Introduction
Lung cancer stem cells (LCSCs) have an important
role in the development of lung cancer; therefore, LCSCs have been
the subject of numerous recent studies (1–3). According
to certain studies, LCSCs originate primarily from normal tissue
stem cells or the de-differentiation of normal cancer cells
(3,4).
Prior investigations of LCSCs have been conducted using cancer cell
lines or patient primary tumor tissue samples, of which, the former
is more often used due to easier access. LCSCs are promising
targets for lung cancer therapy, and it is crucial to improve
available techniques for their identification from normal cancer
cells. The current methods of LCSC collection include fluorescent
activated cell sorting (FACS), magnetic activated cell sorting
(MACS), sphere-forming assay, the establishment of lung cancer cell
lines with stem cell properties and bacterial surface display
library screening. Among these, FACS is the most commonly used
technique. Consequently, the specific markers for isolating LCSCs
became the subject of a number of studies (5–11). ATP
binding cassette superfamily G member 2 (ABCG2), acetaldehyde
dehydrogenase (ALDH), CD133, CD166, CD44, C-X-C chemokine receptor
type 4 (CXCR4) and interleukin-6 receptor (IL-6R) and other
proteins (as shown in Table I) have
all previously been identified as markers of LCSCs, and these
markers may be utilized in further studies to identify LCSCs.
 | Table I.Ratios of LCSCs in lung cancer cell
lines reported in various studies. |
Table I.
Ratios of LCSCs in lung cancer cell
lines reported in various studies.
| Cancer type | Cell line | Marker | Proportion of cell
line (%) | (Refs.) |
|---|
| NSCLC | A549 | SP | 18.1 | (48) |
|
|
|
| 16.82 | (47) |
|
|
|
| 2.55 | (84) |
|
|
|
| ~4 | (49) |
|
|
|
| 0.9 | (50) |
|
|
|
| 24 | (51) |
|
|
|
| 2–4 | (40) |
|
|
|
| 4–10 | (65) |
|
|
| IL-6R | 1.4 | (47) |
|
|
| D133 | 0.3 | (47) |
|
|
|
| ~0.5 | (70) |
|
|
|
| 3.9 | (27) |
|
|
|
| 2 | (71) |
|
|
|
| 0.2 | (72) |
|
|
|
| 0.3–1 | (65) |
|
|
|
CD133+CD328+ | / | (73) |
|
|
|
| 0.93 | (49) |
|
|
|
CD133+CD326+ | 0.14 | (74) |
|
|
|
| 0.25 | (75) |
|
|
| ALDH | / | (76) |
|
|
|
| 4.2 | (49) |
|
|
|
| 2–8 | (65) |
|
|
|
CD44+CD24− | 27.92 | (49) |
|
|
|
CD166+CD44+ | 62.5 | (77) |
|
|
|
CD166+CD326+ | 9.8 | (77) |
|
| H1650 | SP | 7 | (78) |
|
|
|
CD44+CD24− | 0.76 | (78) |
|
|
| CD133 | ~2.5 | (78) |
|
| H460 | SP | / | (67) |
|
|
|
| 24.19 | (47) |
|
|
|
| 5.2 | (79) |
|
|
|
| 5.6 | (69) |
|
|
|
| 3.5 | (80) |
|
|
|
| 2–4 | (40) |
|
|
| IL-6R | 0.04 | (47) |
|
|
| CD133 | 0.82 | (47) |
|
|
|
| 1.1 | (81) |
|
|
| ALDH | ~3 | (42) |
|
| H23 | SP | 77.25 | (47) |
|
|
|
| ~1.5 | (69) |
|
|
| IL-6R | 0.74 | (69) |
|
|
| CD133 | 0.17 | (69) |
|
| H522 | ALDH | 29.66 | (26) |
|
| H1299 | CD44 | 81.3 | (56) |
|
| H322 | ALDH | 0.6 | (42) |
|
| H125 | ALDH | 2.9 | (42) |
|
| H358 | ALDH | 2.82 | (42) |
|
| HCC827 |
ALDH+CD44+ | 3.04 | (43) |
|
|
| SP | 1.9 | (82) |
|
| H1975 | SP | / | (67) |
|
| H441 | SP | ~6.1 | (68) |
|
|
|
| 0.5–3 | (65) |
|
|
| CD133 | 0.1–0.5 | (65) |
|
|
| ALDH | 0.5–2 | (65) |
|
| H661 | CD133 | ~5.3 | (81) |
|
| H2170 |
| ~2.5 | (69) |
|
| PC-9 | SP | 2.6 | (68) |
|
| LHK2 | SP | 2.8 | (50) |
|
| COR L23 |
CD44+CD24− | 0 | (83) |
| SCLC | Lc817 | SP | 0.4 | (50) |
|
| H146 | SP | ~0.8 | (40) |
|
| H526 | SP | ~0.9 | (40) |
| SCC | HTB58 | SP | ~4.5 | (69) |
|
| 1–87 | SP | 0.8 | (50) |
|
| H2170 |
CD166+CD44+ | 0 | (77) |
|
|
|
CD166+CD326+ | 3.1 | (77) |
Lung cancer and LCSCs
Lung cancer is the most frequent cause of
cancer-associated mortality (12) and
~1.4 million individuals succumb to the disease annually (13). There are two major types of lung
cancer: Small cell lung cancer (SCLC), which accounts for ~15% of
all types of lung cancer (14) and
non-small cell lung cancer (NSCLC), which accounts for ~80% of all
lung cancer cases (15). NSCLC maybe
further divided into two subtypes: Squamous cell carcinoma (30% of
NSCLC cases) and adenocarcinoma (70% of NSCLC cases) (13). A good prognosis is typically predicted
if the patients are diagnosed at a relatively early tumor stage
[according to the National Comprehensive Cancer Network (NCCN)
guidelines] (16,17). However, it is well-known that a number
of cases are diagnosed at the third or fourth NCCN stage and are
associated with a poor survival rate due to inefficacious treatment
options. Studies on poor clinical outcomes identified that the
complex tumor microenvironment, particularly of CSCs, is involved
in the promotion of tumor metastasis (18), drug resistance (19) and resistance to radiotherapy (20).
