Introduction
Hepatocellular carcinoma (HCC) is the third most
common cause of cancer mortality worldwide. A variety of risk
factors may promote HCC genesis, including hepatitis B virus (HBV)
infection, hepatitis C virus infection, heavy alcohol consumption
and non-alcoholic fatty liver disease (1,2). In the
last decade, a marked increase in the incidence of HBV-associated
HCC has been observed, particularly in China. Since the prognosis
of patients with HCC is markedly dependent on the stage of the
disease, strategies for early detection of HCC have been
investigated.
MicroRNAs (miRNAs) are short 20–22-nucleotide
single-stranded RNA molecules, which have been associated with
epigenetic regulation in a range of diseases, including
tumorigenesis (3–5). miRNAs regulate gene expression by
altering the stability or the translational efficiency of target
mRNAs. A previous study demonstrated that miRNAs are detectable and
stable in serum (6); therefore,
research has focused on the possibility of using miRNAs as
biomarkers for predicting the diagnosis and prognosis of several
diseases.
Two human miRNAs, miRNA-143 (miR-143) and miR-145
have been investigated as biomarkers for several types of cancer.
miR-143 and miR-145 are stably expressed homologous miRNAs located
within the same host gene, MIR143HG (7). The first study to examine the
miR-143/145 cluster focused on HBV-associated HCC. This study
demonstrated that miR-143 expression was increased in
HBV-associated HCC tumors and was associated with invasive and
metastatic behavior of liver tumor cells (8). Conversely, evaluation of the miR-143/145
cluster expression in other types of tumor, including colonic
carcinoma, pulmonary carcinoma, esophageal carcinoma and prostatic
carcinoma (9–12), demonstrated that the miR-143/145
cluster expression level was decreased. This raises the question of
why the miR-143/145 cluster is overexpressed in HBV-associated
HCC-derived tumors, but is underexpressed in other types of tumor.
However, previous studies have produced contradictory results
(13–15), demonstrating that the miR-143/145
expression level was decreased in HBV-associated HCC-derived tumors
which contradicts the results of another previously mentioned study
(8). Furthermore, a previous study
demonstrated that the expression of the miR-143/145 cluster was
negligible in liver tissues, including normal liver tissue or
HBV-associated HCC tissue (7). This
controversy was investigated further in the present study.
In the present study, the expression profile of
miR-143 and miR-145 in HBV-associated HCC tissues and non-tumor
tissues was investigated using chromatin immunoprecipitation (ChIP)
data from The Cancer Genome Atlas (TCGA) and the Gene Expression
Omnibus (GEO) datasets. Their expression in HBV-associated HCC
tissue was also validated using the reverse
transcription-quantitative polymerase chain reaction (RT-qPCR). The
association between miR-143/145 expression and specific clinical
features including tumorigenesis, tumor progression and prognosis
was subsequently examined. Finally, the serum expression of miR-143
and miR-145 was determined to evaluate their potential clinical
applications. The receiver-operating characteristic (ROC) curves
were used to explore their potential value as biomarkers for
predicting HBV-associated HCC tumorigenesis.
Materials and methods
TCGA miRNA dataset
The miRNA expression profile of 372 patients with
HCC and 50 non-tumor distal liver samples was obtained from the
TCGA dataset (cancergenome.nih.gov). According to the clinical
information, 102 patients with HBV infection were selected as
negative control.
GEO dataset
The GEO database (16)
was accessed to obtain the normalized miRNA microarray dataset
GSE22058 (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE22058),
which represents the miRNA expression profile of tumor and adjacent
non-tumor tissues of 96 patients with HCC. From these patients,
~88% were confirmed positive for the HBV antigen (17).
Identification of differentially
expressed miRNAs
For each dataset, the Wilcoxon signed-rank test was
used to identify the differentially expressed genes between tumor
and non-tumor samples. The null hypothesis was that a gene was not
differentially expressed between the two groups and rejection of
this hypothesis indicated that the gene was differentially
expressed. A miRNA was considered upregulated if P<0.01 and its
fold increase was ≥1.5. Downregulated miRNAs were similarly
defined.
Clinical samples
HBV-associated HCC tissues, matched distal
non-cancerous liver tissues and 5 ml venous peripheral blood were
obtained from 85 patients with HBV-associated HCC from February
2012 to January 2013 at the Provincial Hospital Affiliated to
Shandong University (Jinan, China). The mean patient age was
53.2±9.3 years (range, 25–75 years), and 70.6% of the patients were
male. Ten tissue samples from patients with hepatic hemangioma and
5 ml venous peripheral blood from 50 patients with chronic
hepatitis B were collected as negative controls. All tissue samples
were histologically confirmed by a pathologist using hematoxylin
and eosin staining of cryosectioned specimens. The criteria used
for distal non-cancerous liver tissues were absence of tumor cells
and 5 cm distance from the tumors. Blood samples were collected in
Vacutainer® serum separating tubes (BD Biosciences,
Franklin Lakes, NJ, USA). Serum was collected by centrifugation at
2,000 × g for 10 min at 4°C after allowing the blood to clot for 30
min. Written informed consent was obtained from each patient, and
the consent procedure and study protocol were approved by the
Medical Institutional Ethical Committee of the Provincial Hospital
Affiliated to Shandong University.
