Cancer is a multifactorial disease and is considered
as a major public health issue in both developing and developed
countries (1). Due to the recent
improvements in medical tools and techniques cancer death rates
seem to be declining, at least for some types of cancer. However,
if the bigger picture is to be considered, cancer incidence and
mortality rates are still very high. Furthermore, the increased
adoption of unhealthy lifestyle behaviors, such as smoking, alcohol
abuse, poor diet and physical inactivity, further enhance the risk
of cancer occurrence. Hence, the medical burden of cancer is
increasing at a significant rate, particularly in the less
economically developed countries. According to latest statistics,
an estimated 14.1 million new cancer incidences and 8.2 million
deaths from across the world were reported in 2012 (1).
As far as treatment and management of cancer is
concerned, application of chemotherapy is quite customary.
Cisplatin (DDP), which essentially functions by damaging the
Deoxyribonucleic acid (DNA) of cancerous cells, had been widely
used for many years in clinical settings and has shown satisfactory
results. To date, cisplatin-based multidrug chemotherapy regimens
remain a standard treatment method for many cancers. In the
meantime, other chemotherapeutic drugs, like doxorubicin,
paclitaxel, irinotecan (CPT), oxaliplatin, vincristine,
hydroxyurea, rapamycin, have emerged as potential anticancer
agents. In the recent years, targeted drugs such as cetuximab,
trastuzumab, panitumumab and imatinib have also been identified as
candidate drugs for the treatment of diverse types of cancers.
However, it is noteworthy that the application of many such
chemotherapeutic agents in clinical settings is limited, especially
due to the development of drug resistance against them. Such
conditions often lead to treatment failure and local recurrence of
the disease. Considering the severity of the implications,
chemoresistance in cancer has received a lot of attention by
researchers and medical experts. It is not surprising that the
study of chemoresistance is now considered as important as new
anticancer drug development.
Drug resistance can be classified into two main
categories: intrinsic and acquired (2,3). To devise
methods and drugs that can overcome the effects of drug resistance,
researchers have been focusing on elucidating the molecular
mechanisms underlying the development of drug resistance which can
further help in elucidating chemoresistance-related mechanisms and
devising methods of preventing it. The various mechanisms that were
found to contribute toward the development of chemoresistance are
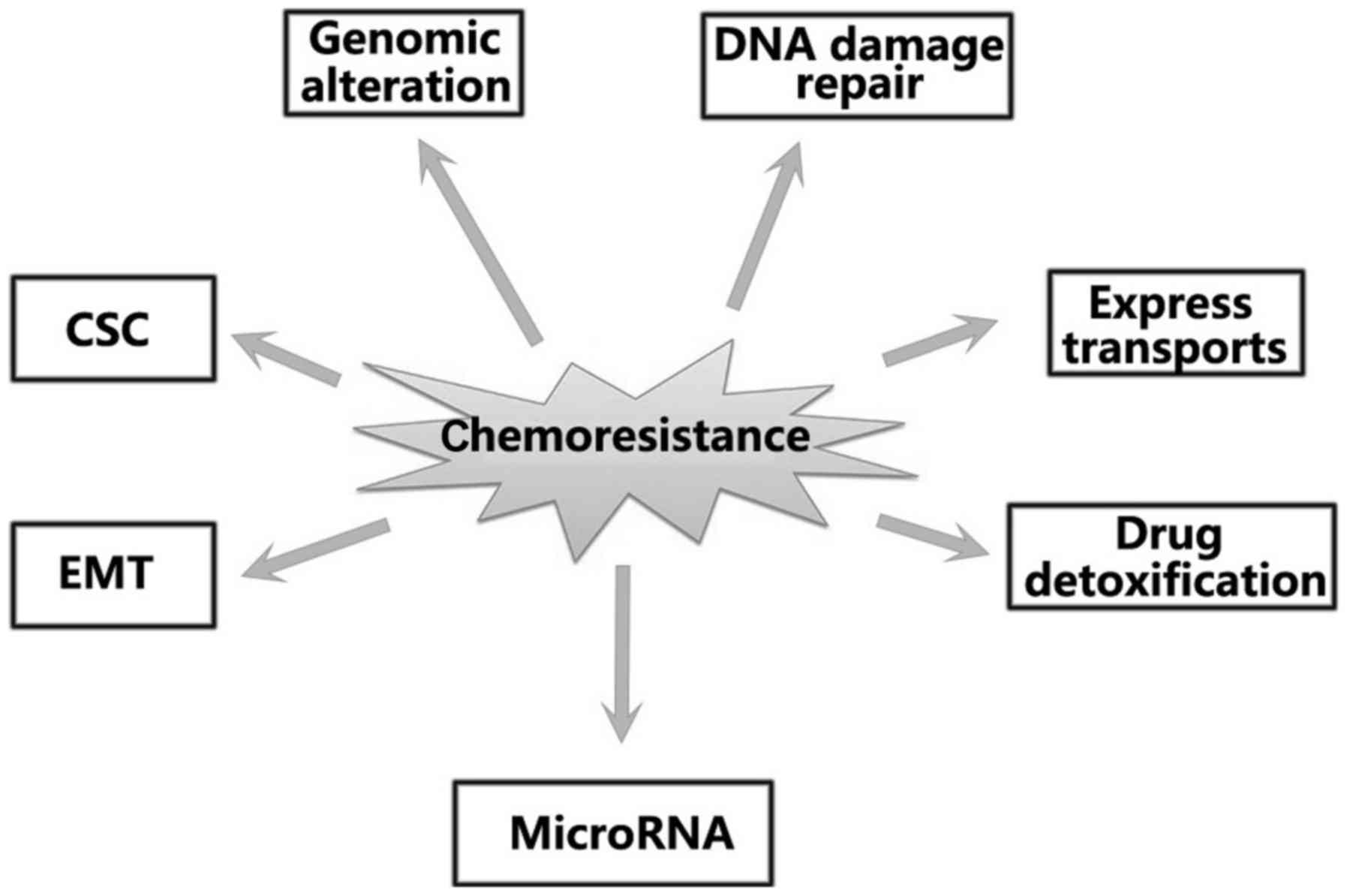
described in Fig. 1. It is evident
that factors, such as energy-dependent transporters (4), enhanced deoxyribonucleic acid repair
abilities (4), drug-detoxification
mechanisms (5),
epithelial-mesenchymal transition (6), apoptosis evasion (7–9), cancer
stem cells (CSCs) (10) as well as
microRNAs (11,12), play a major role in inducing
chemoresistance. However, the exact mechanisms that link these
factors for bringing about resistance are yet to be elucidated.
Almost all current studies on the mechanisms of chemoresistance in
cancer are still in infant stage. Therefore, overcoming
chemoresistance may possibly open new avenues for better treatment
outcome in cancer patients (13).
Protein degradation plays a key role in maintaining
cellular homeostasis. It has already been established that
ubiquitin-proteasome system is crucial for protein degradation
(14). It is also known to be
involved in various physiological responses, like cell cycle
control, DNA replication, transcription and cell signaling
(15,16). Ubiquitin (Ub) is a small protein which
remains covalently conjugated to lysine residues (17). Furthermore, ubiquitination and
deubiquitination are complex reactions whose cellular roles are
considered analogous to phosphorylation (18). The degradation of proteins by the UPS
is a multi-step enzymatic process (Fig.
