Introduction
Aberrant epigenetic regulations have been found in
various types of cancer and numerous types of human disease. The
cancer epigenome is characterized by global changes in DNA
methylation and altered histone modification patterns (1). The global pattern of histone
modifications may serve as a predictor for the risk of recurrence
of human cancers (2,3). It seems that epigenetics have reached
the mainstream in pathogenesis research of numerous types of human
disease, particularly in cancer. Colorectal cancer (CRC) is the
third most commonly diagnosed cancer in the world (4). Cumulative evidence has proved that the
disruption of epigenetic regulation may drive the initiation and
progression of CRC, such as histone modifications (5–7). Emerging
evidences reveal that altered expression of histone
methyltransferases (HMTs) and histone demethylases (HDMs) may be
involved in cancer progression of CRC, such as Enhancer of zeste
homolog 2 (EZH2) and lysine demethylase 4C (5,8,9). The functions of HMTs and HDMs in the
pathogenesis of CRC require further investigation.
G9A is the primary HMT for mono- and dimethylation
of H3K9 in vivo (10). Among
various best-studied histone methylations, H3K9 methylation is
thought to be associated with gene repression (11). Recently, G9A has been reported to
perform critical roles in a number of biological progresses, such
as behavior plasticity, lymphocyte development, stem cells
differentiation and tumor cell growth (12–17). It
has been found that the expression level of G9A is increased in
numerous types of cancer as compared with their corresponding
normal tissues, such as melanoma, lung cancer, neuroblastoma,
leukemia and hepatocellular carcinoma (HCC) (17–19). It
has been demonstrated that decreasing G9A expression level or
inhibiting its activity reduces cellular proliferation and induces
autophagy related cell death in colon cancer cells, breast cancer
cells and neuroblastoma cells (17,20,21). A
recent study also reported that G9A suppression induces DNA damage
in colorectal cancer cells (21).
Although G9A has been reported to be crucial in
numerous types of cancer, the function in colorectal cancer
progression remains unknown. In the present study, the expression
profile of G9A in CRC was examined to explore the function in CRC
progression. The immunohistochemistry analysis of 100 pairs of
tumor specimens revealed that G9A protein expression was markedly
increased in CRC and the high expression may be related to distant
metastasis. A significant elevated mRNA expression level of G9A in
various colorectal carcinomas compared with normal colon and rectum
tissues by Oncomine database analysis was also identified.
Furthermore, the present study investigated the association between
G9A expression level and the clinicopathological features of CRC
using 6 publicly available datasets from Gene Expression Omnibus
(GEO), and found that G9A expression was associated with American
Joint Committee on Cancer (AJCC) staging, tumor differentiation,
and tumor relapse. The present findings provide novel evidence to
further understand the crucial role of G9A in tumorigenesis, and
also offers significant ideas for CRC therapy.
Materials and methods
Patients
In total, 100 patients were diagnosed with CRC (53
with colon cancer and 47 with rectal cancer; Table I), which was classified with the 7th
edition of the International Union against Cancer TNM staging
system (classified into T1, T2, T3 and T4 based on the size and the
extension of the primary tumor; classified into N0, N1 and N2 based
on the degree of spread to regional lymph nodes; classified into M0
and M1 based on the presence of distant metastasis), the Dukes'
staging system (classified into A, B, C and D) and histological
grading (classified into well, moderately, or poorly
differentiated) (22–24). All patients underwent surgical
resection of tumors at the Renmin Hospital of Wuhan University in
Wuhan, China. Ethical approval for the study was granted by the
Renmin Hospital's ethics committee. Informed written consent was
obtained from all participants involved in the study. The key
clinical characteristics of the patients are summarized in Table I. Normal specimens were obtained from
adjacent, grossly normal-appearing tissue taken at least 10 cm away
from the cancer. None of the patients included in this study had
chemotherapy or radiotherapy prior to surgery. There was no
follow-up information available for these patients.
 | Table I.Association between G9A protein
expression and clinicopathological features of CRC. |
Table I.
Association between G9A protein
expression and clinicopathological features of CRC.