Clinical features
Clinical features including histological grade,
metastasis, cirrhosis, and complete tumor capsule were reported by
pathologists following post-surgical pathological examination of
HBV-associated HCC samples.
miRNA/mRNA extraction and RT-qPCR
The mRNAiso Plus and microRNAiso Plus kits (Takara
Biotechnology Co., Ltd., Dalian, China) were used for mRNA and
miRNA extraction from liver tissue, respectively. The MiRCURY RNA
kit (Exiqon A/S, Vedbaek, Denmark), a specific kit for extracting
microRNA from the serum, was used to extract miRNA from serum
samples in order to guarantee the validation of the concentration
of microRNAs. The PrimeScript™ RT Reagent kit and SYBR
Premix Ex Taq (Takara Biotechnology Co., Ltd.) were used for
first-strand cDNA synthesis and RT-qPCR, respectively, according to
the manufacturer's protocols. β-actin was used for mRNA
normalization. β-actin forward, 3′-GGCACCACACCTTCTACAATG-5′ and
reverse, 3′-TAGCACAGCCTGGATAGCAAC-5′. RT-qPCR for microRNAs was
performed on a 480 PCR system (Roche Diagnostics, Basel,
Switzerland), at 95°C for 1 min followed by 45 cycles of 95°C for 5
sec, 65°C for 30 sec and 72°C for 30 sec. The expression of miRNAs
in serum samples was calculated using the comparison
2−∆∆Cq method (18)
relative to RNU6B (U6). The following specific forward and reverse
primers were used in pairs when amplifying miRNA: miR-143 forward,
3′-GTGCAGTGCTGCATCTCTGGT-5′; 143HG forward,
3′-GTGAAGGCAGAGGACACACCT-5′ and reverse, 3′-AAAACCGTGCATTTGGCTG-5′.
The forward primers for miR-145 and U6 and the reverse primers for
all microRNAs were purchased as finished products from Takara
Biotechnology Co., Ltd. (cat. nos. SJD14025 and SJD14078). PCR
product sizes were validated by electrophoresis using 2% agarose
and 12% polyacrylamide, 8 M urea gel (19). All kits were used according to the
manufacturer's protocol. All the experiments were performed >3
times generating similar results and error bars represent the
standard error of the mean of 6 repeats.
Statistical analysis
SPSS software (version 16.0; SPSS, Inc., Chicago,
IL, USA) was used to analyze the data. Comparisons between matched
HBV-associated HCC tissue and non-tumor tissue was performed by
paired Student's t-test and Wilcoxon signed-rank test. Unmatched
continuous data were compared using an independent two-tailed
t-test. The association between the expression of miR-143, miR-145
and MIR143HG is analyzed by Pearson correlation in HBV-associated
HCC tissues. The diagnostic value of serum miR-143/145 expression
was assessed by the area under the ROC curves (AUC). The diagnostic
accuracy was assessed by sensitivity, specificity, positive
predictive value and negative predictive value. P<0.05 was
considered to indicate a statistically significant difference.
Results
miRNA expression profile analysis in
patients with HBV-associated HCC reveals that miR-143 and miR-145
are among the 63 differentially expressed miRNAs
The HBV-associated HCC and non-tumor tissue miRNA
