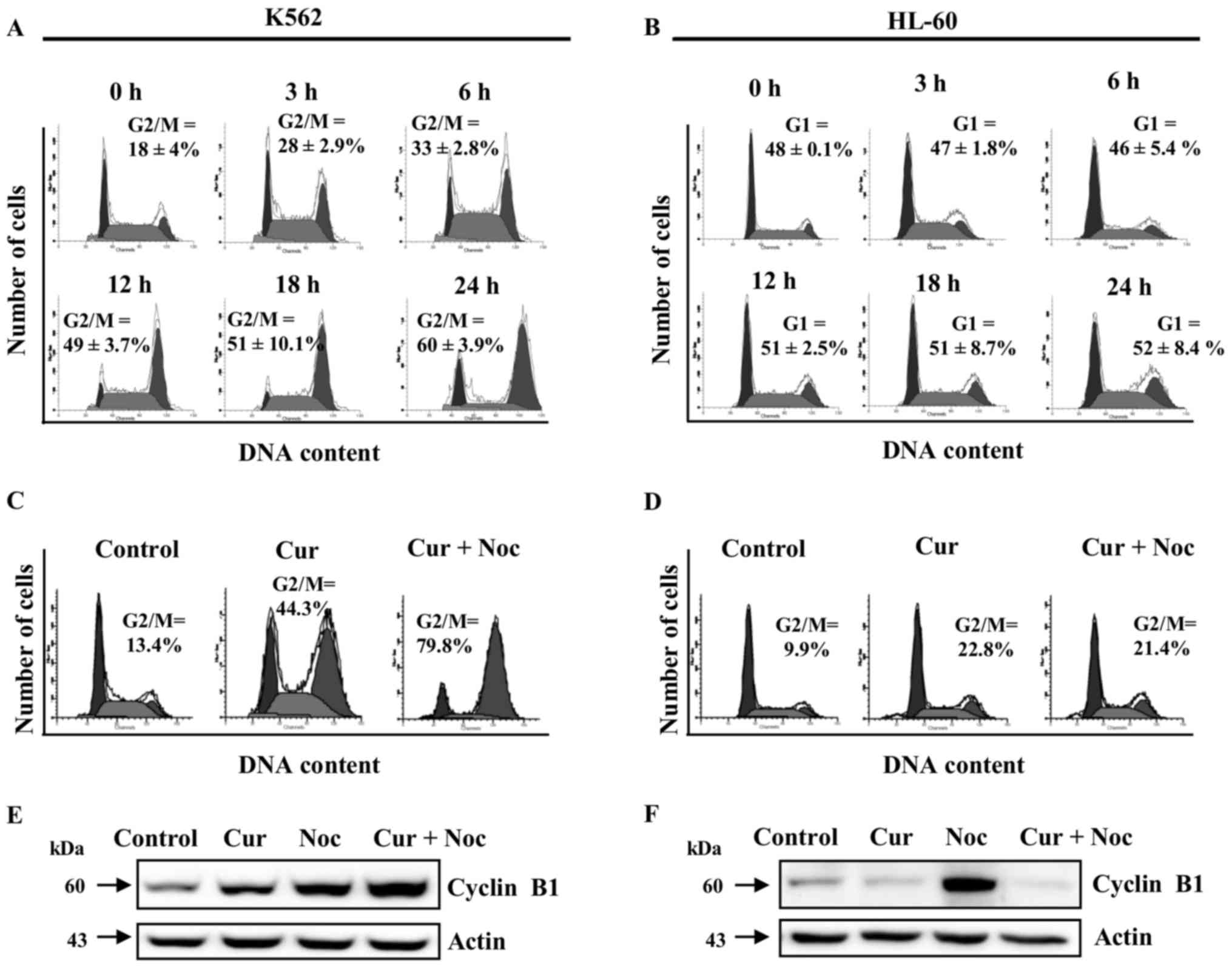
We next investigated the effect of curcumin on cell
cycle progression in K562 and HL-60 cell lines in time-response
assays, using 20 µM of the phytochemical to avoid the excessively
toxic effects observed at higher doses. We found an increase in the
percentage of K562 cells at the G2/M phase in a time-dependent
manner; whereas the HL-60 cell line had a small level of cells
accumulated in G1 phase, which may suggest the presence of a G1
arrest (Fig. 2A and B). These
observations indicate that HL-60 and K562 cells were affected by
curcumin at different points in cell cycle progression.