|
|
| G9A expression, n
(%) |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
features | Case size, n | Low | High |
P-valuea |
|---|
| Diagnosis age
(years) |
|
|
| 0.028 |
|
≤60 | 49 | 22 (44.90) | 27 (55.10) |
|
|
>60 | 51 | 34 (66.67) | 17 (33.33) |
|
| Sex |
|
|
| 1.000 |
|
Male | 50 | 28 (56.00) | 22 (44.00) |
|
|
Female | 50 | 28 (56.00) | 22 (44.00) |
|
| Size
(diameter) |
|
|
| 0.349 |
| <5
cm | 47 | 24 (51.06) | 23 (48.94) |
|
| ≥5
cm | 53 | 32 (60.38) | 21 (39.62) |
|
| Depth of
invasion |
|
|
| 0.106 |
| T1 | 13 | 8 (61.54) | 5 (38.46) |
|
| T2 | 42 | 24 (57.14) | 18 (42.86) |
|
| T3 | 35 | 22 (62.86) | 13 (37.14) |
|
| T4 | 10 | 2 (20.00) | 8 (80.00) |
|
| Nodal
metastasis |
|
|
| 0.799 |
| N0 | 57 | 33 (57.89) | 24 (42.11) |
|
| N1 | 23 | 12 (52.17) | 11 (47.83) |
|
| N2 | 18 | 9 (50.00) | 9 (50.00) |
|
| Distant
metastasis |
|
|
| 0.043 |
| M0 | 89 | 53 (59.55) | 36 (40.45) |
|
| M1 | 11 | 3 (27.27) | 8 (72.73) |
|
| TNM stage |
|
|
| 0.106 |
| I | 13 | 8 (61.54) | 5 (38.46) |
|
| II | 42 | 24 (57.14) | 18 (42.86) | 0.035b |
|
III | 35 | 22 (62.86) | 13 (37.14) | 0.020b |
| IV | 10 | 2 (20.00) | 8 (80.00) |
|
| Dukes stage |
|
|
| 0.209 |
| A | 14 | 9 (64.29) | 5 (35.71) |
|
| B | 41 | 23 (56.10) | 18 (43.90) |
|
| C | 34 | 21 (61.76) | 13 (38.24) |
|
| D | 11 | 3 (27.27) | 8 (72.73) | 0.049c |
|
Differentiation |
|
|
| 0.631 |
|
Well | 20 | 13 (65.00) | 7 (35.00) |
|
|
Moderately | 57 | 30 (52.63) | 27 (47.37) |
|
|
Poorly | 23 | 13 (56.52) | 10 (43.48) |
|
Immunohistochemistry
Immunostaining was performed as described previously
(25). Briefly, deparaffinized
sections were treated with 3% H2O2 and
subjected to antigen retrieval by citric acid (pH 6.0). Subsequent
to overnight incubation with primary antibody of G9A (1:100; cat.
no. ab133482; Abcam, Cambridge, UK) at 4°C, the sections were
incubated for 15 min at room temperature with horseradish
peroxidase-labeled polymer conjugated with secondary antibody
(MaxVision™ HRP-Polymer anti-Rabbit IHC Kit; Maixin-Bio, Fuzhou,
China) and incubated for 1 min with diaminobenzidine. The sections
were then lightly counterstained with hematoxylin. The sections
without primary antibody served as negative controls. The positive
brown staining was visualized and then photographed using a light
microscope at ×200 or ×400 magnification (BX51, Olympus
Corporation, Tokyo, Japan).
Evaluation of immunohistochemical
staining
The immunohistochemical staining results of all
sections were evaluated by two independent observers (Dr Zhi Zeng
and Dr Jian Qin, the co-authors), who were unaware of the disease
outcome. Expression levels were ascertained according to the two
observers' evaluations. As G9A is mainly expressed in the cell
nucleus, the percentage of nucleus staining-positive cells were
graded as 0 (<10%), 1 (≥10%, and <25%), 2 (≥25%, and
<50%), 3 (≥50%, and <75%), 4 (≥75%) (Fig. 1A-F). For analysis the G9A protein
expression levels were divided into two groups: Low expression
level group (score value ≤2) and high expression level group (score
value ≥3).
Oncomine database analysis
The Oncomine database (https://www.oncomine.org) (26), which is a cancer microarray database
and web-based data-mining platform, was interrogated to validate
the expression status of G9A mRNA in various types of CRC. Filter
indexes were set in Oncomine based on research interests. Primary
filters were set as differential analysis (cancer vs. normal) and
cancer type (colorectal cancer). Dataset filters were set as data
type (mRNA). Datasets were ordered by overexpression with P-value.
Datasets were set by P-value (1E-6) and fold change (1.5+). The
datasets were then sorted to analyze the expression of G9A mRNA
associated with different cancer types vs. normal tissue.
Publicly available data analysis
The data sets of the patients with CRC and the
corresponding clinical data were downloaded from the publicly
available Gene Expression Omnibus (GEO) datasets (http://www.ncbi.nlm.nih.gov/gds/; National Center
for Biotechnology Information, Bethesda, USA). A total of 6
independent data sets from GSE38832 (27) (n=122), GSE37892 (28) (n=130), GSE28722 (29) (n=125), GSE17536 (30,31)
(n=177), GSE18088 (32) (n=53), and
GSE33113 (33,34) (n=96) were utilized to analyze the
expression level of G9A in CRC. The patients were divided into two
groups according to their G9A expression level (top 50%, high vs.
bottom 50%, low). For GSE38832, GSE37892, GSE17536, GSE18088 and
GSE33113, Log2 intensity of probe 202326 were used to represent the
expression level of G9A. For GSE28722 and GSE6988, Log2 intensity
of probe 10023819278 and AA434117 were used to represent the
expression level of G9A, respectively. The significance was defined
as P<0.05.
Gene set enrichment analysis
(GSEA)
JAVA program for GSEA (http://www.broadinstitute.org/gsea) (35,36) was
utilized to analyze the potential genes influenced by G9A high
expression. CRC patient gene profiling data (GSE37892 and GSE18088)
was obtained from the GEO site. The patients were divided into two
groups according to their G9A expression level (top 50%, high vs.
bottom 50%, low) and GSEA was carried out to assess the effects of
G9A expression level on various biological gene sets. MsigDB c5 (GO
gene sets, 1,454 gene sets) was used. Gene sets with a false
discovery rate value (FDR) of <0.25 and normal values of
P<0.05 subsequent to performing 1,000 permutations were regarded
as having significant enrichment.