ChIP data were obtained from the TCGA and the GEO datasets
(16,17). It was identified that in the
HBV-associated HCC samples, 24 miRNAs were increased >1.5-fold
(P<0.01) and 39 miRNAs were decreased >1.5-fold compared with
the matched adjacent non-tumor samples. miR-143 and miR-145 were
among the 39 significantly decreased miRNAs in the HBV-associated
HCC samples (Fig. 1). The expression
of miR-145 was significantly downregulated in the HBV-associated
HCC samples compared with the non-tumor samples from the GSE22058
(P<0.001) and the TCGA datasets (P<0.001). Furthermore, the
expression of miR-143 was significantly downregulated in the
HBV-associated HCC samples compared with the non-tumor samples from
the GSE22058 (P<0.001) and the TCGA datasets (P=0.004).
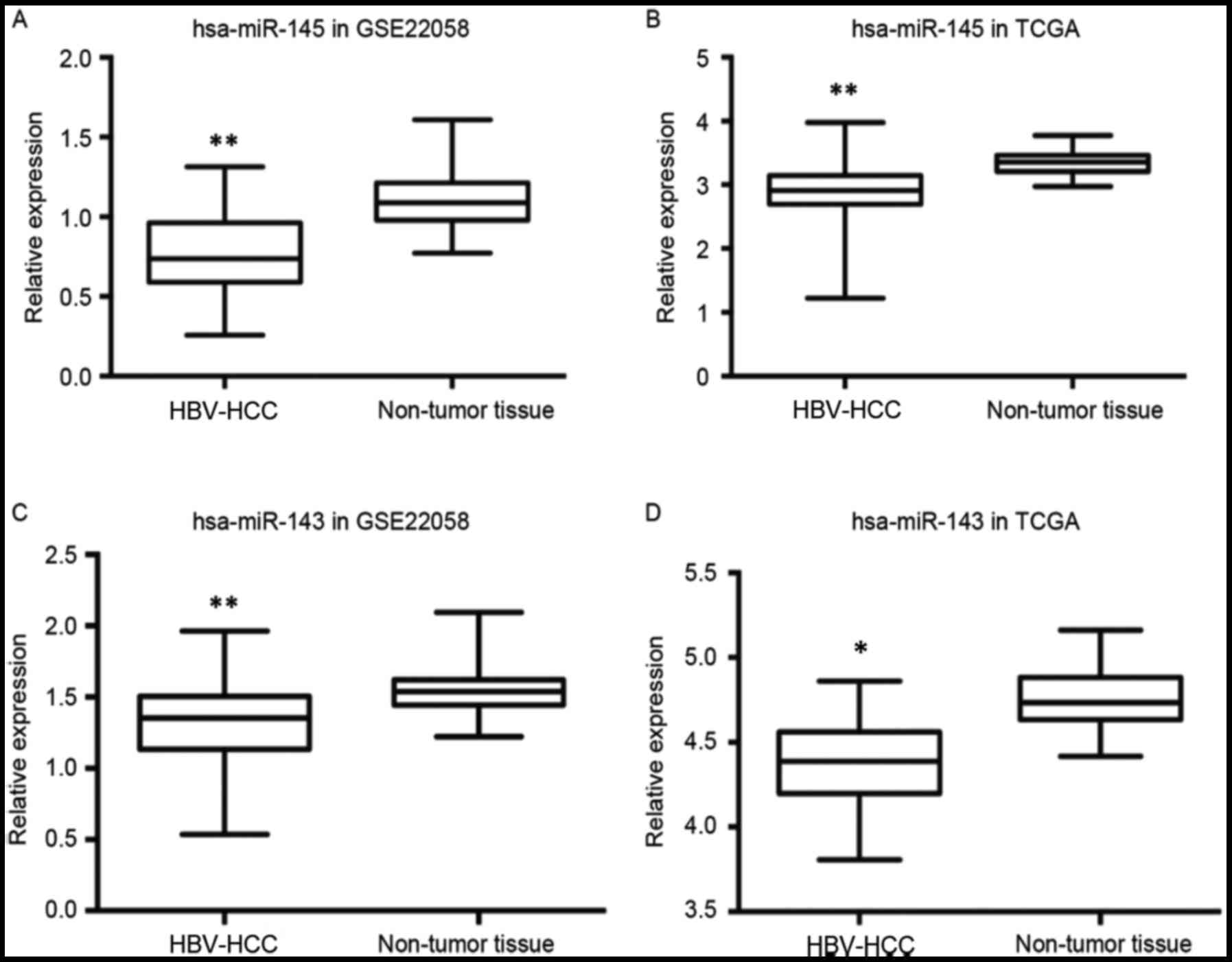 | Figure 1.hsa-143/145 expression analysis using
miRNA ChIP data from the GEO and the TCGA databases. (A) The
expression of miR-145 was decreased in the HBV-associated HCC
samples (n=96, 88% positive for HBV antigen) compared with the
matched adjacent non-tumor tissues (n=96) in the GSE22058 database
(**P<0.001, Wilcoxon signed-rank test). (B) The expression of
miR-145 was significantly decreased in the HBV-associated HCC
samples (n=372) compared with the non-tumor liver samples (n=50) in
the TCGA dataset (**P<0.001; Wilcoxon signed-rank test). (C) The
expression of miR-143 was decreased in the HBV-associated HCC
samples (n=96, 88% positive for HBV antigen) compared with the
matched adjacent non-tumor tissues (n=96) in the GSE22058 database
(**P<0.001, Wilcoxon signed-rank test). (D) The expression of
miR-143 was decreased in the HBV-associated HCC samples (n=102)
compared with the non-tumor liver samples (n=50) in the TCGA
dataset (P=0.004, Wilcoxon signed-rank test). *P<0.01. hsa,
Homo sapiens; miRNA/miR, microRNA; GEO, Gene Expression
Omnibus; TCGA, The Cancer Genome Atlas; ChIP, chromatin
immunoprecipitation; HBV, hepatitis B virus; HCC, hepatocellular
carcinoma. |
miR-143 and miR-145 are downregulated
in HBV-associated HCC samples and their expression pattern is
consistent with the expression pattern of the host gene,
MIR143HG
To validate the expression profiles of miR-143 and
miR-145 and investigate the association with their host gene
MIR143HG (Fig. 2A), the expression of
the miRNAs and the host gene MIR143HG long non-coding RNA (lncRNA)
were examined using RT-qPCR in HCC and normal tissues. As presented
in Fig. 2B, miR-143 expression was
significantly decreased in HCC tissue compared with matched distal
non-cancerous liver tissue (NT) (HCC=0.075 vs. NT=0.205; P<0.05,
Student's t-test). Furthermore, miR-145 expression was
significantly decreased in HCC tissue when compared with NT
(HCC=0.055 vs. NT=0.232; P<0.01, Student's t-test). No
significant differences in miR-143 and miR-145 expression were
identified between NT and negative control samples (NC). These
results suggest that decreased miR-143 and miR-145 expression may
be an indicator of HBV-associated HCC tumorigenesis.