2) (19) that includes
ubiquitin-activating enzyme (E1), ubiquitin conjugating enzyme (E2)
and ubiquitin-protein ligase (E3). Human genes encode over 600 E3
ligases that participate in the ubiquitylation of their individual
targets (20). The SCF complex
(21,22), which consists of four principal
components: Skp1, Cul1/Cdc53, Roc1/Rbx1/Hrt1 and a F-box protein,
is one of the major categories of E3 ligases. UPS regulates an
array of biological processes, including tumor progression and
chemoresistance. Evidently, its defective functioning is often
manifested in human beings as diseases.
Ubiquitination/deubiquitination is closely related to the
occurrence of a wide variety of tumors.
F-box protein has the protein-protein interaction
site that determines the substrate specificity of SCF complex
(23,24). It has been identified that there are
69 FBPs in the human genome. They are known to contribute to
cancers since they can recognize individual targets (25). The recognizable domains beyond the
F-box domain can be organized into three categories viz. WD 40
repeats (FBXW), leucine-rich repeats (FBXL) and other diverse or
unknown domain-containing proteins (FBXO) (10). Then again, there are 10 members of
FBXW in the human genome. FBXW1 (or β-TrCP) and FBXW7 (Cdc4) are
well known FBXW. In addition, the FBXL subfamily has 21 members.
FBXL1 (also known as SKP2) is a typical FBXL family protein. As far
as FBXO are concerned, 38 members have been identified till yet. It
is important to note that FXBO has no common substrate recognition
motif. A given F-box protein can recognize more than one substrate
(e.g. Skp2 targets FOXO1, RASSF1, ATF4). Similarly, one substrate
can be targeted by many F-box proteins (e.g. cyclin D1 can be
targeted by FBXW8 or FBXO31).
Aberrant activation of FBPs has been extensively
reported in numerous cancer types, particularly in digestive system
tumors (26,27). Out of the 69 FBPs, notably, only a few
have been studied extensively. It has also been established that
FBPs can recognize and degrade a number of oncoproteins and tumor
suppressor proteins, such as p27, c-Myc and cyclin D1, including
key regulators of cell death and DNA damage response. The
overexpressed or downregulated FBPs can further contribute to the
dysregulation of their target proteins. Therefore, the possible
roles of FBPs in inducing drug resistance are just beginning to
emerge. In the next section, we will discuss the recent research
advances pertaining to the role of FBPs in chemoresistance and how
the same can be used for directing drug discovery.
FBXW7, a well-established FBXW subfamily protein,
was first identified in budding yeasts in the year 1973 (28). FBXW7 gene resides on human beings
chromosome 4 (4q31.3) which has already been identified as a key
player in the occurrence of many forms of cancers. FBXW7 mutation
has been observed in 6% primary human tumors (29). Mutations in FBXW7 lead to the rapid
accumulation of degradable proteins, which in turn facilitates
tumor progression (30). It is
therefore obvious that FBXW7 plays crucial tumor suppressor
functions in many tumors. FBXW7 primarily exerts its antitumor
functions by regulating the degradation of an entire network of
proteins, including cyclin E (31),
c-Myc (32), c-Jun (33), Notch (33), presenilin (34), (myeloid cell leukemia-1) Mcl-1
(35), many of which have oncogenic
functions. Mutated FBXW7 is also known to mediate the stabilization
of oncoproteins in tumors and thus causes induction of
chemoresistance (36,37). Therefore, the downregulation of FBXW7
protein levels may contribute to the tumor progression and
chemoresistance.
FBXW7 has already been categorized as a
chemoresistance-related gene. Mounting evidences show that FBXW7
genetic status has an intricate relationship with chemotherapeutic
drug resistance in cancer patients. Therefore, FBXW7 is proposed as
a promising therapeutic target to improve sensitivity and efficacy
of chemotherapeutic drugs. Wertz et al (37) have already reported that loss of FBXW7
led to increased resistance of colon cancer cells towards taxol. On
the other hand, inhibition of Mcl-1 was found to restore the cancer
cells' sensitivity toward taxol- and vincristine-induced cell
death. Mcl-1, a key pro-survival BCL2 family member, is known to be
involved in mitotic arrest. This further indicates that there
exists a molecular link that forms the basis of antitubulin agent
resistance and chemotherapy induced polyploidy. Hence, Mcl-1
degradation can be implied for the development of targeted
therapeutic methods intended towards the eradication of colon
cancer cells (38). Detailed analysis
has shown that FBXW7 mutations in colorectal cancer cells are
responsible for blocking Mcl-1 degradation and thus mediating the
development of resistance of against targeted therapies
(regorafenib) (39). In consistent
with the results observed in colon cancer cell lines, FBXW7
ablation in ovarian cancer cells was also found to inhibit c-Myc
degradation. This makes the cells more resistant to
vincristine-induced cell death. The above mentioned research
studies on the one hand showcased the key roles of FBXW7 in
inducing therapeutic effects of the chemotherapeutic drugs, while
on the other hand, they also provided the much needed information
for identifying the possible ways to increase the sensitivity of
cancer cells to vincristine. Inuzuka et al (35) also found that E3 ubiquitin ligase SCF
(FBXW7) plays key regulatory roles in cellular apoptosis by
targeting Mcl-1 for ubiquitylation in a manner which depends on
phosphorylation by glycogen synthase kinase 3. Notably, loss of
FBXW7 and the resultant subsequent higher levels of Mcl-1 were
found to increase the sensitivity of cancer cells to the targeted
therapy drug sorafenib. However, it was observed that these cells
acquired resistance to the Bcl-2 family inhibitor ABT-737 in
T-acute lymphoblastic leukaemia (T-ALL) cells.
In addition, multidrug resistance-associated protein
(MRP) is also found to be closely related to FBXW7 in
nasopharyngeal carcinoma (NPC) cells. MRP makes FBXW7-deficient
cells more resistant against antitumor drug DDP. Conversely,
upregulation of FBXW7 expression restores CDDP chemosensitivity in
these cells (40). Cryptochrome 2
(CRY2), a circadian clock protein, is another such protein which is
overexpressed in chemoresistant colorectal cancer cells (40). Fang et al (41) proved that CRY2 is one of the direct
targets of FBXW7 in DLD-1 and SW480-colorectal cancer cell lines
too. Furthermore, cells with low levels of expression of CRY2 were
found to be more sensitive to oxaliplatin. It has also been proved
that FBXW7 mutation in T cell acute lymphoblastic leukemia (T-ALL)
causes stabilization of c-Myc, which is the Notch-1 target. Due to
these reasons these cells were found to be more resistant to
γ-secretase inhibitor treatment (42). The findings of the study conducted by
O'Neil et al (43) in 2007
were also found to be in agreement with the report of Thompson
et al (42), which indicated
that FBXW7 ablation increased resistance to γ-secretase inhibitors
in leukemic cells via Notch pathway activation. Both the studies
indicated that patients harboring FBXW7 mutation were more
resistant to treatment with GSI. It can thus be concluded that
FBXW7 inactivation had relationship with chemotherapeutic drugs by
its diverse targets. Although many chemoresistance targets of FBXW7
have been identified in different types of cancer cells, it still
remains enigmatic that which one is the most relevant.