The CSC model was first proposed in 1997 following
the identification of leukemia stem cells (21), and evidence for the existence of LCSCs
was presented by Giangreco et al (22) in 2009. The properties of LCSCs
include: Drug resistance, self-renewal and the capacity to form
tumors in xenograft mouse models. These features are the current
gold standard for the identification of human LCSCs (23). Other criteria used to identify CSCs
are as follows: CSCs sorted by FACS are able to generate spheres in
non-adherent cultures; more aggressive metastatic properties
determined using a Transwell assay; the formation of CSC colonies
is efficient, as compared with normal cancer cells; certain mRNAs
and proteins, including octamer-binding transcription factor 4
(OCT4), homeobox protein NANOG and sex-determining region Y HMG-box
2 (SOX2) that are associated with cancer stem cells are
overexpressed (24).
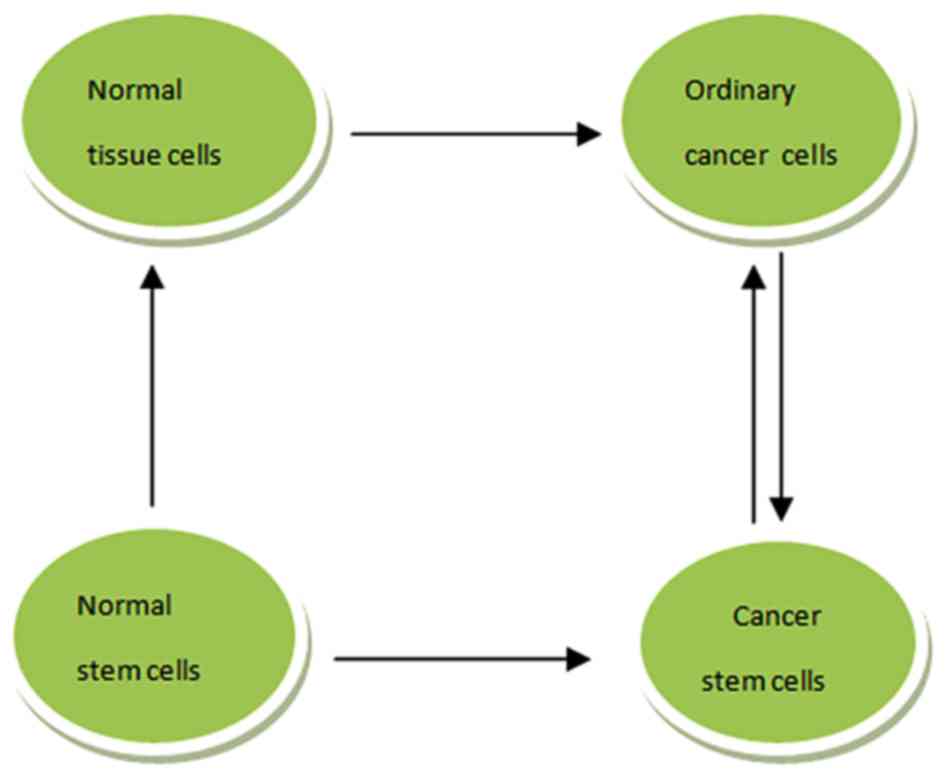
The origin of CSCs is still a matter of debate; the
two hypotheses of CSC sources are presented in Fig. 1. For LCSCs, the possibility of their
origin from normal tissue stem cells was proposed by a previous
study (25). A pulmonary stem cell
population was initially identified at the bronchio alveolar duct
junction and termed bronchio alveolar stem cells (BASCs), on
account of the stem cell-like features (26). Transformation of BASCs to normal lung
cancer stem cells is induced by various carcinogenic factors; it is
also possible for BASCs to transform to LCSCs (26). Additionally, it has been reported that
LCSCs may develop from normal cancer cells that have regained a
capacity for self-renewal following de-differentiation to a
progenitor-like state (27).
Lung cancer cell lines are used for studies
of LCSCs
Prior investigations into LCSCs have been primarily
conducted using various cancer cell lines or primary patient tissue
samples (28,29). Studies of LCSCs using patient tissue
samples are the gold standard; however, these are difficult to
regularly obtain (30) and tissue
samples from patients with early NCCN stage cancer often possess
varying quantities of non-malignant cells (31). Therefore, lung cancer cell lines are
more often used to study LCSCs, rather than patient tumor
tissues.
In the present study, the American Type Culture
Collection (ATCC, Manassas, VA, USA) was searched for lung cancer
cell lines, with a total of 213 subsequently presented. The number
of individual lung cancer cell lines is almost the largest amongst
epithelial cancer cell types, and 20% of cancer cell lines in the
Sanger database (www.sanger.ac.uk) are of lung cancer origin (32). The establishment of cancer cell lines
begins with the dissociation of tumor tissues using trypsin,
following which the dispersed cells are cultured on plates
(33). A minority of cells are able
to proliferate and form colonies followed by several divisions,
whereas other cells undergo apoptosis (34). These colony-forming cells undergo a
limited number of further divisions and subsequently lose their
ability to undergo mitosis. A few cells, however, are able to
overcome the Hayflick limit and become immortal cancer cell line
(34).
Methods of the identification and collection
of LCSCs in lung cancer cell lines
FACS and MACS
FACS is the most widely used technique for the
identification and sorting of LCSCs (5). Specific monoclonal antibodies, each of
which is highly specific for the target antigen and are readily
coupled to fluorescein, phycobiliproteins or other fluorochromes
(6), combine with LCSC protein
markers on the cell surface or in the cytoplasm. Subsequently, the
cells that express these markers can be selected (5). Single-cell sorting using FACS is the
most efficient method currently available, particularly when the
cells are present in low quantities (6). MACS is also utilized for the sorting of
LCSCs. Compared with FACS, MACS is typically faster for sorting
low-abundance cell populations (7).
As the aim is to identify cancer stem cells that account for low
proportion of all cancer cells, MACS maybe more efficient compared
with FACS despite the higher cost (to use MACS, more tools are
necessary compared with FACS, such as the expensive magnetic
bead).
