Introduction
Colorectal cancer is cancer of the colon or rectum,
with ~1,000,000 cases reported in 2012 (1). It is the third most prevalent from of
cancer. The American Cancer Society estimates 135,430 individuals
are newly diagnosed with colorectal cancer with 50,260 deaths in
2017 (1), making it a significant
health burden that requires immediate attention. Currently, the
treatment methods for colon cancer involve various therapies,
notably chemotherapy and radiation therapy. However, for improved
prognosis and early detection, the underlying mechanisms of colon
cancer progression and metastasis require elucidation (2).
Accumulating evidence in the past decade has
established an association between microRNAs (miRNAs/miRs) and
cancer (3–5). miRNAs are short non-coding RNAs of 22–24
nucleotides that are involved in cellular growth, differentiation,
regulation and apoptosis (3). Often
deregulated, miRNAs have been shown to be involved in the
progression of different types of cancer (6).
Several studies have revealed that the miRNA
expression patterns are altered in colorectal cancer (7–10). One of
the earliest studies that identified a correlation between miRNA
expression patterns and colorectal cancer found deregulation of
miR-143 and miR-145 in colorectal cancer, suggesting a tumor
suppressor role of these miRNAs (7).
Subsequently, a variety of miRNAs were identified that either play
an oncogenic or a tumor suppressor role in colorectal cancer. For
example, miR-21 has been shown to play an oncogenic role in
colorectal cancer (8,9). Several studies have shown altered
expression levels of miR-17-92 cluster, miR-31, miR-224 and miR-183
in colorectal cancer (10).
Identification of altered expression of miRNAs in colorectal cancer
is important, as it may act as an early detection marker and
therapeutic targets in colorectal cancer progression and
treatment.
The present study investigates one such miRNA,
miR-137, which has been frequently associated with different types
of cancer. miR-137 has also been shown to be downregulated in
gastric cancer and negatively regulates cell division cycle 42
(Cdc42) (11). A previous study
demonstrated that ectopic expression of miR-137 in lung cancer
cells downregulated Cdc42 and cyclin dependent kinase 6 (12). Similarly, ectopic expression of
miR-137 in breast cancer cells downregulated estrogen-related
receptor α expression at both mRNA and protein levels (13). However, the association between
miR-137 and colon cancer is poorly understood. At present, there
have been a small number of studies that found an association
between miR-137 and colorectal cancer. In one such study, the
expression of miR-137 in colorectal cell lines was inversely
correlated with Cdc42 expression (14). In another similar study, miR-137 was
found to act as a tumor suppressor in the colon by targeting
lysine-specific demethylase 1 (15).
However, other targets of miR-137 that regulate colon cancer cell
migration and invasion remain to be identified and substantiated.
The present study attempted to investigate the role of miR-137 in
colon cancer progression, the downstream targets and the mode of
regulation.
Materials and methods
Cell lines, cell culture and
transfection
The colon cancer COLO205, HCT116 and SW480 cell
lines were purchased from Shanghai Cell Bank, Chinese Academy of
Sciences (Shanghai, China). The cell lines were maintained and
cultured according to the manufacturer's instructions. Dulbecco's
modified Eagle's medium (Thermo Fisher Scientific, Inc., Waltham,
MA, USA) supplemented with 10% fetal bovine serum (Thermo Fisher
Scientific, Inc.) was used for cell culture. All cell lines were
cultured in the aforementioned media, and incubated at 37°C in a 5%
CO2 humidified incubator. Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) was
used for all transfection experiments, and the transfections were
performed according to the manufacturer's protocol. Cells were
transfected with miR-137 (sequence, UUAUUGCUUAAGAAUACGCGUAG;
Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). The negative
control was a validated random sequence known to exhibit no effects
on miRNA function, purchased from Thermo Fisher Scientific, Inc.,
(cat. no. 4464058).
Cell proliferation, migration and
invasion assays
The MTT method was used to determine cell
proliferation. The cells were transfected with miR-137 mimics or
controls and were seeded onto 96-well plates (4×105
cells/well). Cell viability was documented at regular intervals,
every 24 h for 6 days, according to the manufacturer's protocol.