Statistical analysis
Experimental data were analyzed with SPSS 13.0
statistical software (SPSS, Inc., Chicago, IL, USA). The
χ2 and Fisher's exact tests were used to analyze the
statistical significance of the relationship between G9A expression
and the clinicopathological features. For univariate recurrence
analysis, recurrence curves were obtained with the Kaplan-Meier
method and compared using the log-rank test. P<0.05 was
considered to represent a statistically significant difference.
Results
G9A expression was increased in CRC
tumor tissues
To investigate the role of G9A in colorectal cancer,
we examined G9A protein expression level in 100 pairs of colorectal
cancer tissues and the corresponding non-cancerous tissues by
immunohistochemistry. G9A positively staining cells exhibited brown
particles that localized in nuclei (Fig.
1A-G). Results showed that G9A protein expression was
significantly increased in tumor samples (44%, n=100) in comparison
to that in the adjacent non-cancer tissue samples (8.54%, n=82)
(Fig. 1H). The difference was
significant (P<0.05). In Fig. 1G,
the represented specimen was from the junction between tumoral and
non-tumoral tissue. As illustrated in Fig. 1G, G9A protein is strongly stained in
the tumoral region, whereas the non-tumoral region is weakly
stained. The present results indicate that G9A expression in CRC
tumor samples is significantly higher than that in adjacent
noncancerous tissue samples (Fig. 1H;
P<0.05).
Association between G9A expression and
clinicopathological features of CRC
The present study analyzed the association of G9A
protein expression level with patient age, gender, size, TNM stages
(primary tumor status (T1-T4), nodal metastasis (N0-N2), distant
metastasis), Dukes stages (A-D), and histological grade (well,
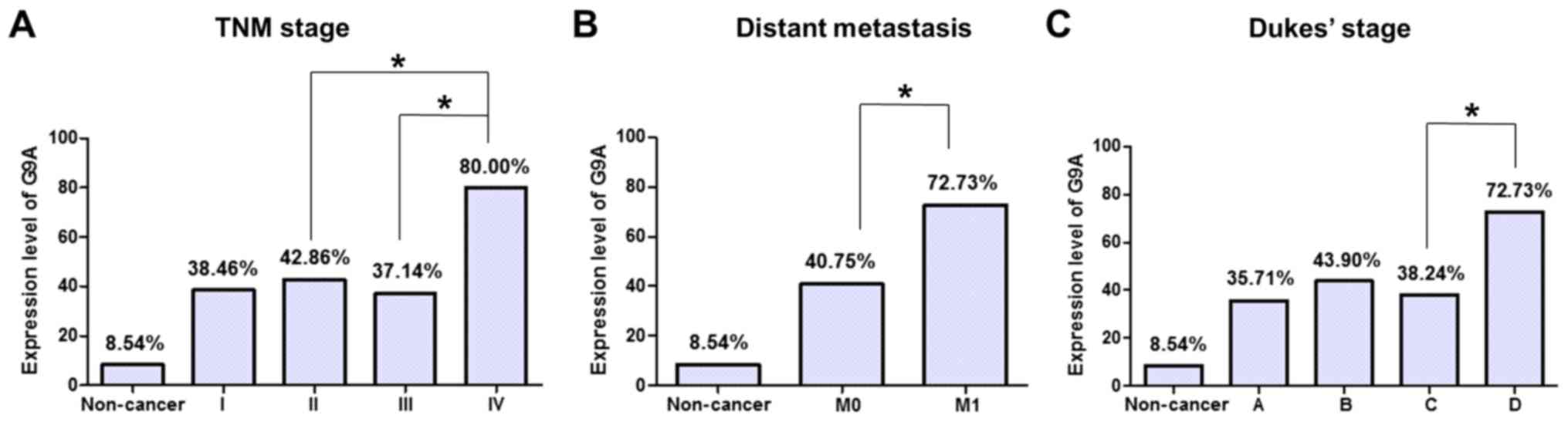
moderately, or poorly differentiated) in the CRC samples (Table I). There were significant differences
in the G9A protein expression levels between later the TNM stage
and the other TNM stages (P=0.035, P=0.020; IV compared with II and
III, respectively; Table I, Fig. 2A). In addition, expression of G9A in
tumor with distant metastasis was increased compared with that
without distant metastasis (P=0.043, Table I, Fig.
2B). It also demonstrated a weak association with Dukes Stage
that G9A expression in Dukes stage D tumor was significantly
increased compared with that in Dukes stage C tumor (P=0.049;
Table I, Fig. 2C). There was no significant
correlation of G9A expression with other clinicopathological
features, such as patient age, gender, tumor size, invasion and
nodal metastasis.