Sphere-forming assay
As aforementioned, sphere forming is one of the
properties of LCSCs, and the enrichment of LCSCs using this assay
method has been used (8). When
cultured in serum-free medium (DMEM/F12 medium) supplemented with
basic fibroblast growth factor and epidermal growth factor, lung
cancer stem cells are able to form floating spheres, and CSCs are
enriched inside these spheres (8).
Establishment of lung cancer cell
lines with stem cell properties
Human bronchial epithelial cells retain the capacity
to differentiate into basal, mucin-producing and columnar ciliated
epithelial cells; therefore, they may be used for the study of CSCs
(9). A previous study established a
cell line as a CSC line by sorting CD133+ cells from
fresh human lung cancer tissue samples using MACS and subsequently
culturing them in serum-free medium (10).
Bacterial surface-display library
screening
For this method, CSCs are enriched using the
aforementioned sphere-forming assay. The spheres are dissociated
into single cells for incubation with a bacterial library, and are
then able to express green fluorescent protein (GFP) and not bind
to the differentiated cells. FACS is used to identify cells
expressing GFP, which facilitates the selection of cells with
fluorescein isothiocyanate (FITC) fluorescence (11).
Protein markers used for the identification
and sorting of LCSCs
As aforementioned, FACS is the major method
currently used for LCSC identification and sorting; the markers
used for their isolation have been the focus of various studies
(33,35–37). It is
possible to identify LCSC by staining cells using antibodies
against these specific markers, and then to sort them using flow
cytometry (35–37). These markers include: CD133, CD166,
CD44, CXCR4 and IL-6R. Combined markers are also used, including
CD133 and CD44, CD44 and CD24, CD166 and Lin, CD133 and CD326,
CD133 and CD338 (Table I). These are
markers expressed on the surface of lung cancer cells or inside
these cells; the majority are functional enzymes, including ALDH,
HO and GLDC (Table I). The present
review discusses three primary LCSC markers, including ATP-binding
cassette sub-family G member 2 (ABCG2), aldehyde dehydrogenase
(ALDH) and CD133.
ABCG2
A side population (SP) is a group of cells with ABC
transporters responsible for the efflux of drugs and drug-like dyes
(Hoechst 33342), which are able to protect the CSCs from certain
cytotoxic agents (38). The cells
with ABC transporter pumps were initially isolated by flow
cytometry as an SP (39), and
numerous studies have demonstrated that SPs exhibit stemness
(Table I). A previous study on SCLC
revealed that SP cells are highly tumorigenic, exhibiting drug
resistance features and the expressing certain stem cell genes
(40). However, SP cells may not
consist entirely of stem cells as other types of cells are also
able to eliminate the Hoechst dye, and the SP phenotype is affected
by staining time, dye concentration and cellular concentrations
(41). In addition, cytometry gating
strategies used to isolate SP cells lack the consistency of gating
strategies used when staining with markers (41).
ALDH
ALDH has been used as a protein marker of LCSCs by
large number of recent studies (Table
I). Cells with high levels of ALDH are considered to be LCSCs
due to a number of their characteristics including self-renewal,
differentiation capacity, drug resistance, expression of other LCSC
markers and positive results following xenotransplantation into
mice; a small number of ALDHhigh cells, identified and
separated with FACS, was demonstrated to lead to successful growth
of cancer (42). Certain prior
studies have also used a combination of ALDH and other surface
markers to identify LCSC, including
ALDH+CD44+ (43). However, Okudela et al (44) demonstrated the presence of an inverse
association between ALDH1A1 expression levels and tumor
aggressiveness. ALDH1A1 expression levels were identified to be
decreased amongst smokers and patients with poorly differentiated
adenocarcinoma and large cell carcinoma, which suggests that the
functional properties of LCSCs may not be sufficient as independent
markers for identifying stem cells (1).
CD133
To the best of our knowledge, CD133 was the first
LCSC marker identified; however, whether CD133 may be utilized as a
marker of LCSCs is under debate. Meng et al (45) revealed that CD133+ and
CD133− and subpopulations of A549 and H446 cells contain
cancer-initiating cells. Furthermore, CD133+ cells have
been identified in normal tissues, including bone marrow (46).
In fact, a well-accepted marker for a certain cell
line, such as A549, does not exist. It has been demonstrated that
≥3 specific stem cell markers, alone or combined, including
ALDH+, CD133+, CD90+ (2), IL-6R (24), OCT-4 (47), CD44+CD24,
CD133+CD44+ (48),
CD133+CD326+/CD133+EpCAM+,
CD133+CD338+/CD133+
ABCG2+ may be useful candidate markers for
identification of LCSCs (Table
I).
Once specific markers have been identified as useful
for classifying stem cells in other types of cancer, they may
subsequently be used for LCSC sorting. It must be considered
whether the cells that were sorted with various markers are
actually the stem-like cells sought if they exhibit characteristics
previously assumed to be exclusively associated with stem cells. If
so, it is essential to identify those markers that may be the most
useful as this will form the foundation for further studies.
Significant differences in the proportion of
LCSCs in lung cancer cell lines
In the present review, markers of LCSCs previously
presented by various studies were evaluated (Table I). The present study identified that
the proportions of LCSCs in the same lung cancer cell line sorted
with different markers are not the same, and the proportion of
LCSCs differs in the same cell line even the same marker used,
after analysis of a number of studies. Specifically, the proportion
of LCSC is ≤7% with ‘SP’, 0. 76% with
CD44+CD24− and 2.5% with CD133.
CD133+LCSCs counts for ~0.3% as reported by Yi et
al (47); however, Tirino et
al (27) demonstrated that the
proportion of LCSCs observed in the same cell line was ≥10 times
higher, at ~3.9%.
Roudi et al (49) studied expression of several CSC
markers of A549 cells and their result showed that ~68.16 or 54.46%
of the cells expressed CD44 or CD24, 27.92% cells express
CD44+CD24−/low. This study also evaluated
that the proportion of ALDH1+ cells was ~4.20%, while
the proportion of ABCG2+ or CD133+cells was
~0.93%. CD44+/133+ populations were rare. It
must be investigated why the proportion of LCSCs within cell lines
can vary so significantly.