Formazan crystals were dissolved in dimethylsulfoxide
(Sigma-Aldrich; Merck KGaA). Subsequently, 25 µl of MTT solution
was added to each well, followed by incubation at 37°C for 6 h. The
plates were centrifuged at 1,000 × g, and the absorbance of the
formazan product was quantified at 490 nm in an ELISA reader
(Bio-Rad Laboratories, Inc., Hercules, CA, USA). Cell migration was
assessed in two 24-well 8-µm pore Transwell plates (BD Biosciences
Franklin Lakes, NJ, USA), according to the manufacturer's
instructions. The plates were then incubated for 16 h at 37°C in 5%
CO2 humidified incubator. Cell invasion was assessed
using the Transwell apparatus. For the invasion assay,
~1×106 cells were added to Matrigel Invasion Chambers in
two 24-well 8-µm pore plates (BD Biosciences), according to the
manufacturer's protocol. Following addition, the plates were
incubated at 37°C for 36 h in 5% CO2 in a humidified
incubator. The cells on the upper surface of the membrane were
gently scraped off subsequent to incubation. Cells that migrated to
the bottom surface were fixed in 100% methanol for 5 min and
stained with 0.4% crystal violet staining solution for 2 min. The
cells were counted from different microscopic fields with a light
microscope at ×300 magnification, and the values were averaged.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
A kit-based approach was used to isolate the total
RNA. Using RNeasy kit (Qiagen, Inc., Valencia, CA, USA), total RNA
from the cells was extracted according to the manufacturer's
protocol at 42°C for 30 min. The final volume of the PCR mixture
was 50 µl, and it contained 0.3 µM probe, 0.3 µM of forward and
reverse primers, 1X PCR buffer, 0.4 mM dNTPs and Taq DNA
polymerase, all purchased from New England BioLabs, Inc. (Ipswich,
MA, USA). The thermocycling conditions included initial
denaturation at 95°C for 5 min, followed by 35 cycles of 95°C for
20 sec, 58°C for 30 sec and 68°C for 40 sec, with a final extension
step of 72°C for 10 min. Specific RNA transcripts were quantified
by SYBR-Green qPCR (Thermo Fisher Scientific, Inc.). Reactions were
performed and analyzed using an ABI Prism 7700 Sequence Detection
System (Applied Biosystems; Thermo Fisher Scientific, Inc.).
β-actin mRNA (Macrogen, Inc., Seoul, Republic of Korea) or GAPDH
mRNA (Macrogen, Inc.) was used as an internal control. Sequences of
primers are presented in Table I.
Data was quantified using ImageJ software (version 1.47; National
Institutes of Health, Bethesda, MD, USA).
 | Table I.Sequences of primers used in the
present study. |
Table I.
Sequences of primers used in the
present study.
| Primer | Sequence |
|---|
| β-actin |
|
| Forward |
5′-GGGACCTGACTGACTACCTCA-3′ |
| Reverse |
5′-TGACTCGTCATACTCCTGCTTG-3′ |
| GAPDH |
|
| Forward |
5′-TGCACCACCAACTGCTTAGC-3′ |
| Reverse |
5′-GGCATGGACTGTGGTCATGAG-3′ |
| TCF4 |
|
| Forward |
5′-GCAAGTTGGACGCCCGCAAGATC-3′ |
| Reverse |
5′-TAGTCAGCCATGGGGCGGAGA-3′ |
Bioinformatical analysis
TargetScan (www.targetscan.org) is a webserver prediction tool
that has been routinely used to predict the biological targets of
miRNAs with improved accuracy compared to other prediction
programs. To identify the biological target of miR-137, miR-137 was
run in TargetScan. From the top 10 targets, TCF4 was finalized
based on a combination of Aggregate PCT, context scores
and based on whether or not miR-137 and the mRNA are expressed in
cells of interest.