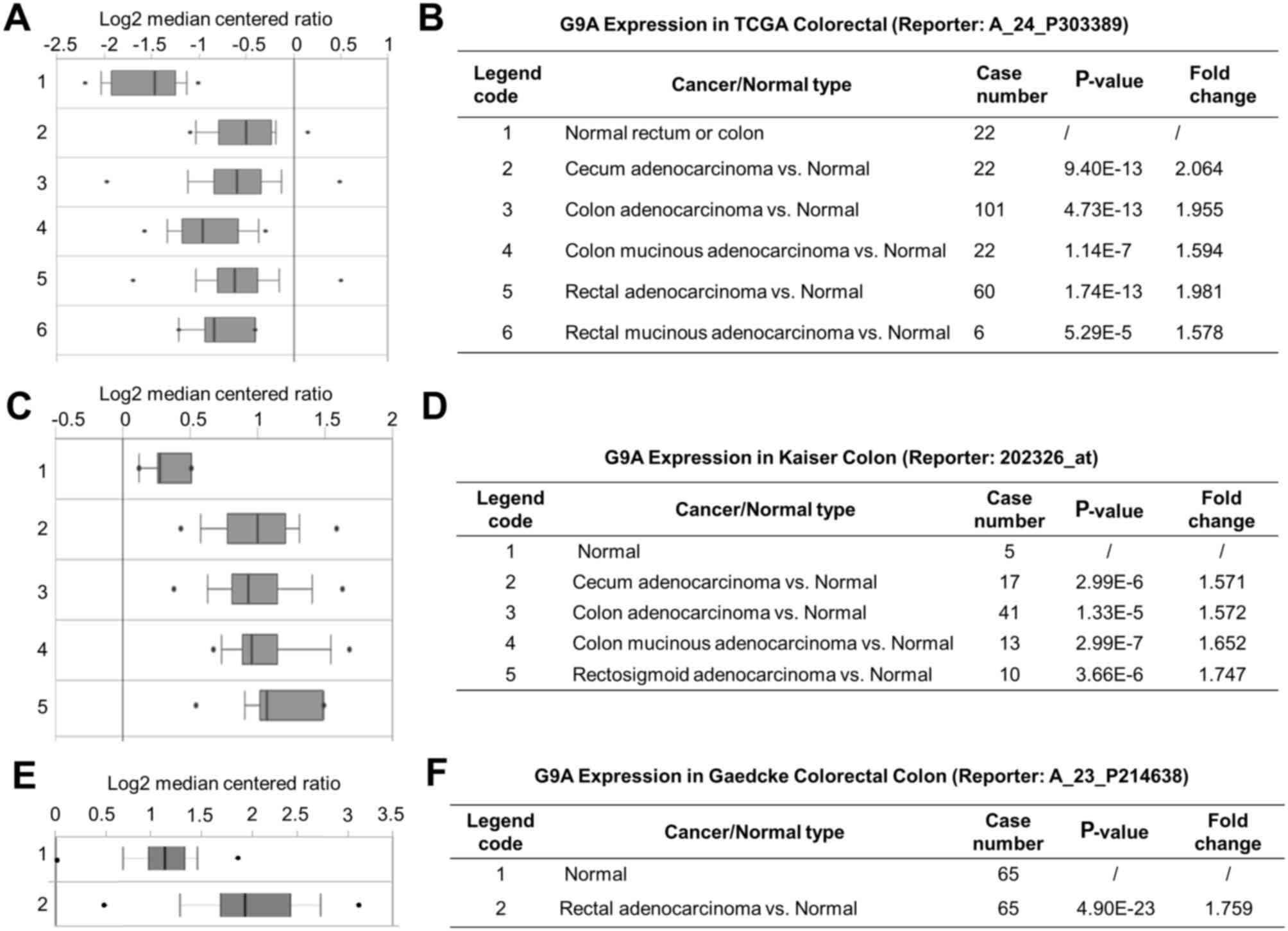
Expression of G9A using online
analysis platform
The present study explored the mRNA expression level
of G9A in CRC. Data mining and analysis of G9A mRNA expression
level from the publicly available Oncomine database was performed.
The threshold was set by the P-value (1E-6) and by fold change
(1.5+). There were 4 data sets that met the requirements, which
were the TCGA (The Cancer Genome Atlas) colorectal cancer dataset
consisting of 237 clinical samples (37), the Kaiser colon dataset consisting of
105 clinical samples (38), the
Gaedcke colorectal dataset consisting of 130 clinical samples
(39) and the Hong colorectal dataset
consisting of 82 samples (40). The
results from Oncomine further confirmed the significantly higher
expression level of G9A identified in various colorectal
carcinomas, such as cecum adenocarcinoma, colon adenocarcinoma,
colon mucinous adenocarcinoma and rectal adenocarcinoma (Fig. 3A-C). The corresponding P-values and
fold changes are exhibited in Fig.
3D-F, respectively. These results indicated that G9A mRNA
expression was increased in CRC tumor tissues.