Proportions of LCSCs in certain cell lines
identified with the same markers may vary, partly due to the
distinct environment of each cell population. Furthermore, the LCSC
proportion within specific cells lines may also vary. Cell lines
are established through single cell separation and cultivation;
during the cell lifetime, the manifestations of its phenotype and
protein expression profile must be observed as once a cell line is
grown it acquires mutations, including chromosome loss or gain.
Although cell lines used in the laboratory are originally
identical, they may experience significant differences in cell
passage number, the medium used, the microenvironment, which may
specifically affect the stem cell proportion, and how they are
maintained; therefore, numerous cell features, including the cell
charge, which may affect the electrophoresis potential, and the
chromosome number, may be inconsistent. The SP cell ratio may range
from 0. 9–24% in the A549 cell line according to the results of two
independent studies (50,51). Genetic or epigenetic changes in cancer
cells are necessary, in vivo and in vitro, in order
to allow the cells to adapt to the conditions to which they are
exposed (52). For example, hypoxic
conditions lead to the selection of a hypoxia-resistance phenotype
for cancer stem cells (53). The high
proportion of LCSCs identified in A549 cells identified by Sung
et al (51) may be due to the
A549 cell lines they employed, which are likely to undergo more
rigorous conditions of existence, such as less supplements and
hypoxia. Studies using other types of cancer cells have also
indicated that, while the microenvironment conditions can lead to
the loss of stemness in CSCs, glioma cell lines may lose their
multi-potential differentiation capability and the expression of
certain stem cell markers when cultured even under standard
conditions (54).
CSCs are considered to be a small subpopulation of
cells, typically ≤1% within a given tissue (55); however, numerous studies have
identified a varying proportion of normal cancer cells. According
to a study conducted by Meng et al (45), ≥45% of A549 and H446 cells are
cancer-initiating cells. Using flow cytometry, Leung et al
(56) investigated the expression of
specific proteins considered to be markers of LCSCs in ten lung
cancer cell lines, including CD34, CD44, CD133, BMI1 and OCT4
(56). The results indicated that
CD133+ cancer cells were rare in those cell lines, and
CD133+ cancer cells were only identified in the HCC1833
cell line. CD44+ cancer cells, as determined by the
authors (56), ‘are enriched for stem
cell-like properties’ and counted for 61.7–95.9% in H1650, H1299,
HCC827 and H23 cells. Therefore, a high proportion of cells in
these lines have stem cell-like properties, which indicates that
the majority of these cells are LCSCs.
Possible explanations for the higher proportion of
CSCs observed in certain studies may include the aforementioned
harsh living conditions and an extended culture duration. As
previously established, when cancer cells are cultured for a
relatively long time, the majority will die and a fraction will
survive and become immortal cancer cells (53). The current definition of cancer stem
cells refers them as a subpopulation of cells within tumors that do
not have a growth limit and are the only tumorigenic cells among
all other tumor cell subsets. The longer the culture duration, the
higher the probability that stem cells will be selected to survive,
and a subsequently higher percentage of stem cells within a cell
population may be observed (57).
Controversies within the study of LCSCs in
lung cancer cell lines
Whether lung cancer cells are suitable and
dependable for the study of LCSCs is a matter of debate. Certain
studies have hypothesized that distinction between lung cancer cell
lines and lung cancer tissues still remains. The results of
clinical trials of treatments for patients with lung cancer may
occasionally differ from the results obtained in studies on lung
cancer cell lines. For example, cell lines are utilized in studies
evaluating treatments targeting cancer stem cells; those agents
that were observed to be effective during experiments using cancer
cell lines did not exhibit a corresponding efficacy in vivo
in patients with cancer, due to the differences between cancer cell
lines and cancer tissues in vivo. Although complete response
(CR) has been observed in a proportion of patients, a large
quantity of diverse cancer cells may still remain, often
≥1×109 (58). The mutation
rate in intra-tumoral types of cancer is usually between
1×10−8-1×10−9 per base per cell division, and
this can result in mutations arising in numerous cancer cells
(59). For example, genes concerning
stemness in specific cells are activated and this is the source of
the changes via which normal cancer cells become cancer stem cells.
Cancer cell lines are typically more stable, with a mutation rate
relatively lower than that identified in cancer tissues; this may
partly explain the failure of clinical agents targeting cancer stem
cells that originated and were developed from studies that utilized
cancer cell lines.
Shortcomings of using lung cancer cell lines for the
study of LCSCs originate from the differences between cancer cell
lines and cancer tissues. As compared with tumor cells in tissues,
tumor cell lines are usually cultured in flasks or dishes in 2D,
which may result in changes in gene expression, cell morphology and
chromatin condensation (60). As
reviewed by Gazdar et al (61), there are four principal disadvantages
of using lung cancer cell lines for the study of lung cancer,
including the following: The possible selection of minor tumor cell
subpopulations that may not be representative of the original
population; the potential acceleration of genomic instability; the
absence of stromal, immune and inflammatory cells; the absence of
vascularization; difficulty in evaluating the metastatic potential.
Other limitations include alterations in the DNA (62) and mRNA (63). Standard culture protocols may lead to
the selective growth of rapidly growing cells that may have more
molecular abnormalities than other tumor cells (61). For example, lung carcinoma cell lines
have extensive chromosomal rearrangements, oncogene mutations and
multiple sites of allelic loss and gene amplification (64).
Through reviewing and summarizing a number of
studies on LCSCs conducted during recent years, it was revealed
that the majority of studies utilized adenocarcinoma cell lines,
particularly the A549 cell line, to investigate LCSCs (presented in
Table I). Therefore, our current
understanding of LCSCs has been derived from information obtained
using only a select few cell lines, which cannot be assumed to
represent the various cancer cells present in lung cancer
tissues.
Are protein markers dependable tools for the
identification of LCSCs?