Western blot analysis
The proteins were extracted by lysing the cells in
sample loading buffer. The composition of the loading buffer was
1.5% SDS, 10% glycerol, 5 mM β-mercaptoethanol, bromphenol blue and
75 mM Tris (pH 7). Whole cell lysates were separated by SDS-PAGE
(12% gel) and the proteins were transferred onto a polyvinylidene
fluoride membrane. Subsequently, the membranes were incubated with
rabbit anti-TCF4 (GTX54531; 1:200; GeneTex International
Corporation, Hsinchu, Taiwan) and anti-β-actin primary antibodies
(cat. no. GTX109639; 1:500; GeneTex International Corporation) at
4°C overnight. Subsequent to incubation, horseradish
peroxidase-conjugated rat anti-mouse IgG1 secondary antibodies
(cat. no. GTX83207; 1:500; GeneTex, Hsinchu City, Taiwan) were used
to detect specific bands.
Statistical analysis
For statistical analysis, SigmaPlot (version 13;
SysStat Software, Inc., San Jose CA, USA) and Microsoft Excel
(version 15.33; Microsoft Corporation, Redmond, WA, USA) were used.
Student's t-test was used to determine significant differences
between groups. In all statistical analyses, two sided tests were
applied. P<0.05 was considered to indicate a statistically
significant difference. All data are shown as the mean ± standard
deviation.
Results
Ectopic expression of miR-137 in colon
cancer COLO205, HCT116 and SW480 cell lines
Firstly, the endogenous levels of miR-137 in colon
cancer COLO205, HCT116 and SW480 cell lines were assessed by
RT-qPCR. The levels of miR-137 in these cell lines were low (data
not shown). Following the transfection of miR-137, the expression
levels of miR-137 in the cell lines were monitored at time
intervals of 24 h. The levels of miR-137 were highest following the
first 24 h period in all the cell lines. However, the expression
levels of miR-137 gradually decreased between 24 and 144 h
(Fig. 1).
miR-137 suppresses cell proliferation
in colon cancer cell lines
Subsequently, the effect of miR-137 on cell
proliferation following transfections in all cell lines was
investigated. Based on the results from the MTT assay,
overexpression of miR-137 in the cell lines significantly
suppressed cell proliferation (P=0.009, P=0.007 and P=0.01;
Fig. 2). Analysis of the results
indicated that the inhibition rate of miR-137 in colon cancer
COLO205, HCT116 and SW480 cell lines was 35.34±3.46, 29.78±4.82 and
32.9±2.8%, respectively. Overall, these results suggest that
miR-137 suppresses cell proliferation in colon cancer cell
lines.
miR-137 suppresses migration and
invasion in colon cancer COLO205, HCT116 and SW480 cell lines
The role of miR-137 in the progression and
metastasis of colon cancer was then investigated using migration
and invasion assays performed with Transwell apparatus. The
transfected cells from log-phase growth were cultured on the
Transwell apparatus. The cells were incubated for 12 h for
migration analysis. Analysis of the results revealed that the cell
migration was significantly decreased in miR-137-transfected cells
compared with corresponding negative controls (P=0.005; Fig. 3). The effect of miR-137 on cell
invasiveness was then determined by incubating the
miR-137-transfected cells for 24 h and monitoring the cellular
movement through the extracellular matrix. The results of the
invasion assay indicated that miR-137-transfected cells exhibited a
significantly reduced invasiveness with respect to their
corresponding controls (P=0.007; Fig.
3). Combining the analyses from the migration and invasion
assays, it is clear that miR-137 suppresses migration and invasion
in COLO205, HCT116 and SW480 colon cancer cell lines.
Transcription factor 4 (TCF4) is a
putative target of miR-137
Subsequently, the downstream targets of miR-150 were
identified to obtain information on the regulatory mechanisms. The
potential targets of miR-150 were predicted using TargetScan
bioinformatics software. Initial runs provided a wide range of
targets regulated by miR-137; however, a more specific range of
best scoring targets was selected using stringent filters; the
predicted targets were shortlisted based on the efficacy of
targeting or the probability of conserved targeting. Among the best
scoring targets, TCF4 appeared to be of particular interest, since
it has been implicated as a direct target of different miRNAs in
various types of cancer, as reported by previously published
studies (16–18). Therefore, experimental validation of
TCF4 as a direct target of miR-137 was performed in colon cancer
cell lines.