Association between G9A mRNA
expression and clinicopathological features of colorectal
cancer
To figure out the relationships between elevated G9A
expression level and the clinicopathological features of CRC, the
present study chose 5 GEO datasets with corresponding clinical
information for further analysis (Table
II). The results demonstrated that G9A expression was
associated with AJCC staging (P=0.027) and tumor cell
differentiation (P=0.012) in GSE17536 (n=177), and it was also
related with cancer relapse in GSE18088 (P=0.045, n=53). In
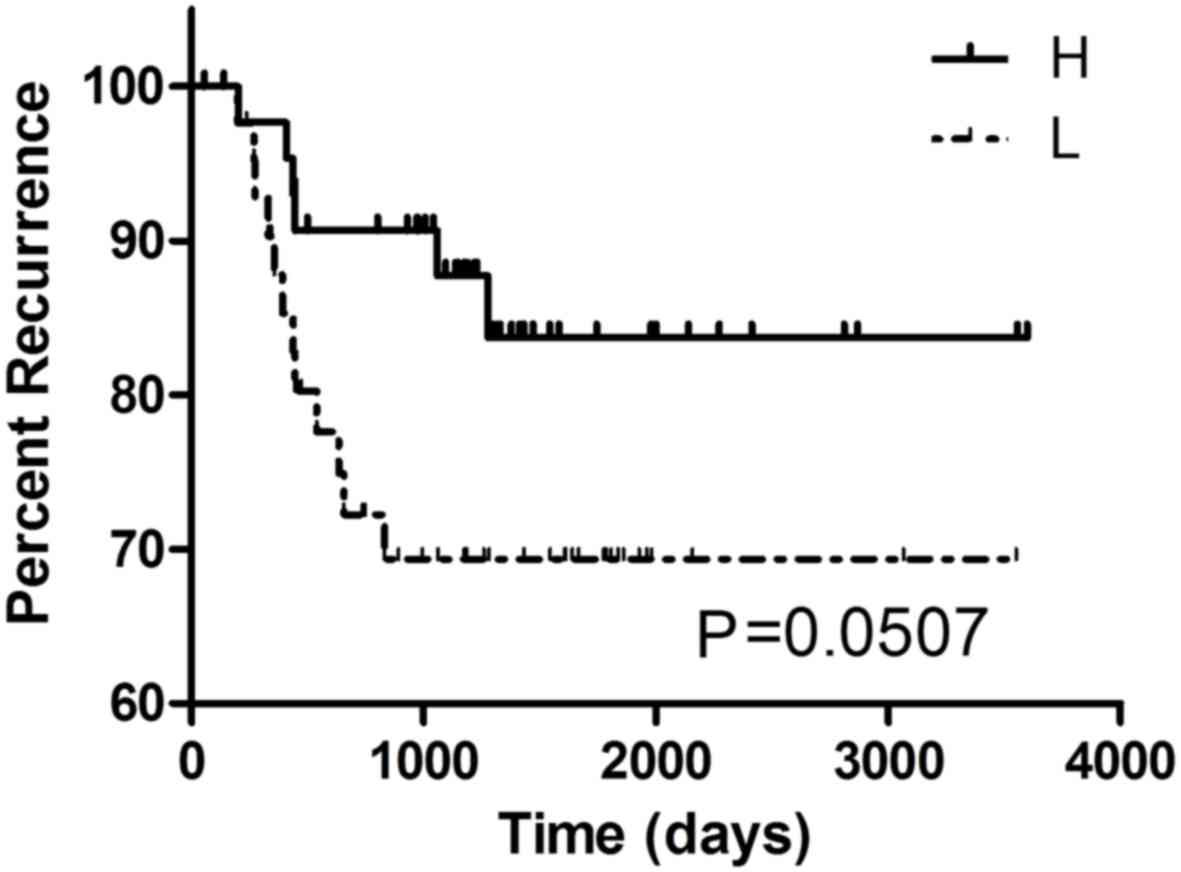
GSE33113 (n=96), a marked association between G9A expression and
the recurrence rate in CRC was found (P=0.0507, Fig. 4). It might suggest that CRC patients
with high expression level of G9A were more likely to suffer
relapse. However, cause the result based on GSE33113 was
borederline signficant (P=0.0507), this result still requires
further investigation to further verify, as the association in this
dataset did not prove to be statistically significant. No other
associations between G9A expression and clinicopathological
features in any other CRC datasets we found, including metastasis,
staging, location or survival rate.
 | Table II.Correlation between G9A mRNA
expression and the clinicopathological features of the CRC. |
Table II.
Correlation between G9A mRNA
expression and the clinicopathological features of the CRC.
|
|
|
| G9 expression |
|
|---|
|
|
|
|
|
|
|---|
| Datasets | Characteristic | Case size | High | Low |
P-valuea |
|---|
| GSE38832 | Ajcc staging |
|
|
| 0.259 |
| Probe: 202326 | 1 | 18 | 7 | 11 |
|
|
| 2 | 35 | 17 | 18 |
|
|
| 3 | 39 | 25 | 14 |
|
|
| 4 | 30 | 14 | 16 |
|
| GSE28722 | Duke's staging |
|
|
| 0.618 |
| Probe:
10023819278 | A | 3 | 1 | 2 |
|
|
| B | 83 | 38 | 45 |
|
|
| C | 34 | 12 | 22 |
|
|
| D | 5 | 3 | 2 |
|
|
| Metastasis |
|
|
| 0.916 |
|
| Y | 33 | 14 | 19 |
|
|
| N | 92 | 40 | 52 |
|
| GSE17536 | Ajcc staging |
|
|
| 0.162 |
| Probe: 202326 | 1 | 24 | 7 | 17 | 0.027b |
|
| 2 | 57 | 32 | 25 |
|
|
| 3 | 57 | 29 | 28 |
|
|
| 4 | 39 | 18 | 21 |
|
|
| Differentiated |
|
|
| 0.012 |
|
| Well | 16 | 2 | 14 |
|
|
| Moderately | 134 | 69 | 65 |
|
|
| Poorly | 27 | 14 | 13 |
|
|
| Recurrence |
|
|
| 0.313 |
|
| Y | 36 | 20 | 16 |
|
|
| N | 109 | 50 | 59 |
|
| GSE18088 | Relapse |
|
|
| 0.045 |
| Probe: 202326 | Y | 13 | 10 | 3 |
|
|
| N | 40 | 18 | 22 |
|
|
| Location |
|
|
| 0.506 |
|
| Proximal | 28 | 16 | 12 |
|
|
| Distal | 25 | 12 | 13 |
|
|
|
Differentiation |
|
|
| 0.327 |
|
| Well | 2 | 1 | 1 |
|
|
| Moderate | 35 | 21 | 14 |
|
|
| Low | 16 | 6 | 10 |
|
G9A expression level associated with
proliferation of CRC cells
The G9A expression level was found consistently
increased in both public available datasets and in the present CRC
tumor samples. A GSEA analysis was conducted to investigate the
potential biological processes that G9A high expression may
influence. The results from GSEA using GSE37892 demonstrated that
gene sets differences in G9A high vs. low patients indicated that
G9A regulated gene sets mainly associated with DNA replication
(P=0.016, FDR=0.160, Fig. 5A). The
result by analyzing GSE18088 also indicated that high expression of
G9A regulated gene sets associated with mitosis, although the FDR
value was weak (M phase, P=0.042, FDR=0.433; mitosis, P=0.040,
FDR=0.442; Fig. 5B and C). The
present study concluded that G9A may be important for proliferation
of CRC cells.