A more recent study showed that inhibitor of growth
5 (ING5)-mediated chemoresistance is also positively associated
with FBXW7 hypoexpression (44). It
was found that ING5 overexpression increased U87 cells'
chemosensitivity towards cisplatin, MG132, paclitaxel and
suberoylanilide hydroxamic acid (SAHA). Further studies indicated
that ING5-associated chemoresistance may be the result of
dysfunction of Akt and NF-κB pathways. Furthermore,
epithelial-mesenchymal transition (EMT) is one of the main factors
responsible for the induction of chemoresistance. Notably, FBXW7 is
known to play a central role in controlling the EMT in cancer cells
(45). Reduced expression of FBXW7 is
proposed to reduce the polarity and adherent junctions of cells,
promote tumor cell invasion and migration, thus resulting in
reduced chemosensitivity of non-small-cell lung cancer cells
towards cisplatin (45).
Cancer stem cells (CSC) are another facet of the
molecular mechanism of chemoresistance. It has been observed that
destruction of the CSCs can be an effective strategy for improving
chemotherapeutic effects of anticancer agents. Some highly
intricate multidimensional studies indicated that FBXW7 also plays
a crucial role in the maintenance of both normal stem cells and
cancer-initiating cells (CICs) (46).
Evidently, FBXW7-deficient leukemia-initiating cells (LICs) were
more found to be resistant against conventional chemotherapy, but
sensitive to imatinib. Studies conducted in cancer mouse models
revealed that combining FBXW7 genetic ablation and imatinib is more
effective than any of these strategies applied alone (47). When all these aspects are considered
together, it becomes apparent that FBXW7 plays prominent role in
the induction of chemoresistance in cancer cells.
Despite the innumerable number of studies conducted
on FBXW7, little is known about the regulatory mechanisms that
control the molecular pathways leading up to FBXW7 induced
chemoresistance. As far as the possibility and scope of
applicability of FBXW7 in devising therapeutic strategies against
cancer is concerned, it has been observed that tumor suppressor
gene p53 directly targets FBXW7 and promotes the transcription of
FBXW7 mRNA (48). Recently, upstream
regulators like EBV Nuclear Antigen (EBNA1)-binding protein 2
(Ebp2) (49), CCAAT/enhancer-binding
protein-δ (C/EBPδ) (50), microRNA
(miRNA)-27a (51) were identified. It
has thus been proposed that all these referred proteins or miRNAs
can be candidate targets that can be implicated to increase drug
sensitivity, particularly by targeting FBXW7. Increasing number of
research studies are indicating that miRNAs have regulatory roles
in the process of tumor progression that are particularly
attributed to chemoresistance or radioresistance. In addition, it
has also been elucidated that FBXW7 is directly regulated by the
dysregulation of microRNA pathway. MicroRNAs, such as miR-25 and
miR-223, play essential role in regulating FBXW7, particularly by
reducing their mRNA levels (52).
miR-223 is an oncogenic microRNA and highly expressed in human
erythroleukemic cell line K562 (52).
miR-223/FBXW7 pathway received increased attention, especially
after the discovery of the possible implications of miR-223/FBXW7
as an efficient therapeutic target for overcoming chemoresistance.
In the past three years alone, three separate research studies have
independently identified miR-223/FBXW7 as a key signal pathway
involved in chemoresistance. The study conducted by Zhou et
al (53) showed that miR-223
could promote DDP resistance in gastric cancer cells by
downregulating FBXW7. Notably, low expression of FBXW7 contributed
to cell cycle arrest and apoptosis. In support of this notion,
miR-223 inhibitor can significantly upregulate the expression of
FBXW7 mRNA and protein in 7901/DDP cells (gastric cancer cell
line). Conversely, miR-223 mimic can downregulate the expression of
FBXW7 mRNA and protein. Further analysis of the same indicated that
the overexpression of miR-223 induced significant increase of the
expression and activity of cyclin E, while reduced miR-223
expression led to increased FBXW7 expression and decreased cyclin E
activity. Yet, another study that was conducted by Li et al
(54) demonstrated that miR-223/FBXW7
axis regulates doxorubicin sensitivity through epithelial
mesenchymal transition in non-small cell lung cancer. miR-223/FBXW7
pathway has also been investigated for drug resistance in cancers
(38,55–57). FBXW7
mutation drives acquired resistance to targeted agents cetuximab or
panitumumab (58). Eto et al
(59) were the first to report that
the overexpression of miR-223 decreases FBXW7 expression and the
sensitivity of gastric cancer cells to trastuzumab. These studies
provided important insights into the possibility of application of
miR-223/FBXW7 as a biomarker that can be used to estimate the
efficacy of DDP-based chemotherapy. However, in order to do so it
is imperative that the upstream stimuli and downstream targets of
the above-mentioned pathway are studied in detail. As of now, it is
said that almost all of the relevant studies indicate that FBXW7 is
a chemosensitizer in cancer cells, which when lost enhances the
risk of chemoresistance.
In 1995, the Beach group discovered that S phase
kinase associated protein 2 (SKP2) gene is located on the 5p13
chromosome (60). Since then,
numerous studies have highlighted it's oncogenic roles (61–64).
Furthermore, the overexpression of SKP2 has been reported in many
tumor types (65–69). These results are a clear indication
that SKP2 has significant impact on the progression of tumors and
also induction of drug resistance in different types of cancer
cells.
SCF SKP2 is a multicomponent RING-type E3 ligase
that targets and degrades many tumor suppressor proteins, such as
p27, p16, p21, p57, E2F-1, TOB1, RBL2, cyclin D/E, BRCA2, FOXO1 and
RASSF1A, and regulates numerous cellular processes. As cell cycle
regulation is a key mechanism by which most chemotherapy agents
exert their cytotoxic effect, their alterations are likely to have
major implications in the drug induced responses (70). When collectively studied, these
studies clearly indicated that SKP2 is an oncoprotein and also the
chief regulator of cell cycle inhibitor p27Kip1, which
is known as a cell cycle protein that is involved in tumor
progression. It was thus proposed that SKP2 may also be associated
with chemoresistance (71). The
present article reviewed the available information regarding the
possible roles of SKP2 towards the induction of chemoresistance.
The authors hope and desire that the information presented in here
is implicated towards achieving a better treatment outcome for the
chemoresistant subset of human cancer patients.
SKP2 supposedly interacts with a variety of
substrates that determine the outcome of chemoresistance in cancer
cells. SKP2 is known to act as an oncoprotein that also has close
correlations with paclitaxel sensitivity, which is especially
brought about in human lung cancer cells by regulating Mad2 via
p27-CDKs-E2F1 signaling axis (72).
Patients with high levels of expression of SKP2 show that small
molecule inhibitors of SKP2 combine paclitaxel and bring about
better lung cancer treatment responses (72). High levels of expression of SKP2 is a
recognized biomarker for poor prognosis in cancer. Davidovich et
al revealed that high levels of SKP2 are known to have poor
response towards preoperative doxorubicin-based chemotherapy in
breast cancer patients in 2008 (73).