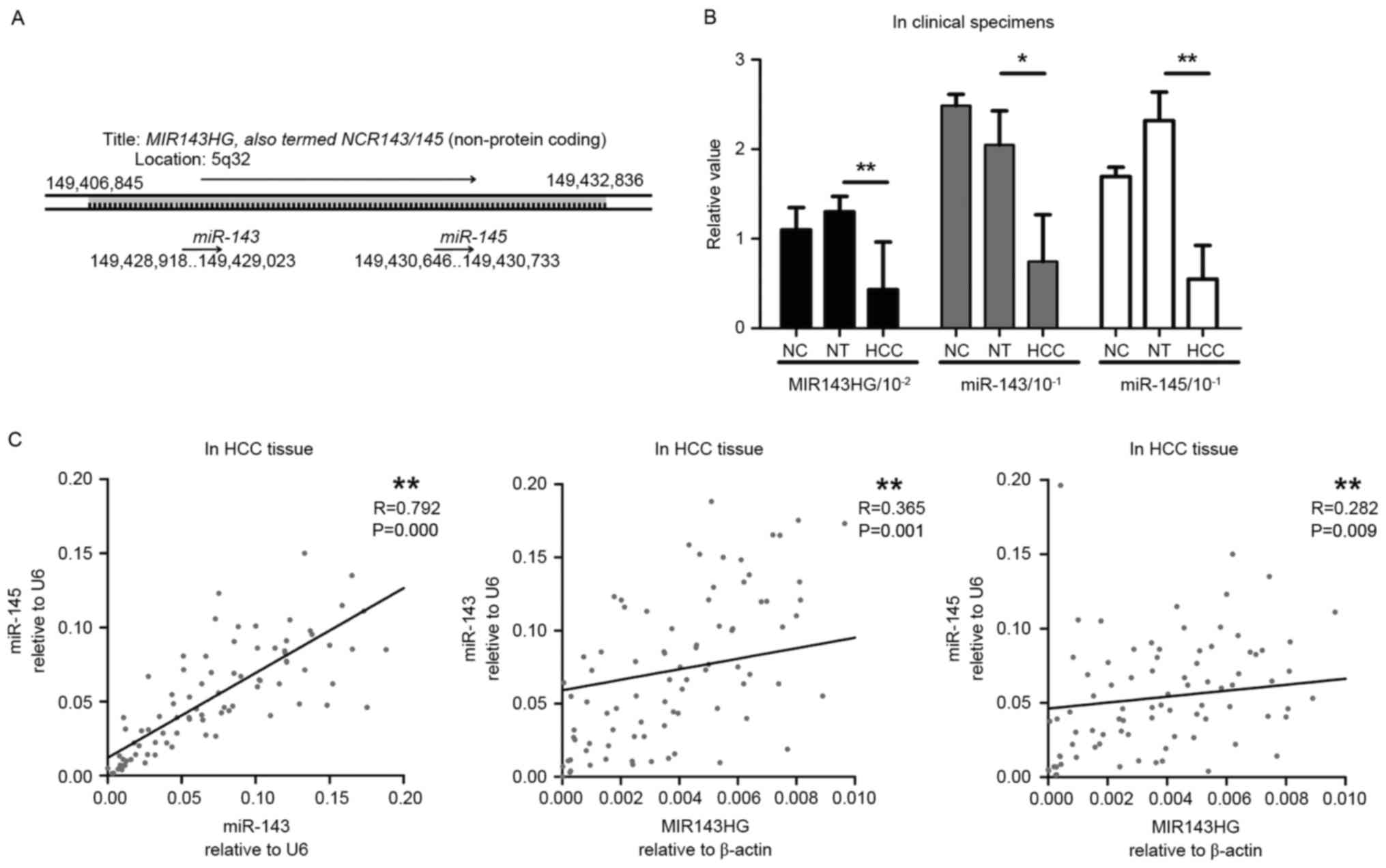 | Figure 2.Expression patterns of MIR143HG,
miR-143 and miR-145 in HBV-associated HCC tissue. (A) The genomic
location of MIR143HG, which encodes miR-143 and miR-145 (7). (B) The expression patterns of MIR143HG,
miR-143 and miR-145 in HBV-associated HCC tissue (n=85), NT (n=85)
and NC (n=10). Expression levels were determined using RT-qPCR. U6
snRNA was used for miRNA normalization. Results are presented as
the mean ± standard error of the mean. *P<0.05, **P<0.01,
Student's t-test. (C) Pearson correlation between miR-143, miR-145
and MIR143HG expression in HBV-associated HCC tissue. **P<0.01.
miRNA/miR, microRNA; HBV, hepatitis B virus; HCC, hepatocellular
carcinoma; NT, non-cancerous liver tissue; NC, negative control;
RT-qPCR, reverse transcription-quantitative polymerase chain
reaction. |
To investigate the association between miR-143,
miR-145 and their host gene MIR143HG, MIR143HG expression was
quantitatively assessed. As presented in Fig. 2B, MIR143HG expression was decreased in
HCC tissue compared with NT (HCC=0.43). Linear regression analysis
indicated that the expression patterns of miR-143 and miR-145 were
consistent with that of their host gene, MIR143HG (Fig. 2C) (miR-145 vs. miR-143; P<0.001,
miR-143 vs. MIR143HG; P=0.001, miR-145 vs. MIR143HG; P=0.009).