Discussion
The present study investigated the
clinicopathological significance of G9A expression in CRC
progression. Immunohistochemistry was performed to explore G9A
protein expression pattern in 100 pairs of CRC samples. Combined
with the results of the bioinformatics analysis, it was
demonstrated that G9A expression was increased in CRC and that it
is therefore involved in the carcinogenesis of CRC.
G9A is the primary histone lysine methyltransferase
of lysine 9 on histone H3. Various studies have demonstrated that
G9A is critical for numerous biological progresses, such as embryo
development, behavior plasticity, lymphocyte development, stem cell
differentiation and tumor cell growth (13,16,17,41).
It has been reported as overexpressed in numerous types of cancer,
including hepatocellular carcinoma, bladder cancer, leukemia and
lung cancer (17,18,42).
Inhibition of G9A represses cellular proliferation by inducing
autophagy related cell death in colon cancer, breast cancer and
neuroblastoma cells (17,20). The present study investigated the
potential functions of G9A in CRC progression.
The results of the present immunohistochemistry
analysis were consistent with Oncomine datasets, demonstrating that
G9A expression was significantly elevated in CRC tumor tissues.
Fig. 1G exhibited clearly that G9A
was strongly stained in the tumor region and weakly stained in the
non-tumor region. The clinical significance of high G9A expression
in CRC was further analyzed. It was found that G9A protein
expression was significantly higher in TNM stage IV tumor than any
other TNM stages in our 100 specimens. In addition, the G9A
expression exhibited a higher expression in Dukes stage D tumor
than that in stage C tumor, and also a higher expression in tumor
with distant metastasis than that without distant metastasis.
According to the principle of all these different types of staging
methods, G9A expression levels may be important for the metastasis
status of CRC. A previous study on lung cancer has stated that G9A
has the ability to promote tumor invasion and metastasis in lung
cancer (43). The present study
provided the evidence that increased expression of G9A was
associated with metastasis in CRC. In accordance, by using publicly
available datasets from GEO, it was found that high G9A expression
was associated with AJCC staging and differentiation of CRC in one
of the datasets analyzed. However, since the results were not
consistent among different datasets used, and the association
between G9A expression and tumor relapse in CRC was borderline
significant, the associations between G9A and tumor metastasis or
tumor recurrence need to be further confirmed.
To find the potential function of G9A in CRC
progression, GSEA analysis was performed. The results demonstrated
that G9A was significantly associated with DNA replication and
mitosis in CRC tumor cells in 2 datasets analyzed. These results
strongly indicated that G9A was important for colorectal cancer
cell proliferation, which was consistent with the previous research
(20). It has been demonstrated that
G9A inhibition represses proliferation in colon cancer HCT116 cells
and numerous other types of cancer cell (20,21). The
present study suggested that high expression level of G9A was
critical for CRC tumorigenesis.
In the present study, the clinicopathological
significance of G9A high expression in CRC was discussed. In
conclusion, the results suggested that G9A expression was increased
in CRC tumor samples and the high expression was important for
tumorigenesis. Evidence that G9A high expression may be important
for distant metastasis in CRC has been provided. The findings
helped to further understand the crucial role of G9A in
tumorigenesis, and also offered significant ideas for CRC
therapy.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant no. 31300609), the
Natural Science Foundation of Hubei Province in China (grant nos.
2012FFB04316 and 2013CFB255) and The Incubator Project of Renmin
Hospital Wuhan University (grant no. 2013RMFH008).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JQ and ZZ analyzed and interpreted the patient data.
ZZ, QL and YH collected the samples. TL contributed to data
analysis and the manuscript drafting. JQ and LC contributed to the
study design and manuscript writing.
Ethics approval and consent to
participate
Ethical approval for the study was granted by the
Renmin Hospital's ethics committee. Informed written consent was
obtained from all participants involved in the study.
Consent for publication
Informed written consent for publication was
obtained from all participants involved in the study.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Rodríguez-Paredes M and Esteller M: Cancer
epigenetics reaches mainstream oncology. Nat Med. 17:330–339. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fraga MF, Ballestar E, Villar-Garea A,
Boix-Chornet M, Espada J, Schotta G, Bonaldi T, Haydon C, Ropero S,
Petrie K, et al: Loss of acetylation at Lys16 and trimethylation at
Lys20 of histone H4 is a common hallmark of human cancer. Nat
Genet. 37:391–400. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wood LD, Parsons DW, Jones S, Lin J,
Sjöblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J, et al: The
genomic landscapes of human breast and colorectal cancers. Science.