Furthermore, SKP2 expression not only contributes to drug
resistance but also extrinsic induction of apoptosis. It has also
been shown that high levels of expression of SKP2 in pancreatic
ductal adenocarcinoma cell lines makes them resistant towards the
tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)
(74). Hence, SKP2 expression levels
can be considered as a key determinant of antitumor responses to
mTOR inhibitors. In addition, overexpression of SKP2 also increases
cellular resistance to rapamycin (75). Furthermore, SKP2 determines
sensitivity of tumor xenograft to rapamycin, which highlights it as
a potential pharmacogenomic marker that can predict therapeutic
sensitivity of the cells towards rapamycin.
Study of SKP2 in relation to target therapy
indicated that SKP2 deficiency enhances herceptin sensitivity in
Her2-positive cancer cells and tumors (76). Except for FBXW7, SKP2 is yet another
F-box family protein that forms the SKP2 SCF complex, which
functions for the regulation of stem cells. Hematopoietic stem
cells, that are the most critical factors in prevention of bone
marrow failure in humans, are also regulated by SKP2.
It has also been studied that SKP2 deficiency leads
to elevated cyclin D1 expression levels, which in turn contribute
to increase hematopoietic stem cell cycling. Doing so enhances
sensitivity of leukemia cells towards other chemotherapeutic
agents, such as cyclophosphamide, 5-FU, and DOX (77). All the above described data highlight
the central role of SKP2 in chemoresistance. It is also proposed
that SKP2 may be used as a biomarker to identify those patients who
are likely to respond to doxorubicin in a more effective
manner.
β-TrCP (FBXW1), β-transducin repeats-containing
proteins, is a representative of the FBXW family. IκBα and
β-catenin are two well-characterized substrates of β-TrCP, thus
β-TrCP is linked closely to tumorigenesis and development (78). IκB is the inhibitor of NF-κB, which
always functions as a tumor suppressor. β-catenin is a downstream
molecule of Wnt signaling pathways. Notably, βTrCP acts as an
oncoprotein in colorectal cancer, but it is known to act as a
suppressor in gastric cancer. The simultaneous elevation of NF-κB
activity with elevated β-TRCP1 expression indicate that it can be
considered as a contributor to chemoresistance towards anticancer
drug etoposide in pancreatic carcinoma cells (79). Furthermore, cyclin D1 overexpression
is also related to chemoresistance in cancer cells.
Berberine (BER) is a traditional Chinese drug, which
is essentially an isoquinoline alkaloid purified from the Berberis
species. It has been reported that the drug possesses multiple
functions including enhancing chemotherapy sensitivity in cancer
(80). Berberine functions by
inhibiting cyclin D1 expression in human hepatoma cells HepG2 and
MHCC97L via the ubiquitin-proteasome signal pathway. Berberine
promotes the phosphorylation of cyclin D1 at the T286 site and then
accelerates binding of β-TrCP to cyclin D1 thus mediating its
degradation. It was thus apparent that knockdown of β-TrCP
expression could reduce this phenomenon (81). On the other hand, Nrf2 is a newly
identified substrate of β-TrCP (82).
Nrf2 is controlled by two distinct β-TrCP recognition motifs in its
Neh6 domain, one of which can be modulated by GSK-3 activity
(83). Furthermore, overexpression of
Nrf2, nuclear factor (erytheroid-derived-2)-like 2, increases
resistance to the chemotherapeutic drug in breast cancer, acute
myeloid leukemia and pancreatic cancer (84–86).
However, the molecular mechanism between them remains further
elucidated.
In multiple myeloma cell lines, DEP-domain
containing mTOR-interacting protein (DEPTOR), another endogenous
mTOR inhibitor, is inversely correlated with cell's responsiveness
to anticancer drugs, such as rapamycin, velcade, paclintaxel, via
modifying the mTOR pathway. The degradation of DEPTOR is mainly
controlled by β-TrCP, which gives us a clue that the dysregulation
of β-TrCP directly affects the response of patients to
chemotherapeutic agents (86). Sp1,
specificity protein 1, is another agent enhances temozolomide
resistance in glioblastoma (87).
Doxorubixin stimulus causes the high expression of β-TrCP in breast
cancer cell line MCF-7, which promoted Sp1 degradation. Therefore,
upregulation of β-TrCP is likely to increase doxorubicin-induced
cell apoptosis (88). The role of
β-TrCP in determining chemoresistance is mainly through its diverse
substrates which are involved cell death.
FBXL5, a critical regulator of iron homeostasis,
also plays crucial roles in cancer. Although little is known about
its role in chemoresistance, Wu et al (89) explored the effects of expression of
FBXL5 on anticancer drug sensitivity. In contrast, Rho GTPase
dissociate inhibitors 2 (RhoGDI2), is known to be associated with
cisplatin resistance, which is mainly infuenced by upregulation of
Bcl-2 expression and reduction of cell apoptosis in gastric cancer
cells (90). It has also been
observed that Cho group use immunoprecipitation assay is highly
efficient in elucidating the interaction between FBXL5 and RhoGDI2.
The exogenous overexpression of FBXL5 increases the sensitivity of
cisplatin through Erk and p38 pathway in RhoGDI2 high-expressed
gastric cancer cells (88).
Furthermore, single nucleotide polymorphisms (SNPs) in the FBXL7
gene are also associated with the increased breast cancer risk
(91). This can be correlated with
the fact that FBXL7 has been shown to induce the ubiquitylation of
Aurora kinase A (92) and survivin
(93) in a cell cycle-independent
manner. Notably, Kamran et al (94) found that overexpression of Aurora
kinase A negatively regulated FBXL7, leading to anti-apoptotic
protein survivin accumulation and doxorubicin resistance in gastric
cancer.
Overexpression of FBXO5 is frequently considered as
a potential oncogenic factor (95,96).
Shimizu et al used small interfering RNA knockdown method to
further explore the mechanism of doxorubicin resistance on FBXO5
accumulation. The results thus obtained indicated that the
downregulation of FBXO5 enhanced the induction of apoptosis by
doxorubicin, but not taxol. However, no synergistic effect of FBXO5
knockdown in combination with doxorubicin treatment was found in
normal cells. Therefore, inhibition of FBXO5 function can be
considered useful for enhancing sensitivity of cancer cells to
chemotherapeutic drugs and ionizing radiations (97).
FBXO6 has been suggested as a potential biomarker
for predicting anticancer drugs responsiveness. Checkpoint kinase 1
(Chk1), one of key components of the replication checkpoint
response to DNA damage response, which was found to be inversely
correlated with FBXO6 in human breast tumor tissues. Zhang et
al (98) proposed that FBXO6
promotes the degradation of Chk1 and a defect in this mechanism may
be responsible for increasing tumor cell resistance to certain
anticancer drugs CPT. It was further suggested that the forced
expression of FBXO6, but not Skp2, in the CPT-resistant cells
promotes the degradation of endogenous levels of Chk1. Such cells
exhibited strong staining for the caspase-3 cleavage product after
CPT treatment. In contrast, depletion of FBXO6 decreased the CPT
sensitivity in lung cancer cell line A549, which was completely
restored by depletion of Chk1.
P-glycoprotein (P-gp), a multidrug resistance
transporter that effluxes chemotherapeutic drugs, is a major cause
of chemotherapy failure. FBXO15 knockdown causing P-gp accumulation
enhanced vincristine resistance (99). CD44 can increase P-gp protein
stability and promote drug resistance. Ravindranath et al
(100) reported the mechanism
between them. CD44 can protect P-gp from FBXO21 mediated
degradation, thus leading to inducing resistance against
valinomycin. In addition, CD147 and type I transmembrane
glycoprotein have positive regulatory effects on P-gp expression.