To further analyze the cytostatic effect of curcumin
in K562 and HL-60 cells, we co-incubated cell cultures with
curcumin (20 µM) and nocodazole (200 nM), an effective inhibitor of
microtubule polymerization and a potent inducer of mitotic arrest.
In the case of K562 cells, co-incubation of nocodazole with
curcumin caused a higher accumulation of cells in G2/M (79.8%),
than those observed in the individual curcumin treatment (44.3%)
(Fig. 2C). On the other hand, HL-60
cultures incubated simultaneously with nocodazole and curcumin
showed no significant difference in the number of cells in the G2/M
phase (21.4%), in comparison with those treated with curcumin
(22.8%) (Fig. 2D).
We also evaluated the protein levels of cyclin B1,
which is normally a high abundance protein in the G2/M transition
phase. In K562 cells, treatment with curcumin or nocodazole alone
induced a significant accumulation of cyclin B1, whereas
co-incubation produced a synergistic effect (Fig. 2E). In contrast, in HL-60 cells protein
levels of cyclin B1 were increased only with the nocodazole
treatment (Fig. 2F). Taken together,
these data indicate that curcumin is able to induce a cell cycle
arrest in both K562 and HL-60 cells, but at different phases of the
cell cycle.
Curcumin-induced activation of
caspases is defective in K562 but complete in HL-60 cells
Additionally, we analyzed the activation of
caspases-9 and −3 in response to curcumin treatment as markers of
apoptotic cell death. Using a curcumin concentration of 30 µM in a
time-response assay, the low molecular weight protein fragments of
the processed forms of caspases-9 and −3 were observed, as well as
the cleavage fragment of PARP, the caspase-3 substrate, in both
K562 and HL-60 cells (Fig. 3A and B).
However, in the K562 cell line, the unprocessed forms of
caspases-9, −3, and PARP (47, 35 and 116 kDa, respectively) were
still present up to the end of the incubation time (Fig. 3A; upper band in each panel). In sharp
contrast, in HL-60 cells the unprocessed forms of the two caspases,
and PARP protein were no longer observable after 18 h of treatment
with curcumin 30 µM (Fig. 3B). In
accordance with these data, activity of caspase-9 and −3 after
incubation with 30 µM of curcumin for 24 h was significantly lower
in K562 as compared to the HL-60 cell line (Fig. 3C). These data suggest that caspases
activation is an essential step in curcumin-induced cell death in
HL-60 cells, but not necessarily in K562.