CSCs enriched using diverse methods are dissimilar
in a number of aspects. One study demonstrated that when
CD133+ and SP cells sorted from A549 cells were cultured
in adherent dishes for 3 weeks with cancer stem cell medium, the
CD133+ cell subpopulation decreased, while the SP
population increased (65). The
author also observed that the SP cells positive for ALDH, or CD133
are also present in low numbers. Furthermore, self-renewal and
metastatic gene expression patterns were revealed to be distinct
between SP, CD133+ and ALDHhigh cells. SP and
CD133+ cells demonstrated increased lung colonization,
as compared with their negative counterparts, while the injection
of ALDHhigh cells into xenograft mice induced numerous
liver metastases (65). Another
phenomenon in which the number of cells sorted within bacterial
surface display library screening are CD133 positive at the same
time the H460 cells as described above is very rare, suggesting
that there may be no common markers for CSCs (66). Akunuru et al (65) revealed the following: SP and non-SP
cells include CD133+ or ALDHhigh members
although non-SP cells express higher levels of each;
ALDHhigh and ALDHlow cells, or SP and non-SP
cells, are capable of forming spheres; non-SP cells can generate
tumors in immune-compromised NSG mice after 8 weeks (3). These results indicate that cells that
are negative for one cancer marker may still be positive for
another.
Hypothetically, if LCSCs may be able to
differentiate to numerous types of progeny cells and express a
variety of markers, and cells with certain markers can be excluded
from the leader cells.
Conclusion
The method most commonly employed to determine if a
specific protein can be used as a marker of LSCs includes the
following steps: First, utilize the fluorescent antibody
corresponding to the target marker to label the cells considered to
be stem cells, then sort them using FACS or MACS; second, culture
the cells to determine whether they can generate spheres, have the
capability of forming clones more effectively compared with normal
cells and are more aggressive; third, transplanting CSCs into
xenograft mice at a relatively small amount to test the
tumorigenicity. Sorting of the CSCs is considered to be the key
step, as this technique can distinguish CSCs from normal cancer
cells directly.
To the best of our knowledge, there is currently no
evidence that any combination of cancer stem-cell markers isolates
any cancer stem-cell population to a high degree of purity
(46). There are certain problems
associated with LCSC studies: Tumor cell lines are used as the
principal model in CSC studies, as aforementioned; marker
expression and cell subsets may vary within the same cell line, so
the results are not directly comparable and cannot be used as a
reference; the proportion of LCSCs identified with certain marker
expression is inconsistent between and within cell lines, which
raises the question of whether those cells are true LCSCs or if
they are other cell types with only specific CSC-associated
characteristics; the gold standard for the identification of LCSCs
includes successful xenotransplantation into immunodeficient mice.
However, immune barriers remain in nude mice despite the lack of T
cells (66). It is possible that
LCSCs have been eliminated by the immune system and the tumor cells
derive from daughter cells of LCSCs; the ability of a cell to form
a tumor is dependent upon the microenvironment, and cells that can
form tumors under one set of conditions may not form tumors under
other conditions (4). Therefore,
further studies are required.
Glossary
Abbreviations
Abbreviations:
|
LCSCs
|
lung cancer stem cells
|
|
BASCs
|
bronchio alveolar stem cells
|
|
FACS
|
fluorescence activated cell
sorting
|
|
MACS
|
magnetic activated cell sorting
|
|
ALDH
|
acetaldehyde dehydrogenase
|
References
|
1
|
Singh S and Chellappan S: Lung cancer stem
cells: Molecular features and therapeutic targets. Mol Aspects Med.
39:50–60. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yan X, Luo H, Zhou X, Zhu B, Wang Y and
Bian X: Identification of CD90 as a marker for lung cancer stem
cells in A549 and H446 cell lines. Oncol Rep. 30:2733–2740. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Akunuru S, Zhai James Q and Zheng Y:
Non-small cell lung cancer stem/progenitor cells are enriched in
multiple distinct phenotypic subpopulations and exhibit plasticity.
Cell Death Dis. 3:e3522012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Meacham CE and Morrison SJ: Tumour
heterogeneity and cancer cell plasticity. Nature. 501:328–337.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dionne LK, Driver ER and Wang XJ: Head and
neck cancer stem cells: From identification to tumor immune
network. J Dent Res. 94:1524–1531. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Herzenberg LA, Parks D, Sahaf B, Perez O,
Roederer M and Herzenberg LA: The history and future of the
fluorescence activated cell sorter and flow cytometry: A view from
Stanford. Clin Chem. 48:1819–1827. 2002.PubMed/NCBI
|
|
7
|
Xu Y, Zhao W, Wu D, Xu J, Lin S, Tang K,
Yin Z and Wang X: Isolation of myeloid-derived suppressor cells
subsets from spleens of orthotopic liver cancer-bearing mice by
fluorescent-activated and magnetic-activated cell sorting:
Similarities and differences. Int J Clin Exp Pathol. 7:7545–7553.
2014.PubMed/NCBI
|
|
8
|
Izumiya M, Kabashima A, Higuchi H,
Igarashi T, Sakai G, Iizuka H, Nakamura S, Adachi M, Hamamoto Y,
Funakoshi S, et al: Chemoresistance is associated with cancer stem
cell-like properties and epithelial-to-mesenchymal transition in
pancreatic cancer cells. Anticancer Res. 32:3847–3853.
2012.PubMed/NCBI
|
|
9
|
Vaughan MB, Ramirez RD, Wright WE, Minna
JD and Shay JW: A three-dimensional model of differentiation of
immortalized human bronchial epithelial cells. Differentiation.
74:141–148. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kucerova L, Feketeova L, Kozovska Z,
Poturnajova M, Matuskova M, Nencka R and Babal P: In vivo
5FU-exposed human medullary thyroid carcinoma cells contain a
chemoresistant CD133+ tumor-initiating cell subset. Thyroid.
24:520–532. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang A, Chen L, Pu K and Zhu Y:
Identification of stem-like cells in non-small cell lung cancer
cells with specific peptides. Cancer Lett. 351:100–107. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Schaefer-Prokop C, Prosch H and Prokop M:
Lung cancer screening: What have we learnt for the practice so far?