FBXO22 mediates the polyubiquitination and degradation of CD147,
thus involving drug resistance. Low level of FBXO22 contributes to
the accumulation of CD147, thereby inducing cisplatin resistance in
non-small-cell lung carcinoma cell line A549/DDP cells (101).
FBXO32 is also well recognized since its
upregulation during skeletal muscle atrophy (102), which is known to negatively regulate
epithelial to mesenchymal transition in platinum
resistant-urothelial carcinoma cells (103). Furthermore, FBXO32 enhances
chemosensitivity to cisplatin by inducing apoptosis in ovarian
cancer cells (104). Although many
FBXO family proteins involved in chemoresistance, for example,
FBXO15 and FBXO21, can regulate P-gp ubiquitination, the question
that remains is that ‘which one of them is the most prominent
determiner of chemoresistance?’ Hence, further investigation that
are intended towards accurate determination of the possible roles
of FBPs in chemoresistance are warranted.
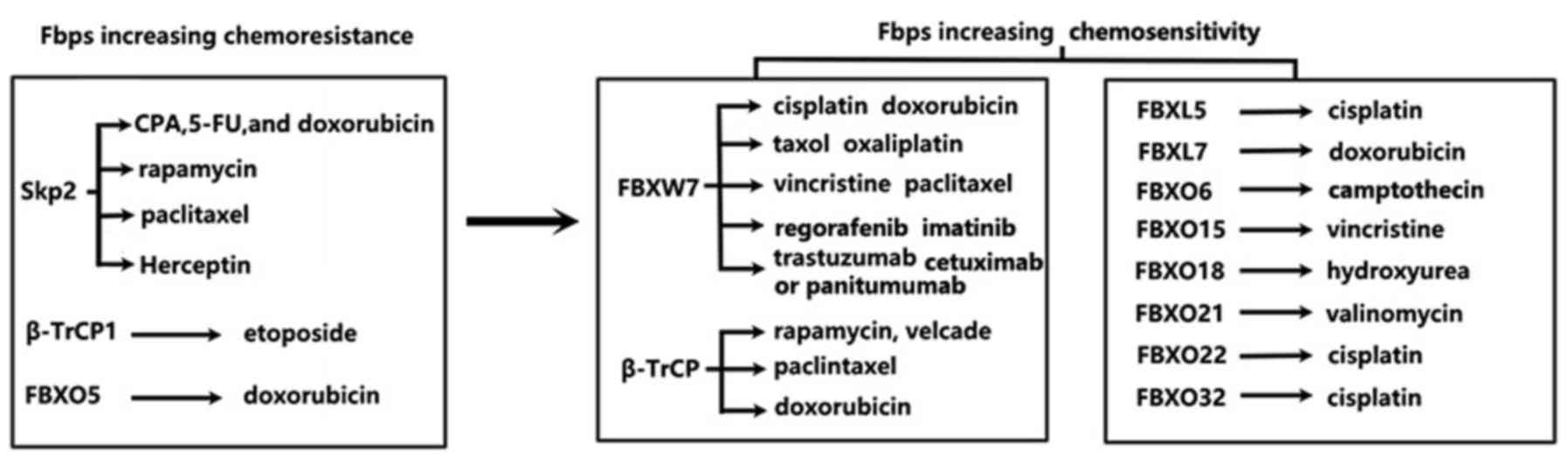
The present article presents an unprecedented
immaculate review of multiple facets of dysregulated expression of
FBPs in human cancer cells, their possible roles in regulation of
substrate turnover, their possible implications in inducing
chemoresistance against anticancer drugs, such as cisplatin,
doxorubicin, paclitaxel, CPT, oxaliplatin, vincristine,
hydroxyurea, rapamycin, trastuzumab and imatinib (Fig. 3). Analyses of the results of multiple
number of studies indicated that reconstitution of the function of
FBPs could restore the sensitivity of tumor cells to the given
chemotherapeutic agents. Organoids models have been used for
testing FBXW7-associated drug response (105). FBXO31, which is considered as a
potential tumor suppressor protein in both liver cancer and gastric
cancer (106,107), is known to induce DNA damage and
promote cyclin D1 degradation. Contrary to the results obtained
from studies conducted on liver cancer and gastric cancer, FBXO31
in esophageal cancer is known to be overexpressed (100). Notably, studies conducted in our
laboratory have demonstrated that FBXO31 is tightly correlated with
incurring drug resistance in esophageal cancer. The data generated
by us have showed that high expression of FBXO31 in esophageal
cancer cell lines promotes cisplatin resistance through MAPK signal
pathway (108). Therefore, FBPs can
be considered as possible targets for reversing the occurrence and
effects of chemoresistance. It is expected that these findings may
contribute to study of chemoresistance in individualized cancer
treatment methods. Although some progress has been made, further
investigations are urgently warranted.
Notably, until now, no compounds that can directly
target FBPs have been reported. Among the 69 FBPs that have been
identified, most studies centered on FBXW7, SKP2 and β-TrCP.
Bortezomib is a proteasome inhibitor which has been approved by the
FDA and successfully used in the treatment of hematologic
malignancies (109,110). It is proposed that it can be used to
design the disease-specific components of UPS inhibitors. FBPs,
such as the ones described in the preceding sections, are beginning
to show therapeutic potential in the field of chemosensitivity.
However, there are still many important questions that need to be
addressed in future studies. For example, many FBPs have opposite
functions in different cancer types. FBXO31 acts as a tumor
suppressor in breast cancer or gastric cancer, but have oncogenic
activity in esophageal cancer. Owing to the vast diversity of the
substrates it interacts with, it is not uncommon that FBPs have
multiple implications in cancer drug resistance. Although many of
these substrates have been identified, the ones that are
specifically related to drug resistance are yet to be elucidated.
Furthermore, such diversity of substrates also raises questions on
whether a given F-box protein can exert both chemoresistant and
chemosensitive roles in the same tissue? Despite extensive studies,
our understanding on the impact of FBPs in drug resistance is still
limited. It is therefore recommended that in-depth investigation on
this must be conducted so that critical players that are involved
in mediating drug resistance can be identified.
JG, YQZ, DLL and JRH are grateful for the
Fundamental Research Funds from the Science And Technology Plan Of
Hunan Province (grant no. 2013SK3033).
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Huang Z, Huang Y, He H and Ni J:
Podocalyxin promotes cisplatin chemoresistance in osteosarcoma
cells through phosphatidylinositide 3-kinase signaling. Mol Med
Rep. 12:3916–3922. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lippert TH, Ruoff HJ and Volm M: Intrinsic
and acquired drug resistance in malignant tumors. The main reason
for therapeutic failure. Arzneimittelforschung. 58:261–264.