Curcumin triggers a defective DNA
damage checkpoint at G2 phase in K562 cells
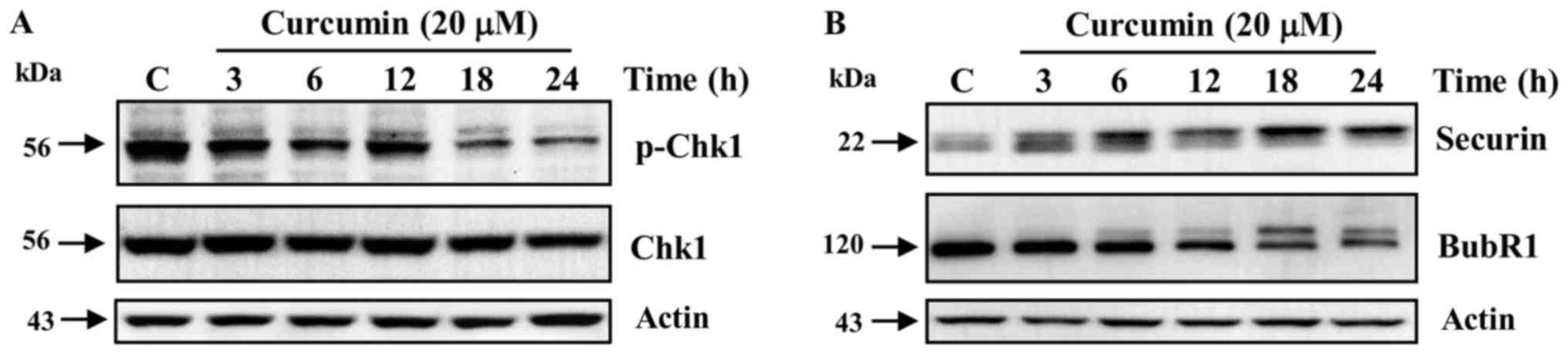
Curcumin is a known inducer of DNA damage in
different types of tumoral cells (26,27). Thus,
we analyzed in K562 cells, the activation of the DNA damage
checkpoints in G2 and mitosis in response to curcumin, by detecting
the phosphorylated status of the checkpoint proteins Chk1, Securin
and BubR1. We observed no activation by phosphorylation in Ser-345
of the G2 checkpoint regulator Chk1 during the entire exposure to
curcumin (Fig. 4A). In sharp
contrast, the level of the phosphorylated and active forms of the
mitotic checkpoint regulators Securin and BubR1 increased in a
time-dependent manner after curcumin exposure (Fig. 4B).
Discussion
Curcumin is a natural bioactive compound which
exerts antiproliferative and pro-apoptotic effects in a wide
variety of cellular and animal cancer models. In fact, curcumin is
being used as a radio and chemo-sensitizer in different types of
cancer (28–30). However, several studies have shown
that curcumin elicits different responses at the level of cell
cycle regulation and cell death, depending upon the dose, time of
exposure, and, perhaps most importantly, the tumor cell type. In
this sense, the effect of curcumin on the regulation of cell cycle
and in cell lines derived from acute and chronic myeloid leukemia
(HL-60 and K562 cell lines, respectively) is highly controversial
(19,25). Moreover, the cytostatic effects of
this phytochemical on cell lines derived from chronic myeloid
leukemia have been poorly studied until now. This highlights the
necessity of comparative studies assessing the cytostatic and
cytotoxic effects of curcumin in these two types of myeloid
leukemia cells.
In the case of the acute myeloid leukemia-derived
cell line HL-60, previous reports have demonstrated the capacity of
curcumin to induce a cell cycle arrest in G1 phase, followed by
caspase-dependent induction of apoptosis (21,31). In
addition, an independent study reported that G1 arrest induced by
curcumin in HL-60 cells was dependent on the pro-oxidant properties
of curcumin (30). In contrast, other
studies described a G2/M arrest in HL-60 cells exposed to curcumin
(19,20). Our assays, using simultaneous
treatment with nocodazole and curcumin (mitotic-trap), clearly
demonstrated that the phytochemical induced a G1 arrest in the
HL-60 cell line. In agreement with our present findings using this
mitotic-trap method, curcumin exposure was also shown to induced a
G1 arrest in other acute myeloid leukemia-derived cell lines such
as MV4-11, KG1a, and Kasumi-1 (32).
We have no clear explanation for the discrepancy between our
findings and those describing G2/M arrest (19,20), but
it is important to note that the curcumin concentration used in the
previous study was lower (10 µM) than that used in our present
study.
Regarding chronic myeloid leukemia-derived cell line
K562, few studies have analyzed the cytostatic effects of curcumin.
However, in agreement with the curcumin-induced G2/M arrest we
observed in the present study, in a mouse lymphoblastoid model
overexpressing the BCR-ABL chimeric protein, curcumin exposure also
induced a G2/M arrest (33). Notably,
arsenic trioxide, a potent pro-oxidant compound used as a therapy
in acute myeloid leukemia, also induced a G2/M arrest in K562
cells, whereas in leukemia-derived cell lines lacking the
BCR-ABL fusion gene (e.g. HL-60 and U937) induced a G1
arrest (34). These data indicate
that the underlying cellular mechanisms, which are responsible for
curcumin-induced cell cycle arrest in HL-60 and K562 cells are
different, and could be related to the presence of the
BCR-ABL fusion.