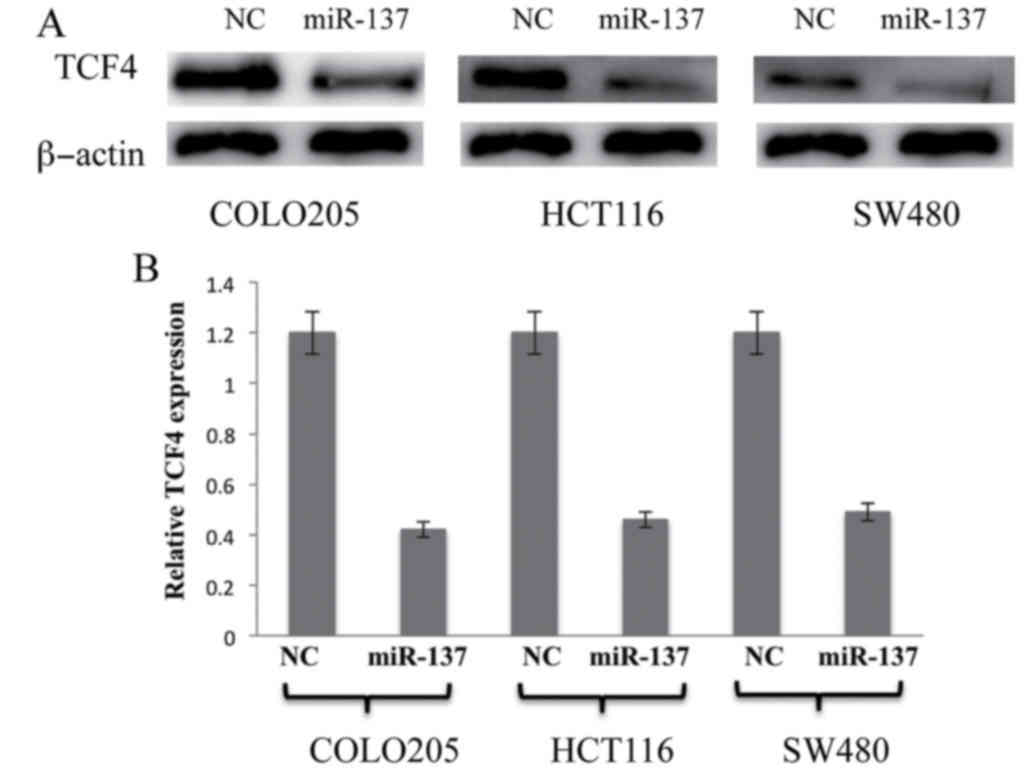
miR-137 negatively regulates TCF4
expression in colon cancer COLO205, HCT116 and SW480 cell
lines
To experimentally validate the association between
miR-137 and the predicted target TCF4, miR-137 was transfected into
the colon cancer cell lines followed by subsequent incubation for
36 h. The levels of TCF4 mRNA and protein expression were detected
and quantified by RT-qPCR and western blot analysis. The results
were analyzed, and as shown in Fig.
4, overexpression of miR-137 significantly suppressed TCF4 in
all the colon cancer cell lines compared with the controls
(P<0.05; Fig. 4). This data
clearly indicates that TCF4 is a direct target of miR-137 in colon
cancer cell lines and overexpression of miR-137 may negatively
regulate TCF4 expression in colon cancer cell lines.