2008.PubMed/NCBI
|
|
4
|
Fletcher JI, Haber M, Henderson MJ and
Norris MD: ABC transporters in cancer: More than just drug efflux
pumps. Nat Rev Cancer. 10:147–156. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Burger H, Loos WJ, Eechoute K, Verweij J,
Mathijssen RH and Wiemer EA: Drug transporters of platinum-based
anticancer agents and their clinical significance. Drug Resist
Updat. 14:22–34. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Singh A and Settleman J: EMT, cancer stem
cells and drug resistance: An emerging axis of evil in the war on
cancer. Oncogene. 29:4741–4751. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li RJ, Zhang GS, Chen YH, Zhu JF, Lu QJ,
Gong FJ and Kuang WY: Down-regulation of mitochondrial ATPase by
hypermethylation mechanism in chronic myeloid leukemia is
associated with multidrug resistance. Ann Oncol. 21:1506–1514.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li SS, Yang S, Wang S, Yang XM, Tang QL
and Wang SH: Latent membrane protein 1 mediates the resistance of
nasopharyngeal carcinoma cells to TRAIL-induced apoptosis by
activation of the PI3K/Akt signaling pathway. Oncol Rep.
26:1573–1579. 2011.PubMed/NCBI
|
|
9
|
Marin JJ, Castaño B, Blazquez AG, Rosales
R, Efferth T and Monte MJ: Strategies for overcoming chemotherapy
resistance in enterohepatic tumours. Curr Mol Med. 10:467–485.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dean M, Fojo T and Bates S: Tumour stem
cells and drug resistance. Nat Rev Cancer. 5:275–284. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fang Y, Fang D and Hu J: MicroRNA and its
roles in esophageal cancer. Med Sci Monit. 18:RA22–RA30. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sarkar FH, Li Y, Wang Z, Kong D and Ali S:
Implication of microRNAs in drug resistance for designing novel
cancer therapy. Drug Resist Updat. 13:57–66. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shapira A, Livney YD, Broxterman HJ and
Assaraf YG: Nanomedicine for targeted cancer therapy: Towards the
overcoming of drug resistance. Drug Resist Updat. 14:150–163. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hershko DD: Oncogenic properties and
prognostic implications of the ubiquitin ligase Skp2 in cancer.
Cancer. 112:1415–1424. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gao M and Karin M: Regulating the
regulators: Control of protein ubiquitinationand ubiquitin-like
modifications by extracellular stimuli. Mol Cell. 19:581–593. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Thrower JS, Hoffman L, Rechsteiner M and
Pickart CM: Recognition of the polyubiquitin proteolytic signal.
EMBO J. 19:94–102. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lee TY, Chen SA, Hung HY and Ou YY:
Incorporating distant sequence features and radial basis function
networks to identify ubiquitin conjugation sites. PLoS One.
6:e173312011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bielskienė K, Bagdonienė L, Mozūraitienė
J, Kazbarienė B and Janulionis E: E3 ubiquitin ligases as drug
targets and prognostic biomarkers in melanoma. Medicina. 51:1–9.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gong J, Cao J, Liu G and Huo JR: Function
and mechanism of F-box proteins in gastric cancer (Review). Int J
Oncol. 47:43–50. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhao Y and Sun Y: Cullin-RING Ligases as
attractive anti-cancer targets. Curr Pharm Des. 19:3215–3225. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Reed SI: Ratchets and clocks: The cell
cycle, ubiquitylation and protein turnover. Nat Rev Mol Cell Biol.
4:855–864. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Peters JM: The anaphase promoting
complex/cyclosome: A machine designed to destroy. Nat Rev Mol Cell
Biol. 7:644–656. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kipreos ET and Pagano M: The F-box protein
family. Genome Biol 1: REVIEWS3002, 2000. Epub 2000 Nov 10.
|
|
24
|
Xie CM, Wei W and Sun Y: Role of
SKP1-CUL1-F-box-protein (SCF) E3 ubiquitin ligases in skin cancer.
J Genet Genomics. 40:97–106. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang Z, Liu P, Inuzuka H and Wei W: Roles
of F-box proteins in cancer. Nat Rev Cancer. 14:233–247. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gong J, Lv L and Huo J: Roles of F-box
proteins in human digestive system tumors (Review). Int J Oncol.
45:2199–2207. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gong J and Huo J: New insights into the
mechanism of F-box proteins in colorectal cancer (Review). Oncol
Rep. 33:2113–2120. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hartwell LH, Mortimer RK, Culotti J and
Culotti M: Genetic control of the cell division cycle in yeast: V.
Genetic analysis of cdc mutants. Genetics. 74:267–286.
1973.PubMed/NCBI
|
|
29
|
Akhoondi S, Sun D, von der Lehr N,
Apostolidou S, Klotz K, Maljukova A, Cepeda D, Fiegl H, Dafou D,
Marth C, et al: FBXW7/hCDC4 is a general tumor suppressor in human
cancer. Cancer Res. 67:9006–9012. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bedford L, Lowe J, Dick LR, Mayer RJ and
Brownell JE: Ubiquitin-like protein conjugation and the
ubiquitin-proteasome system as drug targets. Nat Rev Drug Discov.
10:29–46. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Minella AC, Welcker M and Clurman BE: Ras
activity regulates cyclin E degradation by the Fbw7 pathway. Proc
Natl Acad Sci USA. 102:9649–9654. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yada M, Hatakeyama S, Kamura T, Nishiyama
M, Tsunematsu R, Imaki H, Ishida N, Okumura F, Nakayama K and
Nakayama KI: Phosphorylation-dependent degradation of c-Myc is
mediated by the F-box protein Fbw7. Embo J. 23:2116–2125. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hoeck JD, Jandke A, Blake SM, Nye E,
Spencer-Dene B, Brandner S and Behrens A: Fbw7 controls neural stem
cell differentiation and progenitor apoptosis via Notch and c-Jun.
Nat Neurosci. 13:1365–1372. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Rocher-Ros V, Marco S, Mao JH, Gines S,
Metzger D, Chambon P, Balmain A and Saura CA: Presenilin modulates
EGFR signaling and cell transformation by regulating the ubiquitin
ligase Fbw7. Oncogene. 29:2950–2961. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Inuzuka H, Shaik S, Onoyama I, Gao D,
Tseng A, Maser RS, Zhai B, Wan L, Gutierrez A, Lau AW, et al:
SCFFBW7 regulates cellular apoptosis by targeting MCL1
for ubiquitylation and destruction. Nature. 471:104–109. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Inuzuka H, Fukushima H, Shaik S, Liu P,
Lau AW and Wei W: Mcl-1 ubiquitination and destruction. Oncotarget.
2:239–244. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wertz IE, Kusam S, Lam C, Okamoto T,
Sandoval W, Anderson DJ, Helgason E, Ernst JA, Eby M, Liu J, et al:
Sensitivity to antitubulin chemotherapeutics is regulated by MCL1
and FBW7. Nature. 471:110–114. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Tong J, Tan S, Zou F, Yu J and Zhang L:
FBW7 mutations mediate resistance of colorectal cancer to targeted
therapies by blocking Mcl-1 degradation. Oncogene. 36:787–796.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Tong J, Wang P, Tan S, Chen D,
Nikolovska-Coleska Z, Zou F, Yu J and Zhang L: Mcl-1 degradation is
required for targeted therapeutics to eradicate colon cancer cells.