We also observed differential effects of curcumin on
cell death between HL-60 and K562 cells. In accordance with
previous reports, we found that HL-60 cells exposed to curcumin
showed induction of caspase-dependent apoptosis as a late event.
Nevertheless, in the case of K562 cells, we found that
curcumin-related cell death occurred at the G2/M transition phase,
after a partial activation of caspases-9 and −3. This may suggest
the presence of an alteration in the regulation of apoptosis in
K562 cells that avoids full activation of caspases, and thereby
triggers an alternative form of cell death. Furthermore, in a
recent study, we have demonstrated that curcumin-induced cell death
in K562 was produced by mitotic catastrophe (35). In contrast to our data, other studies
have reported a curcumin-related activation of caspases-3 and −9,
followed by apoptosis in K562 cells (23,24). It is
important to note that a previous report demonstrated that curcumin
exposure is able to induce apoptosis, through the activation of
caspase-9 and −3, but also to promote autophagy, by the induction
of Beclin1 and LC3 proteins (25).
Several previously published experiments highlight
the generation of reactive oxygen species and the alteration of
tubulin metabolism as the two main mechanisms underlying curcumin
effects on regulation of cell cycle progression and cell death
(36–38). Our results suggest that HL-60 cells
may be able to response to the pro-oxidant properties of curcumin,
which induces cell cycle arrest at G1, followed by classical
apoptosis, with full activation of caspases. On the other hand, in
K562 cells, which may be not proficient at inducing cell cycle
arrest at G1 and classical apoptosis, curcumin's effects are likely
due to its ability to disrupt tubulin metabolism, which induces a
G2/M arrest and cell death with partial activation of caspases. It
is worth to mention that in a previous study (35), we demonstrated the phosphorylation of
histone H3 in Ser-10 in K562 cells, after curcumin (20 µM for 24 h)
or nocodazole (200 nM for 24 h) exposure, indicating the arrest of
K562 cells in mitosis. Considering that progression from G2 to
mitosis is promoted by the dephosphorylation of T14/Y15 at Cdc2 by
CDC25C phosphatase, we speculate that in K562 cells, these residues
are dephosphorylated in the treatment with curcumin. In order to
test this hypothesis, in future studies we will investigate the
celular proteins involved in the regulation of cyclin B/Cdc2
activity in response to curcumin in chronic myeloid leukemia
cells.
Supporting the hypothesis that K562 cells are not
able to induce cell cycle arrest in response to curcumin-related
DNA damage, we observed a lack of activation of the DNA damage
checkpoint protein Chk1 in the presence of this phytochemical,
together with the activation by phosphorylation of the mitotic
checkpoint proteins BubR1 and Securin. These data suggest that in
K562 the DNA damage checkpoint in G2 is not functioning correctly
in response to curcumin, allowing cells with DNA damage to progress
into mitosis, where the checkpoint machinery arrest the cell cycle.
To further understand cellular response to curcumin exposure in
both types of leukemia it would be necessary to evaluate the
participation of key molecules in DNA damage response and cell
cycle regulation. An important limitation of the present study is
the absence of experimental data using markers of DNA damage such
as phosphorylation of histone 2AX (γ-H2AX).
In summary, these data represent the first
side-by-side comparison regarding the effects of curcumin on two
leukemic cell lines, K562 and HL-60 cells. Our results clearly
demonstrated that curcumin exposure arrest cell cycle at G1 in
HL-60 cells and at G2/M in K562 cells. We also observed that the
cellular mechanisms underlying curcumin's effects on cell death
regulation in chronic and acute myeloid leukemia-derived cell lines
were mechanistically distinct. The HL-60 cells appeared to be
dependent upon caspase for curcumin-induced cell death, while the
K562 cells underwent cell death in a caspase-independent manner. We
consider that these data could help to further analyzed the
particular alterations in the mechanisms of cell cycle arrest and
cell death in each type of leukemia.