Discussion
In humans, miR-137 is located on the chromosome 1p22
and undergoes transcription and extensive processing to result in a
final mature miRNA 22–25 nucleotides in length. miR-137 has been
associated with various types of cancer, as shown by previous
studies. Studies have shown that miR-137 often functions as a tumor
suppressor in gastric cancer (11),
lung cancer (12), colorectal cancer
(14), neuroblastoma (19), glioblastoma (20) and melanoma (21). Studies have also shown that miR-137
negatively regulates a wide spectrum of downstream targets in
different types of cancers (12,13);
however, a minority of such direct targets has been experimentally
validated. Additionally, there is a lack of data on regulatory
mechanisms involving miR-137, downstream targets and cancer
progression. The association between miR-137 and colorectal/colon
cancer has been established in the last few years (16). One such study established that miR-137
negatively regulates Cdc42 expression, inducing cell cycle arrest
in the G1 phase and resulting in the inhibition of invasion in
colorectal cancer cells (14). Since
miRNAs can target multiple transcripts, the present study
identified and experimentally validated one such important target
of miR-137, which may play crucial role in colon cancer
pathogenesis. In the present study, TCF4 was identified as a direct
target of miR-137 in COLO205, HCT116 and SW480 colon cancer cell
lines. Furthermore, miR-137 suppresses the proliferation, migration
and invasion of colon cancer cell lines by targeting TCF4.
The endogenous levels of miR-137 in colon cancer
COLO205, HCT116 and SW480 cell lines were barely detectable.
However, following the ectopic expression of miR-137, cell
proliferation was significantly suppressed in all the three colon
cancer cell lines. Since migration and invasion assays provide
important insights in progression of cancer, the present study
investigated the effect of miR-137 on cell migration and
invasiveness. miR-137 suppressed cell migration and invasion in all
the three colon cancer cell lines. These results clearly suggested
that miR-137 may act as a tumor suppressor in colon cancer. There
have been several well-documented studies that attribute the role
of tumor suppressor function to miR-137 in various types of cancer
(11–13).
The identification of the downstream targets of
miR-137 was subsequently attempted to gain insights into the
regulatory mechanisms of miR-137. A list of potential targets was
identified by bioinformatics analysis and by adopting a ‘narrow
down approach’; the best scoring targets were selected. The
analysis was primarily focused on one target, TCF4. There have been
few studies that suggest the regulation of TCF4 by miRNAs in
cancer. One such notable study was performed by Chen et al
(22). This study showed that miR-24
acts an oncogene in glioma and regulates cell migration and
invasion via TCF4 signaling (22).
However, to the best of our knowledge, the present study is the
first to identify TCF4 as a direct target of miR-137 in colon
cancer.
To conclusively establish TCF4 as a direct target of
miR-137, subsequent to treatment with miR-137 the levels of TCF4
were detected at the mRNA and protein level in all three colon
cancer cell lines. The mRNA and protein levels of TCF4 were
significantly decreased, which suggests that TCF4 is a direct
target of miR-137 and miR-137 negatively regulates TCF4 in colon
cancer cell lines. Since there were few other best-scoring targets
predicted, it is possible that miR-137 may contribute to colon
cancer progression via regulatory mechanisms using other
targets.
To conclude, the present study showed that miR-137
inhibits cell proliferation, migration and invasion in colon cancer
cell lines by negatively regulating the expression of TCF4.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Van Cutsem E, Nordlinger B and Cervantes
A: ESMO Guidelines Working Group: Advanced colorectal cancer: ESMO
Clinical Practice Guidelines for treatment. Ann Oncol. 21 Suppl
5:v93–v97. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Di Leva G, Garofalo M and Croce CM:
MicroRNAs in cancer. Ann Rev Pathol. 9:287–314. 2014. View Article : Google Scholar
|
|
4
|
Jansson MD and Lund AH: MicroRNA and
cancer. Mol Oncol. 6:590–610. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li M, Li J, Ding X, He M and Cheng SY:
microRNA and cancer. AAPS J. 12:309–317. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Su J, Liang H, Yao W, Wang N, Zhang S, Yan
X, Feng H, Pang W, Wang Y, Wang X, et al: MiR-143 and MiR-145
regulate IGF1R to suppress cell proliferation in colorectal cancer.
PLoS One. 9:e1144202014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Slaby O, Svoboda M, Fabian P, Smerdova T,
Knoflickova D, Bednarikova M, Nenutil R and Vyzula R: Altered
expression of miR-21, miR-31, miR-143 and miR-145 is related to
clinicopathologic features of colorectal cancer. Oncology.