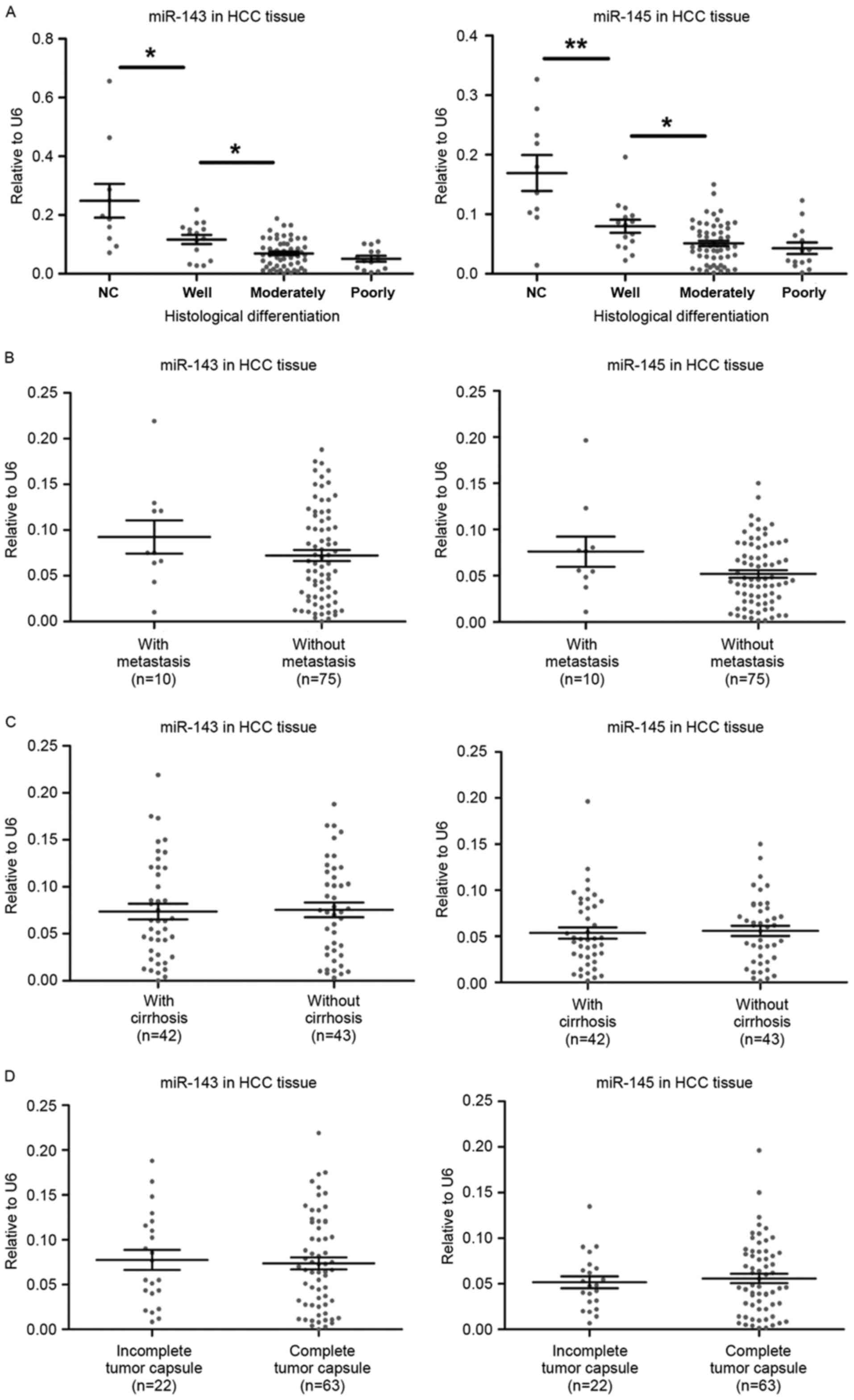
miR-143 and miR-145 expression is
associated with the histological differentiation of HBV-associated
HCC tumors
To investigate the role of miR-143/145 in
HBV-associated HCC, the expression level of the miRNAs was
associated with key clinical features including tumor progression
and disease prognosis. miR-143/145 expression was not associated
with the histological grade of the HBV-associated HCC samples.
However, as presented in Fig. 3A, the
moderately differentiated group had significantly increased miR-143
expression compared with the well-differentiated group (P=0.026).
Similarly, miR-145 expression in the moderately differentiated
group was increased compared with the well-differentiated group
(P=0.012). miR-143 and miR-145 were significantly overexpressed in
the well-differentiated group compared with the negative control
(P=0.004 and P=0.016, respectively). The expression of the miRNAs
was also associated with the presence of metastasis, cirrhosis and
complete tumor capsule (Fig. 3B-D);
however, no statistically significant association was detected.
Similarly, no statistically significant association was identified
with age or sex (data not shown). These results demonstrate that
decreased miR-143 and miR-145 expression is associated with tumor
differentiation and may be a useful indicator of poor prognosis for
patients with HBV-associated HCC.
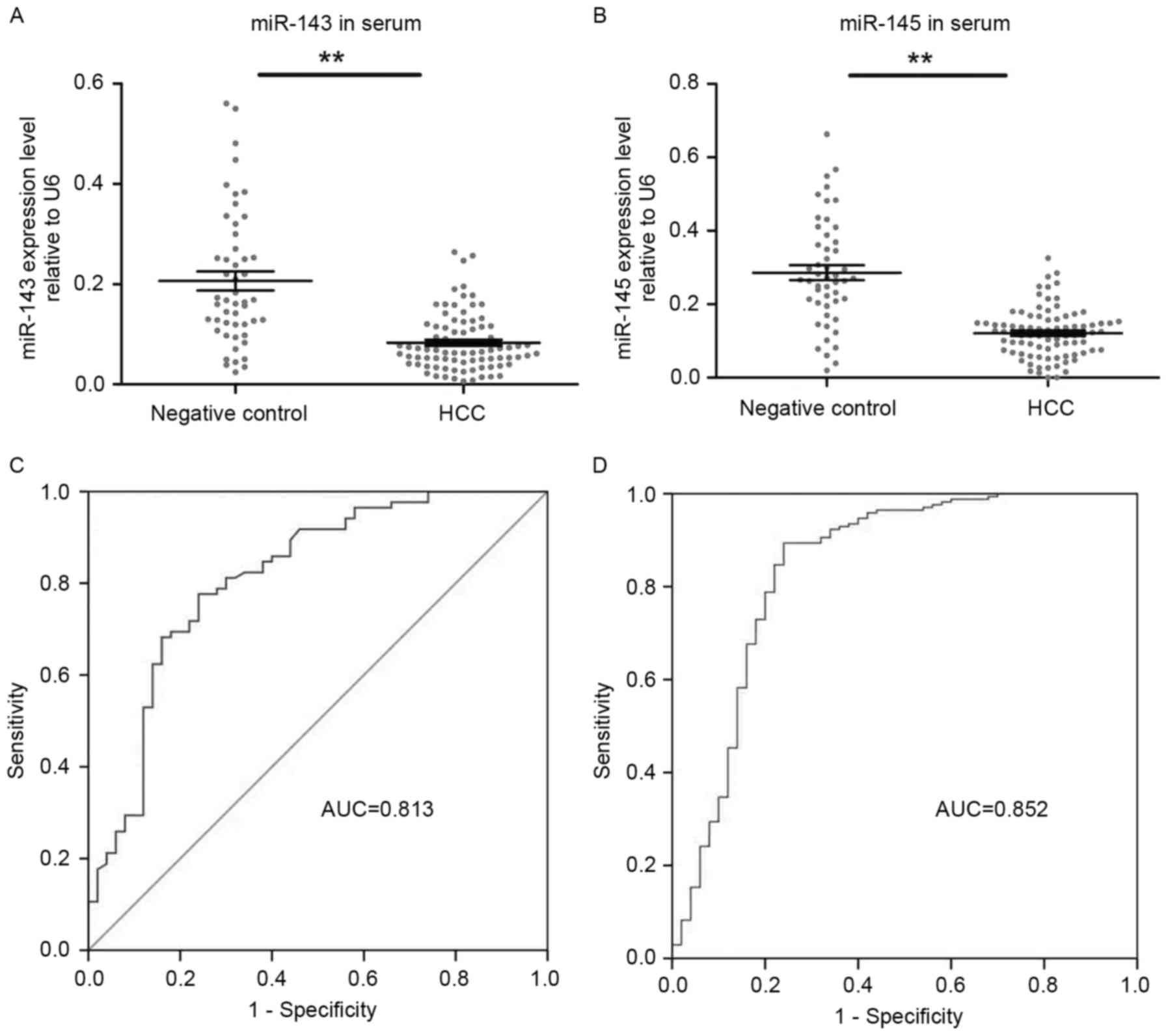
Serum levels of miR-143 and miR-145
are decreased in patients with HBV-associated HCC and may serve as
a potential diagnostic biomarker for HBV-associated HCC
The results of the present study confirmed that
miR-143 and miR-145 were decreased in HBV-associated HCC tissue. To
evaluate their use in clinical applications, their expression was
quantified in serum prior to and following surgical tumor excision.
As presented in Fig. 4A, miR-143
expression was identified to be significantly decreased in the
serum of patients with HBV-associated HCC compared with NC
(HCC=0.06 vs. NC=0.22; P<0.01, independent Student's t-test).
The miR-145 expression was also identified to be significantly
decreased in the serum of patients with HBV-associated HCC compared
with NC (HCC=0.13 vs. NC 0.27; P<0.01; Fig 4B).
Subsequently, the serum miR-143 and miR-145 RT-qPCR
data were used to generate ROC curves for each miRNA and determine
the respective AUC. As presented in Fig.
4C and D, the AUC of the miR-143 ROC curve was 0.813±0.04 (mean