Acknowledgements
The authors would like to thank Dr Jose Luis
Cruz-Colin (National Institute of Genomic Medicine, Clinic
Research, Mexico City, Mexico) for his valuable technical
assistance for cell culture support and Dr Raúl Bonilla Moreno
(Department of Molecular Biomedicine, Center of Studies and Advance
Research, Mexico City, Mexico) for expert technical assistance.
This work was supported by the Consejo Nacional de Ciencia y
Tecnología (CONACyT) grants CB-2014-01-243587, 128686, CINVESTAV
and INMEGEN. Work was also performed under the auspices of the U.S.
Department of Energy by Lawrence Livermore National Laboratory
under Contract DE-AC52-07NA27344.
References
|
1
|
Shakibaei M, Buhrmann C, Kraehe P, Shayan
P, Lueders C and Goel A: Curcumin chemosensitizes 5-fluorouracil
resistant MMR-deficient human colon cancer cells in high density
cultures. PLoS One. 9:e853972014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fan Z, Duan X, Cai H, Wang L, Li M, Qu J,
Li W, Wang Y and Wang J: Curcumin inhibits the invasion of lung
cancer cells by modulating the PKCα/Nox-2/ROS/ATF-2/MMP-9 signaling
pathway. Oncol Rep. 34:691–698. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ferreira LC, Arbab AS, Jardim-Perassi BV,
Borin TF, Varma NR, Iskander AS, Shankar A, Ali MM and Zuccari DA:
Effect of curcumin on pro-angiogenic factors in the xenograft model
of breast cancer. Anticancer Agents Med Chem. 15:1285–1296. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Huang AC, Lin SY, Su CC, Lin SS, Ho CC,
Hsia TC, Chiu TH, Yu CS, Ip SW, Lin TP and Chung JG: Effects of
curcumin on N-bis(2-hydroxypropyl) nitrosamine (DHPN)-induced lung
and liver tumorigenesis in BALB/c mice in vivo. In Vivo.
22:781–785. 2008.PubMed/NCBI
|
|
5
|
Carroll RE, Benya RV, Turgeon DK, Vareed
S, Neuman M, Rodriguez L, Kakarala M, Carpenter PM, McLaren C,
Meyskens FL Jr and Brenner DE: phase IIa clinical trial of curcumin
for the prevention of colorectal neoplasia. Cancer Prev Res.
4:354–364. 2011. View Article : Google Scholar
|
|
6
|
Epelbaum R, Schaffer M, Vizel B, Badmaev V
and Bar-Sela G: Curcumin and gemcitabine in patients with advanced
pancreatic cancer. Nutr Cancer. 62:1137–1141. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sha J, Li J, Wang W, Pan L, Cheng J, Li L,
Zhao H and Lin W: Curcumin induces G0/G1 arrest and apoptosis in
hormone independent prostate cancer DU-145 cells by down regulating
Notch signaling. Biomed Pharmacother. 84:177–184. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sun SH, Huang HC, Huang C and Lin JK:
Cycle arrest and apoptosis in MDA-MB-231/Her2 cells induced by
curcumin. Eur J Pharmacol. 690:22–30. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cheng C, Jiao JT, Qian Y, Guo XY, Huang J,
Dai MC, Zhang L, Ding XP, Zong D and Shao JF: Curcumin induces
G2/M arrest and triggers apoptosis via FoxO1 signaling
in U87 human glioma cells. Mol Med Rep. 13:3763–3770. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lu JJ, Cai YJ and Ding J: Curcumin induces
DNA damage and caffeine insensitive cell cycle arrest in colorectal
carcinoma HCT116 cells. Mol Cell Biochem. 354:247–252. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Guo H, Xu YM, Ye ZQ, Yu JH and Hu XY:
Curcumin induces cell cycle arrest and apoptosis of prostate cancer
cells by regulating the expression of IkappaBalpha, c-Jun and
androgen receptor. Pharmazie. 68:431–434. 2013.PubMed/NCBI
|
|
12
|
Cao A, Li Q, Yin P, Dong Y, Shi H, Wang L,
Ji G, Xie J and Wu D: Curcumin induces apoptosis in human gastric
carcinoma AGS cells and colon carcinoma HT-29 cells through
mitochondrial dysfunction and endoplasmic reticulum stress.