Radiologe. 54:462–469. 2014.(In German). View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Spira A, Halmos B and Powell CA: Update in
lung cancer 2014. Am J Respir Crit Care Med. 192:283–294. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Herbst RS, Heymach JV and Lippman SM: Lung
cancer. N Engl J Med. 359:1367–1380. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Anglim PP, Alonzo TA and Laird-Offringa
IA: DNA methylation-based biomarkers for early detection of
non-small cell lung cancer: An update. Mol Cancer. 7:812008.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ettinger DS, Wood DE, Akerley W, Bazhenova
LA, Borghaei H, Camidge DR, Cheney RT, Chirieac LR, D'Amico TA,
Demmy TL, et al: Non-small cell lung cancer, Version 6. 2015. J
Natl Compr Canc Netw. 13:515–524. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kalemkerian GP, Akerley W, Bogner P,
Borghaei Hossein, Chow LQ, Downey RJ, Gandhi L, Ganti P AK,
Govindan R, Grecula JC, et al: Small cell lung cancer. J Natl Compr
Canc Netw. 11:78–98. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan
A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al: The
epithelial-mesenchymal transition generates cells with properties
of stem cells. Cell. 133:704–715. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bao S, Wu Q, McLendon RE, Hao Y, Shi Q,
Hjelmeland AB, Dewhirst MW, Bigner DD and Rich JN: Glioma stem
cells promote radioresistance by preferential activation of the DNA
damage response. Nature. 444:756–760. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Alison MR, Lin WR, Lim SM and Nicholson
LJ: Cancer stem cells: In the line of fire. Cancer Treat Rev.
38:589–598. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bonnet D and Dick JE: Human acute myeloid
leukemia is organized as a hierarchy that originates from a
primitive hematopoietic cell. Nat Med. 3:730–737. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Giangreco A, Arwert EN, Rosewell IR,
Snyder J, Watt FM and Stripp BR: Stem cells are dispensable for
lung homeostasis but restore airways after injury. Proc Natl Acad
Sci USA. 106:9286–9291. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lathia JD, Mack SC, Mulkearns-Hubert EE,
Valentim CL and Rich JN: Cancer stem cells in glioblastoma. Genes
Dev. 29:1203–1217. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gawlik-Rzemieniewska N and Bednarek I: The
role of NANOG transcriptional factor in the development of
malignant phenotype of cancer cells. Cancer Biol Ther. 17:1–10.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fulawka L, Donizy P and Halon A: Cancer
stem cells-the current status of an old concept: Literature review
and clinical approaches. Biol Res. 47:662014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ucar D, Cogle CR, Zucali JR, Ostmark B,
Scott EW, Zori R, Gray BA and Moreb JS: Aldehyde dehydrogenase
activity as a functional marker for lung cancer. Chem Biol
Interact. 178:48–55. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tirino V, Camerlingo R, Bifulco K, Irollo
E, Montella R, Paino F, Sessa G, Carriero MV, Normanno N, Rocco G
and Pirozzi G: TGF-β1 exposure induces epithelial to mesenchymal
transition both in CSCs and non-CSCs of the A549 cell line, leading
to an increase of migration ability in the CD133+ A549 cell
fraction. Cell Death Dis. 4:e6202013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ahmadipour F, Noordin MI, Mohan S, Arya A,
Paydar M, Looi CY, Keong YS, Siyamak EN, Fani S, Firoozi M, et al:
Koenimbin, a natural dietary compound of murraya koenigii (L)
spreng: Inhibition of MCF7 breast cancer cells and targeting of
derived MCF7 breast cancer stem cells (CD44(+)/CD24 (-/low)): An in
vitro study. Drug Des Devel Ther. 9:1193–1208. 2015.PubMed/NCBI
|
|
29
|
Chang Y, Zhao Y, Gu W, Cao Y, Wang S, Pang
J and Shi Y: Bufalin inhibits the differentiation and proliferation
of cancer stem cells derived from primary osteosarcoma cells
through Mir-148a. Cell Physiol Biochem. 36:1186–1196. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Keysar SB and Jimeno A: More than markers:
Biological significance of cancer stem cell-defining molecules. Mol
Cancer Ther. 9:2450–2457. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
White A and Swanson SJ: Minimally invasive
surgery for early-stage lung cancer: From innovation to standard of
care. Oncology (Williston Park). 30:982–987. 2016.PubMed/NCBI
|
|
32
|
Gazdar AF, Girard L, Lockwood WW, Lam WL
and Minna JD: Lung cancer cell lines as tools for biomedical
discovery and research. J Natl Cancer Inst. 102:1310–1321. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Damhofer H, Ebbing EA, Steins A, Welling
L, Tol JA, Krishnadath KK, van Leusden T, van de Vijver MJ,
Besselink MG, Busch OR, et al: Establishment of patient-derived
xenograft models and cell lines for malignancies of the upper
gastrointestinal tract. J Transl Med. 13:1152015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Rubin H: Multistage carcinogenesis in cell
culture. Dev Biol (Basel). 106(61–67): 143–160. 2001.
|
|
35
|
Jones MF, Hara T, Francis P, Li XL, Bilke
S, Zhu Y, Pineda M, Subramanian M, Bodmer WF and Lal A: The
CDX1-microRNA-215 axis regulates colorectal cancer stem cell
differentiation. Proc Natl Acad Sci USA. 112:E1550–E1558. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang K, Zeng J, Luo L, Yang J, Chen J, Li
B and Shen K: Identification of a cancer stem cell-like side
population in the HeLa human cervical carcinoma cell line. Oncol
Lett. 6:1673–1680. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Van den Broeck A, Vankelecom H, Van Delm
W, Gremeaux L, Wouters J, Allemeersch J, Govaere O, Roskams T and
Topal B: Human pancreatic cancer contains a side population
expressing cancer stem cell-associated and prognostic genes. PLoS
One. 8:e739682013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chen X, Chen D, Yang S, Ma R, Pan Y, Li X
and Ma S: Impact of ABCG2 polymorphisms on the clinical outcome of
TKIs therapy in Chinese advanced non-small-cell lung cancer
patients. Cancer Cell Int. 15:432015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Goodell MA, Brose K, Paradis G, Conner AS
and Mulligan RC: Isolation and functional properties of murine
hematopoietic stem cells that are replicating in vivo. J Exp Med.