± standard error of the mean; 95% confidence interval, 0.735–0.892;
P<0.01) and that of the miR-145 ROC curve was 0.852±0.039 (mean
± standard error of the mean; 95% confidence interval, 0.776–0.928;
P<0.05).
A threshold for each miRNA was selected to predict
HCC between patients with HBV infection. The 1.194 miR-145
threshold identified 88.2% of patients with HBV-associated HCC with
a comparatively lower specificity of 78%. The 0.118 miR-143
threshold was characterized by 77.6% sensitivity and 86%
specificity (Fig. 5). The results of
the present study indicate that the two miRNAs may potentially be
used to predict HCC in patients infected with HBV, with a
sensitivity and specificity of ~0.80, according to the
recommendations of the International Mesothelioma Interest Group
for MPM biomarkers (20).
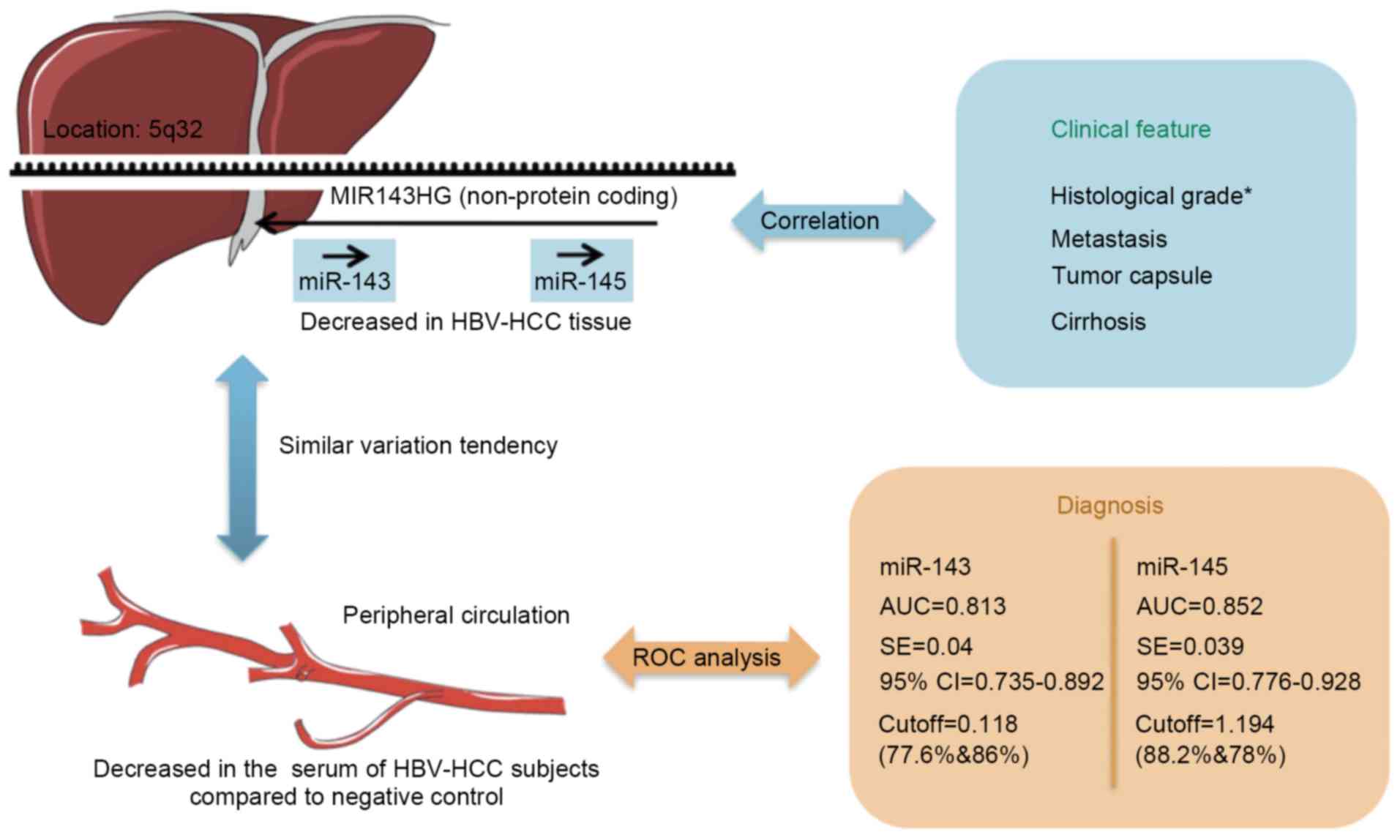 | Figure 5.Consolidated model for the role of
miR-143 and miR-145 in HBV-associated HCC. In this model, the
expression of miR-143, miR-145 and their host gene MIR143HG is
decreased in HBV-associated HCC tissue, and their expression is
associated with the histological grade of the tumor (*P<0.05).
Serum miR-143 and miR-145 are decreased in patients with
HBV-associated HCC compared with NC, and may serve as potential
diagnostic biomarkers to predict tumorigenesis in patients infected
with HBV. miRNA/miR, microRNA; HBV, hepatitis B virus; HCC,
hepatocellular carcinoma; ROC, receiver-operating characteristic;
AUC, area under the ROC curve; SE, standard error; CI, confidence
interval. |
Discussion
According to GenBank, miR-143 and miR-145 are
encoded by the MIR143HG gene, which is a non-protein coding gene.
miR-143 and miR-145 have distinct sequences, and are located in
proximity within the MIR143HG gene. Previous studies have
demonstrated decreased miR-143/145 expression in colonic carcinoma,
pulmonary carcinoma, esophageal carcinoma and prostatic carcinoma
(9–12). However, their expression profile in
HCC remains controversial. A previous study demonstrated increased
expression (8), whereas decreased
expression was demonstrated in others (14,15). By
contrast, a previous study demonstrated that miR-143/145 expression
was markedly decreased in liver tissue (7). To investigate this further, miR-143 and
miR-145 expression was examined in tissue and serum samples of
patients with HBV-associated HCC. In addition, serum miRNA data