Apoptosis. 18:1391–1402. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yao Q, Lin M, Wang Y, Lai Y, Hu J, Fu T,
Wang L, Lin S, Chen L and Guo Y: Curcumin induces the apoptosis of
A549 cells via oxidative stress and MAPK signaling pathways. Int J
Mol Med. 36:1118–1126. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Subramaniam D, Ramalingam S, Linehan DC,
Dieckgraefe BK, Postier RG, Houchen CW, Jensen RA and Anant S: RNA
binding protein CUGBP2/CELF2 mediates curcumin-induced mitotic
catastrophe of pancreatic cancer cells. PLoS One. 6:e169582011.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
O'Sullivan-Coyne G, O'Sullivan GC,
O'Donovan TR, Piwocka K and McKenna SL: Curcumin induces
apoptosis-independent death in oesophageal cancer cells. Br J
Cancer. 101:1585–1595. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhao G, Han X, Zheng S, Li Z, Sha Y, Ni J,
Sun Z, Qiao S and Song Z: Curcumin induces autophagy, inhibits
proliferation and invasion by downregulating AKT/mTOR signaling
pathway in human melanoma cells. Oncol Rep. 35:1065–1074. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tork OM, Khaleel EF and Abdelmaqsoud OM:
Altered cell to cell communication, autophagy and mitochondrial
dysfunction in a model of hepatocellular carcinoma: Potential
protective effects of curcumin and stem cell therapy. Asian Pac J
Cancer Prev. 16:8271–8279. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tan TW, Tsai HR, Lu HF, Lin HL, Tsou MF,
Lin YT, Tsai HY, Chen YF and Chung JG: Curcumin-induced cell cycle
arrest and apoptosis in human acute promyelocytic leukemia HL-60
cells via MMP changes and caspase-3 activation. Anticancer Res.
26:4361–4371. 2006.PubMed/NCBI
|
|
19
|
Sarkar R, Mukherjee A, Mukherjee S, Biswas
R, Biswas J and Roy M: Curcumin augments the efficacy of antitumor
drugs used in leukemia by modulation of heat shock proteins via
HDAC6. J Environ Pathol Toxicol Oncol. 33:247–263. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang JR, Lu F, Lu T, Dong WH, Li P, Liu
N, Ma DX and Ji CY: Inactivation of FoxM1 transcription factor
contributes to curcumin-induced inhibition of survival,
angiogenesis, and chemosensitivity in acute myeloid leukemia cells.
J Mol Med. 92:1319–1330. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu Y, Chang RL, Cui XX, Newmark HL and
Conney AH: Synergistic effects of curcumin on all-trans retinoic
acid- and 1 alpha,25-dihydroxyvitamin D3-induced differentiation in
human promyelocytic leukemia HL-60 cells. Oncol Res. 9:19–29.
1997.PubMed/NCBI
|
|
22
|
Chen J, Wang G, Wang L, Kang J and Wang J:
Curcumin p38-dependently enhances the anticancer activity of
valproic acid in human leukemia cells. Eur J Pharm Sci. 41:210–218.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Duvoix A, Morceau F, Schnekenburger M,
Delhalle S, Galteau MM, Dicato M and Diederich M: Curcumin-induced
cell death in two leukemia cell lines: K562 and Jurkat. Ann N Y
Acad Sci. 1010:389–392. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chakraborty S, Ghosh U, Bhattacharyya NP,
Bhattacharya RK and Roy M: Inhibition of telomerase activity and
induction of apoptosis by curcumin in K-562 cells. Mutat Res.