183:1797–1806. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Salcido CD, Larochelle A, Taylor BJ,
Dunbar CE and Varticovski L: Molecular characterization of side
population cells with cancer stem cell-like characteristics in
small-cell lung cancer. Br J Cancer. 102:1636–1644. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wu C and Alman BA: Side population cells
in human cancers. Cancer Lett. 268:1–9. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Jiang F, Qiu Q, Khanna A, Todd NW, Deepak
J, Xing L, Wang H, Liu Z, Su Y, Stass SA and Katz RL: Aldehyde
dehydrogenase 1 is a tumor stem cell-associated marker in lung
cancer. Mol Cancer Res. 7:330–338. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Liu J, Xiao Z, Wong SK, Tin VP, Ho KY,
Wang J, Sham MH and Wong MP: Lung cancer tumorigenicity and drug
resistance are maintained through ALDH(hi)CD44(hi) tumor initiating
cells. Oncotarget. 4:1698–1711. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Okudela K, Woo T, Mitsui H, Suzuki T,
Tajiri M, Sakuma Y, Miyagi Y, Tateishi Y, Umeda S, Masuda M and
Ohashi K: Downregulation of ALDH1A1 expression in non-small cell
lung carcinomas-its clinicopathologic and biological significance.
Int J Clin Exp Pathol. 6:1–12. 2013.PubMed/NCBI
|
|
45
|
Meng X, Wang X and Wang Y: More than 45%
of A549 and H446 cells are cancer initiating cells: Evidence from
cloning and tumorigenic analyses. Oncol Rep. 21:995–1000.
2009.PubMed/NCBI
|
|
46
|
Willerson JT: CD133+ stem cells in the
treatment of patients with refractory angina. Circ Res.
120:602–603. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Yi H, Cho HJ, Cho SM, Jo K, Park JA, Lee
SH, Chang BJ, Kim JS and Shin HC: Effect of 5-FU and MTX on the
expression of drug-resistance related cancer stem cell markers in
non-small cell lung cancer cells. Korean J Physiol Pharmacol.
16:11–16. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhang HZ, Lin XG, Hua P, Wang M, Ao X,
Xiong LH, Wu C and Guo JJ: The study of the tumor stem cell
properties of CD133+CD44+ cells in the human lung adenocarcinoma
cell lineA549. Cell Mol Biol (Noisy-le-grand). 56
Suppl:OL1350–OL1358. 2010.PubMed/NCBI
|
|
49
|
Roudi R, Madjd Z, Ebrahimi M, Samani FS
and Samadikuchaksaraei A: CD44 and CD24 cannot act as cancer stem
cell markers in human lung adenocarcinoma cell line A549. Cell Mol
Biol Lett. 19:23–36. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Nakatsugawa M, Takahashi A, Hirohashi Y,
Torigoe T, Inoda S, Murase M, Asanuma H, Tamura Y, Morita R,
Michifuri Y, et al: SOX2 is overexpressed in stem-like cells of
human lung adenocarcinoma and augments the tumorigenicity. Lab
Invest. 91:1796–1804. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Sung JM, Cho HJ, Yi H, Lee CH, Kim HS, Kim
DK, Abd El-Aty AM, Kim JS, Landowski CP, Hediger MA and Shin HC:
Characterization of a stem cell population in lung cancer A549
cells. Biochem Biophys Res Commun. 371:163–167. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Curry JM, Sprandio J, Cognetti D,
Luginbuhl A, Bar-ad V, Pribitkin E and Tuluc M: Tumor
microenvironment in head and neck squamous cell carcinoma. Semin
Oncol. 41:217–234. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Kondo T: Stem cell-like cancer cells in
cancer cell lines. Cancer Biomark. 3:245–250. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Sääf AM, Halbleib JM, Chen X, Yuen ST,
Leung SY, Nelson WJ and Brown PO: Parallels between global
transcriptional programs of polarizing Caco-2 intestinal epithelial
cells in vitro and gene expression programs in normal colon and
coloncancer. Mol Biol Cell. 18:4245–4260. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Klonisch T, Wiechec E, Hombach-Klonisch S,
Ande SR, Wesselborg S, Schulze-Osthoff K and Los M: Cancer stem
cell markers in common cancers-therapeutic implications. Trends Mol
Med. 14:450–460. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Leung EL, Fiscus RR, Tung JW, Tin VP,
Cheng LC, Sihoe AD, Fink LM, Ma Y and Wong MP: Non-small cell lung
cancer cells expressing CD44 are enriched for stem cell-like
properties. PLoS One. 5:e140622010. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Valent P, Bonnet D, De Maria R, Lapidot T,
Copland M, Melo JV, Chomienne C, Ishikawa F, Schuringa JJ, Stassi
G, et al: Cancer stem cell definitions and terminology: The devil
is in the details. Nat Rev Cancer. 12:767–775. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Lin WC, Rajbhandari N and Wagner KU:
Cancer cell dormancy in novel mouse models for reversible
pancreatic cancer: A lingering challenge in the development of
targeted therapies. Cancer Res. 74:2138–2143. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Williams AB: Spontaneous mutation rates
come into focus in Escherichia coli. DNA Repair (Amst). 24:73–79.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Le Beyec J, Xu R, Lee SY, Nelson CM, Rizki
A, Alcaraz J and Bissell MJ: Cell shape regulates global histone
acetylation in human mammary epithelial cells. Exp Cell Res.