Cancer Res. 77:2512–2521. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Song Y, Zhou X, Bai W and Ma X: FBW7
increases drug sensitivity to cisplatin in human nasopharyngeal
carcinoma by downregulating the expression of multidrug
resistance-associated protein. Tumour Biol. 36:4197–4202. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Fang L, Yang Z, Zhou J, Tung JY, Hsiao CD,
Wang L, Deng Y, Wang P, Wang J and Lee MH: Circadian clock gene
CRY2 degradation is involved in chemoresistance of colorectal
cancer. Mol Cancer Ther. 14:1476–1487. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Thompson BJ, Buonamici S, Sulis ML,
Palomero T, Vilimas T, Basso G, Ferrando A and Aifantis I: The
SCFFBW7 ubiquitin ligase complex as a tumor suppressor
in T cell leukemia. J Exp Med. 204:1825–1835. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
O'Neil J, Grim J, Strack P, Rao S,
Tibbitts D, Winter C, Hardwick J, Welcker M, Meijerink JP, Pieters
R, et al: FBW7 mutations in leukemic cells mediate NOTCH
pathway activation and resistance to γ-secretase inhibitors. J Exp
Med. 204:1813–1824. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ding XQ, Zhao S, Yang L, Zhao X, Zhao GF,
Zhao SP, Li ZJ and Zheng HC: The nucleocytoplasmic translocation
and up-regulation of ING5 protein in breast cancer: A potential
target for gene therapy. Oncotarget. 8:81953–81966. 2017.PubMed/NCBI
|
|
45
|
Yu HG, Wei W, Xia LH, Han WL, Zhao P, Wu
SJ, Li WD and Chen W: FBW7 upregulation enhances cisplatin
cytotoxicity in non-small cell lung cancer cells. Asian Pac J
Cancer Prev. 14:6321–6326. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Izumi D, Ishimoto T, Miyake K, Eto T,
Arima K, Kiyozumi Y, Uchihara T, Kurashige J, Iwatsuki M, Baba Y,
et al: Colorectal cancer stem cells acquire chemoresistance through
the upregulation of F-Box/WD repeat-containing protein 7 and the
consequent degradation of c-Myc. Stem Cells. 35:2027–2036. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Takeishi S and Nakayama KI: Role of Fbxw7
in the maintenance of normal stem cells and cancer-initiating
cells. Br J Cancer. 111:1054–1059. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Yokobori T, Mimori K, Iwatsuki M, Ishii H,
Onoyama I, Fukagawa T, Kuwano H, Nakayama KI and Mori M:
p53-Altered FBXW7 expression determines poor
prognosis in gastric cancer cases. Cancer Res. 69:3788–3794. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Welcker M, Larimore EA, Frappier L and
Clurman BE: Nucleolar targeting of the fbw7 ubiquitin ligase by a
pseudosubstrate and glycogen synthase kinase 3. Mol Cell Biol.
31:1214–1224. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Balamurugan K, Wang JM, Tsai HH, Sharan S,
Anver M, Leighty R and Sterneck E: The tumour suppressor C/EBPδ
inhibits FBXW7 expression and promotes mammary tumour metastasis.
EMBO J. 29:4106–4117. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Lerner M, Lundgren J, Akhoondi S, Jahn A,
Ng HF, Moqadam Akbari F, Vrielink Oude JA, Agami R, Den Boer ML,
Grandér D and Sangfelt O: MiRNA-27a controls FBW7/hCDC4-dependent
cyclin E degradation and cell cycle progression. Cell Cycle.
10:2172–2183. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Xu Y, Sengupta T, Kukreja L and Minella
AC: MicroRNA-223 regulates cyclin E activity by modulating
expression of F-box and WD-40 domain protein 7. J Biol Chem.
285:34439–34446. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zhou X, Jin W, Jia H, Yan J and Zhang G:
MiR-223 promotes the cisplatin resistance of human gastric cancer
cells via regulating cell cycle by targeting FBXW7. J Exp Clin
Cancer Res. 34:282015. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Li R, Wu S, Chen X, Xu H, Teng P and Li W:
miR-223/FBW7 axis regulates doxorubicin sensitivity through
epithelial mesenchymal transition in non-small cell lung cancer. Am
J Transl Res. 2016:2512–2524. 2016.
|
|
55
|
Ye M, Zhang Y, Zhang X and Zhang J, Jing
P, Cao L, Li N, Li X, Yao L and Zhang J and Zhang J: Targeting FBW7
as a strategy to overcome resistance to targeted therapy in
non-small cell lung cancer. Cancer Res. 77:3527–3539. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Adua D, Di Fabio F, Ercolani G, Fiorentino
M, Gruppioni E, Altimari A, Limpe Rojas FL, Normanno N, Pinna AD
and Pinto C: Heterogeneity in the colorectal primary tumor and the
synchronous resected liver metastases prior to and after treatment
with an anti-EGFR monoclonal antibody. Mol Clin Oncol. 7:113–120.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Zheng M, Xu H, Liao XH, Chen CP, Zhang AL,
Lu W, Wang L, Yang D, Wang J, Liu H, et al: Inhibition of the
prolyl isomerase Pin1 enhances the ability of sorafenib to induce
cell death and inhibit tumor growth in hepatocellular carcinoma.
Oncotarget. 8:29771–29784. 2017.PubMed/NCBI
|
|
58
|
Lupini L, Bassi C, Mlcochova J, Musa G,
Russo M, Vychytilova-Faltejskova P, Svoboda M, Sabbioni S, Nemecek
R, Slaby O and Negrini M: Prediction of response to anti-EGFR
antibody-based therapies by multigene sequencing in colorectal
cancer patients. BMC Cancer. 15:8082015. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Eto K, Iwatsuki M, Watanabe M, Ishimoto T,
Ida S, Imamura Y, Iwagami S, Baba Y, Sakamoto Y, Miyamoto Y, et al:
The sensitivity of gastric cancer to trastuzumab is regulated by
the miR-223/FBXW7 pathway. Int J Cancer. 136:1537–1545. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Demetrick DJ, Zhang H and Beach DH:
Chromosomal mapping of the genes for the human CDK2/cyclin
A-associated proteins p19 (SKP1A and SKP1B) and p45 (SKP2).
Cytogenet Cell Genet. 73:104–107. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Hershko D, Bornstein G, Ben-Izhak O,
Carrano A, Pagano M, Krausz MM and Hershko A: Inverse relation
between levels of p27Kip1 and of its ubiquitin ligase
subunit Skp2 in colorectal carcinomas. Cancer. 91:1745–1751. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Fukuchi M, Masuda N, Nakajima M, Fukai Y,
Miyazaki T, Kato H and Kuwano H: Inverse correlation between
expression levels of p27 and the ubiquitin ligase subunit Skp2 in
early esophageal squamous cell carcinoma. Anticancer Res.
24:777–783. 2004.PubMed/NCBI
|
|
63
|
Yang G, Ayala G and De Marzo A: Tian W,
Frolov A, Wheeler TM, Thompson TC and Harper JW: Elevated Skp2
protein expression in human prostate cancer: Association with loss
of the cyclin-dependent kinase inhibitor p27 and PTEN and with
reduced recurrence-free survival. Clin Cancer Res. 8:3419–3426.
2002.PubMed/NCBI
|
|
64
|
Traub F, Mengel M, Lück HJ, Kreipe HH and
von Wasielewski R: Prognostic impact of Skp2 and p27 in human
breast cancer. Breast Cancer Res Treat. 99:185–191. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Rose AE, Wang G, Hanniford D, Monni S, Tu
T, Shapiro RL, Berman RS, Pavlick AC, Pagano M, Darvishian F, et
al: Clinical relevance of SKP2 alterations in metastatic melanoma.