596:81–90. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jia YL, Li J, Qin ZH and Liang ZQ:
Autophagic and apoptotic mechanisms of curcumin-induced death in
K562 cells. J Asian Nat Prod Res. 11:918–928. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jiang AJ, Jiang G, Li LT and Zheng JN:
Curcumin induces apoptosis through mitochondrial pathway and
caspases activation in human melanoma cells. Mol Biol Rep.
42:267–275. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Blakemore LM, Boes C, Cordell R and Manson
MM: Curcumin-induced mitotic arrest is characterized by spindle
abnormalities, defects in chromosomal congression and DNA damage.
Carcinogenesis. 34:351–360. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mathur A, Abd Elmageed ZY, Liu X,
Kostochka ML, Zhang H, Abdel-Mageed AB and Mondal D: Subverting
ER-stress towards apoptosis by nelfinavir and curcumin coexposure
augments docetaxel efficacy in castration resistant prostate cancer
cells. PLoS One. 9:e1031092014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zeng Y, Weng G, Fan J, Li Z, Wu J, Li Y,
Zheng R, Xia P and Guo K: Curcumin reduces the expression of
survivin, leading to enhancement of arsenic trioxide induced
apoptosis in myelodysplastic syndrome and leukemia stem-like cells.
Oncol Rep. 36:1233–1242. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen J, Wanming D, Zhang D, Liu Q and Kang
J: Water-soluble antioxidants improve the antioxidant and
anticancer activity of low concentrations of curcumin in human
leukemia cells. Pharmazie. 60:57–61. 2005.PubMed/NCBI
|
|
31
|
Yu J, Peng Y, Wu LC, Xie Z, Deng Y, Hughes
T, He S, Mo X, Chiu M, Wang QE, et al: Curcumin down-regulates DNA
methyltransferase 1 and plays an anti-leukemic role in acute
myeloid leukemia. PLoS One. 8:e559342013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Rao J, Xu DR, Zheng FM, Long ZJ, Huang SS,
Wu X, Zhou WH, Huang RW and Liu Q: Curcumin reduces expression of
Bcl-2, leading to apoptosis in daunorubicin-insensitive
CD34+ acute myeloid leukemia cell lines and primary
sorted CD34+ acute myeloid leukemia cells. J Transl Med.
9:712011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wolanin K, Magalska A, Mosieniak G,
Klinger R, McKenna S, Vejda S, Sikora E and Piwocka K: Curcumin
affects components of the chromosomal passenger complex and induces
mitotic catastrophe in apoptosis-resistant Bcr-Abl-expressing
cells. Mol Cancer Res. 4:457–469. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sánchez Y, Simón GP, Calviño E, de Blas E
and Aller P: Curcumin stimulates reactive oxygen species production
and potentiates apoptosis induction by the antitumor drugs arsenic
trioxide and lonidamine in human myeloid leukemia cell lines. J
Pharmacol Exp Ther. 335:114–123. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Martinez-Castillo M, Bonilla-Moreno R,
Aleman-Lazarini L, Meraz-Rios MA, Orozco L, Cedillo-Barron L,
Cordova EJ and Villegas-Sepulveda N: A subpopulation of the K562
cells are killed by curcumin treatment after G2/M arrest and
mitotic catastrophe. PLoS One. 11:e01659712016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Qadir MI, Naqvi ST and Muhammad SA:
Curcumin: A polyphenol with molecular targets for cancer control.
Asian Pac J Cancer Prev. 17:2735–2739. 2016.PubMed/NCBI
|
|
37
|
Chakraborti S, Das L, Kapoor N, Das A,
Dwivedi V, Poddar A, Chakraborti G, Janik M, Basu G, Panda D, et
al: Curcumin recognizes a unique binding site of tubulin. J Med
Chem. 54:6183–6196. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Jackson SJ, Murphy LL, Venema RC,
Singletary KW and Young AJ: Curcumin binds tubulin, induces mitotic
catastrophe, and impedes normal endothelial cell proliferation.
Food Chem Toxicol. 60:431–438. 2013. View Article : Google Scholar : PubMed/NCBI
|