were used to evaluate the potential clinical application of miRNAs
as biomarkers for HBV-associated HCC.
In the present study, a model was developed to
describe the role of the miR-143/145 cluster in HBV-associated HCC.
As Fig. 5 indicates, the expression
of miR-143 and miR-145 was significantly decreased in tumor tissue
of patients with HBV-associated HCC compared with non-tumor distal
tissue. This was particularly evident in patients with moderately
and poorly differentiated tumors. The miR-143 and miR-145 serum
levels were also decreased in patients with HBV-associated HCC in
accordance with the expression profile observed in the tumor
tissues. By contrast, the expression of MIR143HG in the serum
presented no significant alteration in patients with HBV-associated
HCC compared with NC, even though MIR143HG was identified to be
downregulated in tumor tissue. The ROC curves generated from
miR-143 and miR-145 serum expression data indicated that serum
miR-143/145 expression may be a promising biomarker for predicting
tumorigenesis in patients with HBV infection.
When examining the expression of a miRNA, its
expression in tissue samples is first determined. However, when
investigating its potential clinical application, examination of
serum miRNA levels is required. HBV-associated HCC is a serious
condition, characterized by high morbidity and mortality, and early
detection is crucial for effective surgical excision. Since the HCC
biomarkers used currently, including serum α-fetoprotein and the
carcinoembryonic antigen, have limited sensitivity and specificity,
novel biomarkers are required to improve the diagnosis and
management of patients with liver diseases. It has been reported
that tumor-derived miRNAs are frequently released into the
peripheral blood circulation and remain stable, protected from
RNase (21). The results of the
present study support this hypothesis. It was demonstrated that
serum miR-143/145 expression is consistent with the expression in
tumor tissue and the expression levels of miR-143/145 in patients
with HBV-associated HCC were decreased compared with NC. Taking
into consideration the ROC curves as well, the results of the
present study support the hypothesis that serum miR-143/145 may be
a potential diagnostic biomarker for HBV-associated HCC. Further
research is currently underway to validate this hypothesis and
elucidate the underlying molecular mechanism of
miR-143/145-mediated regulation of tumor biology.