318:1108–1113. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Carmona FJ and Esteller M: Moving closer
to a prognostic DNA methylation signature in colon cancer. Clin
Cancer Res. 17:1215–1217. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Irizarry RA, Ladd-Acosta C, Wen B, Wu Z,
Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, et al:
The human colon cancer methylome shows similar hypo- and
hypermethylation at conserved tissue-specific CpG island shores.
Nat Genet. 41:178–186. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jiang X, Tan J, Li J, Kivimäe S, Yang X,
Zhuang L, Lee PL, Chan MT, Stanton LW, Liu ET, et al: DACT3 is an
epigenetic regulator of Wnt/beta-catenin signaling in colorectal
cancer and is a therapeutic target of histone modifications. Cancer
Cell. 13:529–541. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fluge Ø, Gravdal K, Carlsen E, Vonen B,
Kjellevold K, Refsum S, Lilleng R, Eide TJ, Halvorsen TB, Tveit KM,
et al: Expression of EZH2 and Ki-67 in colorectal cancer and
associations with treatment response and prognosis. Br J Cancer.
101:1282–1289. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yamamoto S, Tateishi K, Kudo Y, Yamamoto
K, Isagawa T, Nagae G, Nakatsuka T, Asaoka Y, Ijichi H, Hirata Y,
et al: Histone demethylase KDM4C regulates sphere formation by
mediating the cross talk between Wnt and Notch pathways in colonic
cancer cells. Carcinogenesis. 34:2380–2388. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shinkai Y and Tachibana M: H3K9
methyltransferase G9a and the related molecule GLP. Genes Dev.
25:781–788. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Dawson MA and Kouzarides T: Cancer
epigenetics: From mechanism to therapy. Cell. 150:12–27. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Maze I, Covington HE III, Dietz DM,
LaPlant Q, Renthal W, Russo SJ, Mechanic M, Mouzon E, Neve RL,
Haggarty SJ, et al: Essential role of the histone methyltransferase
G9a in cocaine-induced plasticity. Science. 327:213–216. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Thomas LR, Miyashita H, Cobb RM, Pierce S,
Tachibana M, Hobeika E, Reth M, Shinkai Y and Oltz EM: Functional
analysis of histone methyltransferase g9a in B and T lymphocytes. J
Immunol. 181:485–493. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Katoh K, Yamazaki R, Onishi A, Sanuki R
and Furukawa T: G9a histone methyltransferase activity in retinal
progenitors is essential for proper differentiation and survival of
mouse retinal cells. J Neurosci. 32:17658–17670. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ueda J, Ho JC, Lee KL, Kitajima S, Yang H,
Sun W, Fukuhara N, Zaiden N, Chan SL, Tachibana M, et al: The
hypoxia-inducible epigenetic regulators Jmjd1a and G9a provide a
mechanistic link between angiogenesis and tumor growth. Mol Cell
Biol. 34:3702–3720. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen X, Skutt-Kakaria K, Davison J, Ou YL,
Choi E, Malik P, Loeb K, Wood B, Georges G, Torok-Storb B and
Paddison PJ: G9a/GLP-dependent histone H3K9me2 patterning during
human hematopoietic stem cell lineage commitment. Genes Dev.
26:2499–2511. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ke XX, Zhang D, Zhu S, Xia Q, Xiang Z and
Cui H: Inhibition of H3K9 methyltransferase G9a repressed cell
proliferation and induced autophagy in neuroblastoma cells. PLoS
One. 9:e1069622014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Huang J, Dorsey J, Chuikov S, Pérez-Burgos
L, Zhang X, Jenuwein T, Reinberg D and Berger SL: G9a and Glp
methylate lysine 373 in the tumor suppressor p53. J Biol Chem.
285:9636–9641. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kondo Y, Shen L, Suzuki S, Kurokawa T,
Masuko K, Tanaka Y, Kato H, Mizuno Y, Yokoe M, Sugauchi F, et al:
Alterations of DNA methylation and histone modifications contribute
to gene silencing in hepatocellular carcinomas. Hepatol Res.
37:974–983. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kim Y, Kim YS, Kim DE, Lee JS, Song JH,
Kim HG, Cho DH, Jeong SY, Jin DH, Jang SJ, et al: BIX-01294 induces
autophagy-associated cell death via EHMT2/G9a dysfunction and
intracellular reactive oxygen species production. Autophagy.
9:2126–2139. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang J, He P, Xi Y, Geng M, Chen Y and
Ding J: Down-regulation of G9a triggers DNA damage response and
inhibits colorectal cancer cells proliferation. Oncotarget.
6:2917–2927. 2015.PubMed/NCBI
|
|
22
|
Edge SB and Compton CC: The American joint
committee on cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kyriakos M: The President's cancer, the
Dukes classification, and confusion. Arch Pathol Lab Med.
109:1063–1066. 1985.PubMed/NCBI
|
|
24
|
Barresi V, Bonetti Reggiani L, Ieni A,
Caruso RA and Tuccari G: Histological grading in colorectal cancer:
New insights and perspectives. Histol Histopathol. 30:1059–1067.
2015.PubMed/NCBI
|
|
25
|
Zeng Z, Wu HX, Zhan N, Huang YB, Wang ZS,
Yang GF, Wang P and Fu GH: Prognostic significance of USP10 as a
tumor-associated marker in gastric carcinoma. Tumour Biol.