72:397–402. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Asangani IA, Rasheed SAK, Nikolova DA,
Leupold JH, Colburn NH, Post S and Allgayer H: MicroRNA-21 (miR-21)
post-transcriptionally downregulates tumor suppressor Pdcd4 and
stimulates invasion, intravasation and metastasis in colorectal
cancer. Oncogene. 27:2128–2136. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Motoyama K, Inoue H, Takatsuno Y, Tanaka
F, Mimori K, Uetake H, Sugihara K and Mori M: Over-and
under-expressed microRNAs in human colorectal cancer. Int J Oncol.
34:1069–1075. 2009.PubMed/NCBI
|
|
11
|
Chen Q, Chen X, Zhang M, Fan Q, Luo S and
Cao X: miR-137 is frequently down-regulated in gastric cancer and
is a negative regulator of Cdc42. Dig Dis Sci. 56:2009–2016. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhu X, Li Y, Shen H, Li H, Long L, Hui L
and Xu W: miR-137 inhibits the proliferation of lung cancer cells
by targeting Cdc42 and Cdk6. FEBS Lett. 587:73–81. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhao Y, Li Y, Lou G, Zhao L, Xu Z, Zhang Y
and He F: MiR-137 targets estrogen-related receptor alpha and
impairs the proliferative and migratory capacity of breast cancer
cells. PLoS One. 7:e391022012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu M, Lang N, Qiu M, Xu F, Li Q, Tang Q,
Chen J, Chen X, Zhang S, Liu Z, et al: miR137 targets Cdc42
expression, induces cell cycle G1 arrest and inhibits invasion in
colorectal cancer cells. Int J Cancer. 128:1269–1279. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Balaguer F, Link A, Lozano JJ, Cuatrecasas
M, Nagasaka T, Boland CR and Goel A: Epigenetic silencing of
miR-137 is an early event in colorectal carcinogenesis. Cancer Res.
70:6609–6618. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gu W, Li X and Wang J: miR-139 regulates
the proliferation and invasion of hepatocellular carcinoma through
the WNT/TCF-4 pathway. Oncol Rep. 31:397–404. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lan F, Yue X, Han L, Shi Z, Yang Y, Pu P,
Yao Z and Kang C: Genome-wide identification of TCF7L2/TCF4 target
miRNAs reveals a role for miR-21 in Wnt-driven epithelial cancer.
Int J Oncol. 40:519–526. 2012.PubMed/NCBI
|
|
18
|
Chen L, Zhang A, Li Y, Zhang K, Han L, Du
W, Yan W, Li R, Wang Y, Wang K, et al: MiR-24 regulates the
proliferation and invasion of glioma by ST7L via β-catenin/Tcf-4
signaling. Cancer Lett. 329:174–180. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Althoff K, Beckers A, Odersky A, Mestdagh
P, Köster J, Bray IM, Bryan K, Vandesompele J, Speleman F,
Stallings RL, et al: MiR-137 functions as a tumor suppressor in
neuroblastoma by downregulating KDM1A. Int J Cancer. 133:1064–1073.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen L, Wang X, Wang H, Li Y, Yan W, Han
L, Zhang K, Zhang J, Wang Y, Feng Y, et al: miR-137 is frequently
down-regulated in glioblastoma and is a negative regulator of
Cox-2. Eur J Cancer. 48:3104–3111. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen X, Wang J, Shen H, Lu J, Li C, Hu DN,
Dong XD, Yan D and Tu L: Epigenetics, microRNAs, and
carcinogenesis: Functional role of microRNA-137 in uveal melanoma.
Invest Ophthalmol Vis Sci. 52:1193–1199. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen L, Han L, Zhang K, Shi Z, Zhang J,
Zhang A, Wang Y, Song Y, Li Y, Jiang T, et al: VHL regulates the
effects of miR-23b on glioma survival and invasion via suppression
of HIF-1α/VEGF and β-catenin/Tcf-4 signaling. Neuro Oncol.
14:1026–1036. 2012. View Article : Google Scholar : PubMed/NCBI
|