313:3066–3075. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Gazdar AF, Gao B and Minna JD: Lung cancer
cell lines: Useless artifacts or invaluable tools for medicine
science? Lung Cancer. 68:309–318. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Roschke AV, Tonon G, Gehlhaus KS, McTyre
N, Bussey KJ, Lababidi S, Scudiero DA, Weinstein JN and Kirsch IR:
Karyotypic complexity of the NCI-60 drug-screening panel. Cancer
Res. 63:8634–8647. 2003.PubMed/NCBI
|
|
63
|
Daniel VC, Marchionni L, Hierman JS,
Rhodes JT, Devereux WL, Rudin CM, Yung R, Parmigiani G, Dorsch M,
Peacock CD and Watkins DN: A primary xenograft model of small-cell
lung cancer reveals irreversible changes in gene expression imposed
by culture in vitro. Cancer Res. 69:3364–3373. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Gazdar AF, Bader S, Hung J, Kishimoto Y,
Sekido Y, Sugio K, Virmani A, Fleming J, Carbone DP and Minna JD:
Molecular genetic changes found in human lung cancer and its
precursor lesions. Cold Spring Harb Symp Quant Biol. 59:565–572.
1994. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Akunuru S, Zhai James Q and Zheng Y:
Non-small cell lung cncer stem/progenitor cells are enriched in
multiple distinct phenotypic subpopulations and exhibit plasticity.
Cell Death Dis. 3:e3522012. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Shultz LD, Ishikawa F and Greiner DL:
Humanized mice in translational biomedical research. Nature Rev
Immunol. 7:118–130. 2007. View Article : Google Scholar
|
|
67
|
Perumal D, Singh S, Yoder SJ, Bloom GC and
Chellappan SP: A novel five gene signature derived from stem-like
side population cells predicts overall and recurrence-free survival
in NSCLC. PLoS One. 7:e435892012. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Yeh CT, Su CL, Huang CY, Lin JK, Lee WH,
Chang PM, Kuo YL, Liu YW, Wang LS, Wu CH, et al: A preclinical
evaluation of antimycin a as a potential antilung cancer stem cell
agent. Evid Based Complement Alternat Med. 2013:9104512013.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Ho MM, Ng AV, Lam S and Hung JY: Side
population in human lung cancer cell lines and tumors is enriched
with stem-like cancer cells. Cancer Res. 67:4827–4833. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Liu J and Mao Z, Huang J, Xie S, Liu T and
Mao Z: Blocking the NOTCH pathway can inhibit the growth of
CD133-positive A549 cells and sensitize to chemotherapy. Biochem
Biophys Res Commun. 444:670–675. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Xu Y, Jiang Z, Zhang Z, Sun N, Zhang M,
Xie J, Li T, Hou Y and Wu D: HtrA1 downregulation induces cisplatin
resistance in lung adenocarcinoma by promoting cancer stem
cell-like properties. J Cell Biochem. 115:1112–1121. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Bertolini G, Roz L, Perego P, Tortoreto M,
Fontanella E, Gatti L, Pratesi G, Fabbri A, Andriani F, Tinelli S,
et al: Highly tumorigenic lung cancer CD133+ cells exhibit
stem-like features and are spared by cisplatin treatment. Proc Natl
Acad Sci USA. 106:16281–16286. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Yao Q, Sun JG, Ma H, Zhang AM, Lin S, Zhu
CH, Zhang T and Chen ZT: Monitoring microRNAs using a molecular
beacon in CD133+/CD338+ human lung adenocarcinoma-initiating
A549cells. Asian Pac J Cancer Prev. 15:161–166. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Lin S, Sun JG, Wu JB, Long HX, Zhu CH,
Xiang T, Ma H, Zhao ZQ, Yao Q, Zhang AM, et al: Aberrant microRNAs
expression in CD133+/CD326+ human lung
adenocarcinoma initiating cells from A549. Mol Cells. 33:277–283.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Li Z, Xiang Y, Xiang L, Xiao Y, Li F and
Hao P: ALDH maintains the stemness of lung adenoma stem cells by
suppressing the notch/CDK2/CCNE pathway. PLoS One. 9:e926692014.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Kim IG, Kim SY, Choi SI, Lee JH, Kim KC
and Cho EW: Fibulin-3-mediated inhibition of
epithelial-to-mesenchymal transition and self-renewal of ALDH+ lung
cancer stem cells through IGF1R signaling. Oncogene. 33:3908–3917.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Zakaria N, Yusoff NM, Zakaria Z, Lim MN,
Baharuddin PJ, Fakiruddin KS and Yahaya B: Human non-small cell
lung cancer expresses putative cancer stem cell markers and
exhibits the transcriptomic profile of multipotent cells. BMC
Cancer. 15:842015. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Ghosh G, Lian X, Kron SJ and Palecek SP:
Properties of resistant cells generated from lung cancer cell lines
treated with EGFR inhibitors. BMC Cancer. 12:952012. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Levina V, Marrangoni AM, DeMarco R,
Gorelik E and Lokshin AE: Drug-selected human lung cancer stem
cells: Cytokine network, tumorigenic and metastatic properties.
PLoS One. 3:e30772008. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Shi Y, Fu X, Hua Y, Han Y, Lu Y and Wang
J: The side population in human lung cancer cell line NCI-H460 is
enriched in stem-like cancer cells. PLoS One. 7:e333582012.
View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Liu YP, Yang CJ, Huang MS, Yeh CT, Wu AT,
Lee YC, Lai TC, Lee CH, Hsiao YW, Lu J, et al: Cisplatin selects
for multidrug-resistant CD133+ cells in lung adenocarcinoma by
activating Notch signaling. Cancer Res. 73:406–416. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Shien K, Toyooka S, Yamamoto H, Soh J,
Jida M, Thu KL, Hashida S, Maki Y, Ichihara E, Asano H, et al:
Acquired resistance to EGFR inhibitors is associated with a
manifestation of stem cell-like properties in cancer cells. Cancer
Res. 73:3051–3061. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Jaggupilli A and Elkor E: Significance of
CD44 and CD24 as cancer stem cell markers: An enduring ambiguity.
Clin Dev Immunol. 2012:7080362012. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Teng Y, Wang X, Wang Y and Ma D:
Wnt/beta-catenin signaling regulates cancer stem cells in lung
cancer A549 cells. Biochem Biophys Res Commun. 392:373–379. 2010.
View Article : Google Scholar : PubMed/NCBI
|