35:3845–3853. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rhodes DR, Yu J, Shanker K, Deshpande N,
Varambally R, Ghosh D, Barrette T, Pandey A and Chinnaiyan AM:
ONCOMINE: A cancer microarray database and integrated data-mining
platform. Neoplasia. 6:1–6. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tripathi MK, Deane NG, Zhu J, An H, Mima
S, Wang X, Padmanabhan S, Shi Z, Prodduturi N, Ciombor KK, et al:
Nuclear factor of activated T-cell activity is associated with
metastatic capacity in colon cancer. Cancer Res. 74:6947–6957.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Laibe S, Lagarde A, Ferrari A, Monges G,
Birnbaum D and Olschwang S: COL2 Project: A seven-gene signature
aggregates a subgroup of stage II colon cancers with stage III.
OMICS. 16:560–565. 2012.PubMed/NCBI
|
|
29
|
Loboda A, Nebozhyn MV, Watters JW, Buser
CA, Shaw PM, Huang PS, Van't Veer L, Tollenaar RA, Jackson DB,
Agrawal D, et al: EMT is the dominant program in human colon
cancer. BMC Med Genomics. 4:92011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Smith JJ, Deane NG, Wu F, Merchant NB,
Zhang B, Jiang A, Lu P, Johnson JC, Schmidt C, Bailey CE, et al:
Experimentally derived metastasis gene expression profile predicts
recurrence and death in patients with colon cancer.
Gastroenterology. 138:958–968. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Freeman TJ, Smith JJ, Chen X, Washington
MK, Roland JT, Means AL, Eschrich SA, Yeatman TJ, Deane NG and
Beauchamp RD: Smad4-mediated signaling inhibits intestinal
neoplasia by inhibiting expression of β-catenin. Gastroenterology.
142:562–571.e2. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gröne J, Lenze D, Jurinovic V, Hummel M,
Seidel H, Leder G, Beckmann G, Sommer A, Grützmann R, Pilarsky C,
et al: Molecular profiles and clinical outcome of stage UICC II
colon cancer patients. Int J Colorectal Dis. 26:847–858. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kemper K, Versloot M, Cameron K, Colak S,
de Sousa e Melo F, de Jong JH, Bleackley J, Vermeulen L, Versteeg
R, Koster J and Medema JP: Mutations in the Ras-Raf Axis underlie
the prognostic value of CD133 in colorectal cancer. Clin Cancer
Res. 18:3132–3141. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
de Sousa E, Melo F, Colak S, Buikhuisen J,
Koster J, Cameron K, de Jong JH, Tuynman JB, Prasetyanti PR,
Fessler E, van den Bergh SP, et al: Methylation of
cancer-stem-cell-associated Wnt target genes predicts poor
prognosis in colorectal cancer patients. Cell Stem Cell. 9:476–485.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Mootha VK, Lindgren CM, Eriksson KF,
Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E,
Ridderstråle M, Laurila E, et al: PGC-1alpha-responsive genes
involved in oxidative phosphorylation are coordinately
downregulated in human diabetes. Nat Genet. 34:267–273. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Cancer Genome Atlas Network: Comprehensive
molecular characterization of human colon and rectal cancer.
Nature. 487:330–337. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kaiser S, Park YK, Franklin JL, Halberg
RB, Yu M, Jessen WJ, Freudenberg J, Chen X, Haigis K, Jegga AG, et
al: Transcriptional recapitulation and subversion of embryonic
colon development by mouse colon tumor models and human colon
cancer. Genome Biol. 8:R1312007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Gaedcke J, Grade M, Jung K, Camps J, Jo P,
Emons G, Gehoff A, Sax U, Schirmer M, Becker H, et al: Mutated KRAS
results in overexpression of DUSP4, a MAP-kinase phosphatase, and
SMYD3, a histone methyltransferase, in rectal carcinomas. Genes
Chromosomes Cancer. 49:1024–1034. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hong Y, Downey T, Eu KW, Koh PK and Cheah
PY: A ‘metastasis-prone’ signature for early-stage mismatch-repair
proficient sporadic colorectal cancer patients and its implications
for possible therapeutics. Clin Exp Metastasis. 27:83–90. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Tachibana M, Sugimoto K, Nozaki M, Ueda J,
Ohta T, Ohki M, Fukuda M, Takeda N, Niida H, Kato H and Shinkai Y:
G9a histone methyltransferase plays a dominant role in euchromatic
histone H3 lysine 9 methylation and is essential for early
embryogenesis. Genes Dev. 16:1779–1791. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Cho HS, Kelly JD, Hayami S, Toyokawa G,
Takawa M, Yoshimatsu M, Tsunoda T, Field HI, Neal DE, Ponder BA, et
al: Enhanced expression of EHMT2 is involved in the proliferation
of cancer cells through negative regulation of SIAH1. Neoplasia.
13:676–684. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chen MW, Hua KT, Kao HJ, Chi CC, Wei LH,
Johansson G, Shiah SG, Chen PS, Jeng YM, Cheng TY, et al: H3K9
histone methyltransferase G9a promotes lung cancer invasion and
metastasis by silencing the cell adhesion molecule Ep-CAM. Cancer
Res. 70:7830–7840. 2010. View Article : Google Scholar : PubMed/NCBI
|