Using miRNA ChIP and RT-qPCR data, the results of
the present study confirmed that miR-143/145 and their host gene
MIR143HG are downregulated in tumor tissue of patients with
HBV-associated HCC. It was also demonstrated that the expression of
miR-143/145 was associated with the histological differentiation of
HBV-associated HCC tissues. In addition, serum levels of miR-143
and miR-145 were examined and it was demonstrated that the two
miRNAs were decreased in patients with HBV-associated HCC compared
with NC. On the basis of the ROC curves, the results of the present
study suggest that serum miR-143 or miR-145 expression may be a
valuable diagnostic biomarker for predicting tumorigenesis in
patients with HBV-associated HCC.
Acknowledgements
The present study was supported by the Natural
Science Foundation of Shandong Province (grant nos. ZR2016HB06 and
ZR2017MH035) and the Medical Science and Technology Development
Plan of Shandong Province (grant no. 2016WS0413).
References
|
1
|
Bréchot C, Gozuacik D, Murakami Y and
Paterlini-Bréchot P: Molecular bases for the development of
hepatitis B virus (HBV)-related hepatocellular carcinoma (HCC).
Semin Cancer Biol. 10:211–231. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhao Q, Li T, Qi J, Liu J and Qin C: The
miR-545/374a cluster encoded in the Ftx IncRNA is overexpressed in
HBV-related hepatocellular carcinoma and promotes tumorigenesis and
tumor progression. PLoS One. 9:e1097822014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dimitrova N, Gocheva V, Bhutkar A, Resnick
R, Jong RM, Miller KM, Bendor J and Jacks T: Stromal expression of
miR-143/145 promotes neoangiogenesis in lung cancer development.
Cancer Discov. 6:188–201. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Croce CM: Causes and consequences of
microRNA dysregulation in cancer. Nat Rev Genet. 10:704–714. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Callegari E, Gramantieri L, Domenicali M,
D'Abundo L, Sabbioni S and Negrini M: MicroRNAs in liver cancer: A
model for investigating pathogenesis and novel therapeutic
approaches. Cell Death Differ. 22:46–57. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu H, Zhu L, Liu B, Yang L, Meng X, Zhang
W, Ma Y and Xiao H: Genome-wide microRNA profiles identify miR-378
as a serum biomarker for early detection of gastric cancer. Cancer
Lett. 316:196–203. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Iio A, Nakagawa Y, Hirata I, Naoe T and
Akao Y: Identification of non-coding RNAs embracing
microRNA-143/145 cluster. Mol Cancer. 9:1362010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang X, Liu S, Hu T, Liu S, He Y and Sun
S: Up-regulated MicroRNA-143 transcribed by nuclear factor kappa B
enhances hepatocarcinoma metastasis by repressing fibronectin
expression. Hepatology. 50:490–499. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fan X, Chen X, Deng W, Zhong G, Cai Q and
Lin T: Up-regulated microRNA-143 in cancer stem cells
differentiation promotes prostate cancer cells metastasis by
modulating FNDC3B expression. BMC Cancer. 13:612013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ma Q, Jiang Q, Pu Q, Zhang X, Yang W, Wang
Y, Ye S, Wu S, Zhong G, Ren J, et al: MicroRNA-143 inhibits
migration and invasion of human non-small-cell lung cancer and its
relative mechanism. Int J Biol Sci. 9:680–692. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ni Y, Meng L, Wang L, Dong W, Shen H, Wang
G, Liu Q and Du J: MicroRNA-143 functions as a tumor suppressor in
human esophageal squamous cell carcinoma. Gene. 517:197–204. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Qian X, Yu J, Yin Y, He J, Wang L, Li Q,
Zhang LQ, Li CY, Shi ZM, Xu Q, et al: MicroRNA-143 inhibits tumor
growth and angiogenesis and sensitizes chemosensitivity to
oxaliplatin in colorectal cancers. Cell Cycle. 12:1385–1394. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang R, Wang L and Yang AG: Is
microRNA-143 really a turncoat of tumor suppressor MicroRNA in
hepatitis B virus-related hepatocellular carcinoma? Hepatology.
50:987–988. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Noh JH, Chang YG, Kim MG, Jung KH, Kim JK,
Bae HJ, Eun JW, Shen Q, Kim SJ, Kwon SH, et al: MiR-145 functions
as a tumor suppressor by directly targeting histone deacetylase 2
in liver cancer. Cancer Lett. 335:455–462. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang Y, Hu C, Cheng J, Chen B, Ke Q, Lv Z,
Wu J and Zhou Y: MicroRNA-145 suppresses hepatocellular carcinoma
by targeting IRS1 and its downstream Akt signaling. Biochem Biophys
Res Commun. 446:1255–1260. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Edgar R, Domrachev M and Lash AE: Gene
expression omnibus: NCBI gene expression and hybridization array
data repository. Nucleic Acids Res. 30:207–210. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Burchard J, Zhang C, Liu AM, Poon RT, Lee
NP, Wong KF, Sham PC, Lam BY, Ferguson MD, Tokiwa G, et al:
microRNA-122 as a regulator of mitochondrial metabolic gene network
in hepatocellular carcinoma. Mol Syst Biol. 6:4022010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shi R and Chiang VL: Facile means for
quantifying microRNA expression by real-time PCR. Biotechniques.
39:519–525. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Schetter AJ, Leung SY, Sohn JJ, Zanetti
KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK, et
al: MicroRNA expression profiles associated with prognosis and
therapeutic outcome in colon adenocarcinoma. JAMA. 299:425–436.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sun HH, Vaynblat A and Pass HI: Diagnosis
and prognosis-review of biomarkers for mesothelioma. Ann Transl
Med. 5:2442017. View Article : Google Scholar : PubMed/NCBI
|