Pigment Cell Melanoma Res. 24:197–206. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Xu HM, Liang Y, Chen Q, Wu QN, Guo YM,
Shen GP, Zhang RH, He ZW, Zeng YX, Xie FY, et al: Correlation of
Skp2 overexpression to prognosis of patients with nasopharyngeal
carcinoma from South China. Chin J Cancer. 30:204–212. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Schüler S, Diersch S, Hamacher R, Schmid
RM, Saur D and Schneider G: SKP2 confers resistance of pancreatic
cancer cells towards TRAIL induced apoptosis. Int J Oncol.
38:219–225. 2011.PubMed/NCBI
|
|
68
|
Wang Z, Fukushima H, Inuzuka H, Wan L, Liu
P, Gao D, Sarkar FH and Wei W: Skp2 is a promising therapeutic
target in breast cancer. Front Oncol. 1:pii: 18702. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Shapira M, Ben-Izhak O, Linn S, Futerman
B, Minkov I and Hershko DD: The prognostic impact of the ubiquitin
ligase subunits Skp2 and Cks1 in colorectal carcinoma. Cancer.
103:1336–1346. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Blagosklonny MV: Review Why therapeutic
response may not prolong the life of a cancer patient: Selection
for oncogenic resistance. Cell Cycle. 4:1693–1698. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Zhuang K, Zhang L, Zhang X, Tang H, Zhang
J, Yan Y, Han K and Guo H: Gastrin induces multidrug resistance via
the degradation of p27Kip1 in the gastric carcinoma cell
line SGC7901. Int J Oncol. 50:2091–2100. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Huang T, Yang L, Wang G, Ding G, Peng B,
Wen Y and Wang Z: Inhibition of Skp2 sensitizes lung cancer cells
to paclitaxel. Onco Targets Ther. 10:439–446. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Davidovich S, Ben-Izhak O, Shapira M,
Futerman B and Hershko DD: Over-expression of Skp2 is associated
with resistance to preoperative doxorubicin-based chemotherapy in
primary breast cancer. Breast Cancer Res. 10:R632008. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Schüler S, Diersch S, Hamacher R, Schmid
RM, Saur D and Schneider G: SKP2 confers resistance of pancreatic
cancer cells towards TRAIL-induced apoptosis. Int J Oncol.
38:219–225. 2011.PubMed/NCBI
|
|
75
|
Totary-Jain H, Sanoudou D, Dautriche CN,
Schneller H, Zambrana L and Marks AR: Rapamycin resistance is
linked to defective regulation of Skp2. Cancer Res. 72:1836–1843.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Chan CH, Li CF, Yang WL, Gao Y, Lee SW,
Feng Z, Huang HY, Tsai KK, Flores LG, Shao Y, et al: The Skp2-SCF
E3 ligase regulates Akt ubiquitination, glycolysis, herceptin
sensitivity, and tumorigenesis. Cell. 149:1098–1111. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Wang J, Han F, Wu J, Lee SW, Chan CH, Wu
CY, Yang WL, Gao Y, Zhang X, Jeong YS, et al: The role of Skp2 in
hematopoietic stem cell quiescence, pool size, and self-renewal.
Blood. 118:5429–5438. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Guardavaccaro D, Kudo Y, Boulaire J,
Barchi M, Busino L, Donzelli M, Margottin-Goguet F, Jackson PK,
Yamasaki L and Pagano M: Control of meiotic and mitotic progression
by the F box protein beta-Trcp1 in vivo. Dev Cell. 4:799–812. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Müerköster S, Arlt A, Sipos B, Witt M,
Grossmann M, Klöppel G, Kalthoff H, Fölsch UR and Schäfer H:
Increased expression of the E3-ubiquitin ligase receptor subunit
betaTRCP1 relates to constitutive nuclear factor-kappaB activation
and chemoresistance in pancreatic carcinoma cells. Cancer Res.
65:1316–1324. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Pan Y, Zhang F, Zhao Y, Shao D, Zheng X,
Chen Y, He K, Li J and Chen L: Berberine enhances chemosensitivity
and induces apoptosis through dose-orchestrated AMPK signaling in
breast cancer. J Cancer. 8:1679–1689. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Wang N, Wang X, Tan HY, Li S, Tsang CM,
Tsao SW and Feng Y: Berberine suppresses cyclin D1 expression
through proteasomal degradation in human hepatoma cells. Int J Mol
Sci. 17:pii: E1899. 2016. View Article : Google Scholar
|
|
82
|
Harder B, Jiang T, Wu T, Tao S, de la Vega
Rojo M, Tian W, Chapman E and Zhang DD: Molecular mechanisms of
Nrf2 regulation and how these influence chemical modulation for
disease intervention. Biochem Soc Trans. 43:680–686. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Chowdhry S, Zhang Y, McMahon M, Sutherland
C, Cuadrado A and Hayes JD: Nrf2 is controlled by two distinct
β-TrCP recognition motifs in its Neh6 domain, one of which can be
modulated by GSK-3 activity. Oncogene. 32:3765–3781. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Woo Y, Oh J and Kim JS: Suppression of
Nrf2 activity by chestnut leaf extract increases chemosensitivity
of breast cancer stem cells to paclitaxel. Nutrients. 9:pii: E760.
2017. View Article : Google Scholar
|
|
85
|
Karathedath S, Rajamani BM, Aalam Musheer
SM, Abraham A, Varatharajan S, Krishnamurthy P, Mathews V,
Velayudhan SR and Balasubramanian P: Role of NF-E2 related factor 2
(Nrf2) on chemotherapy resistance in acute myeloid leukemia (AML)
and the effect of pharmacological inhibition of Nrf2. PLoS One.
12:e01772272017. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Duong HQ, You KS, Oh S, Kwak SJ and Seong
YS: Silencing of NRF2 reduces the expression of ALDH1A1 and ALDH3A1
and sensitizes to 5-FU in pancreatic cancer cells. Antioxidants.
6:pii: E52. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Chang KY, Hsu TI, Hsu CC, Tsai SY, Liu JJ,
Chou SW, Liu MS, Liou JP, Ko CY, Chen KY, et al: Specificity
protein 1-modulated superoxide dismutase 2 enhances temozolomide
resistance in glioblastoma, which is independent of
O6-methylguanine-DNA methyltransferase. Redox Biol.
13:655–664. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Liu WH and Chang LS: Fas/FasL-dependent
and -independent activation of caspase-8 in doxorubicin-treated
human breast cancer MCF-7 cells: ADAM10 down-regulation activates
Fas/FasL signaling pathway. Int J Biochem Cell Biol. 43:1708–1719.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Wu WD, Wang M, Ding HH and Qiu ZJ: FBXL5
attenuates RhoGDI2-induced cisplatin resistance in gastric cancer
cells. Eur Rev Med Pharmacol Sci. 20:2551–2557. 2016.PubMed/NCBI
|
|
90
|
Cho HJ, Baek KE, Park SM, Kim IK, Nam IK,
Choi YL, Park SH, Im MJ, Choi J, Ryu J, et al: RhoGDI2 confers
gastric cancer cells resistance against cisplatin-induced apoptosis
by upregulation of Bcl-2 expression. Cancer Lett. 311:48–56. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Wang X, Pankratz VS, Fredericksen Z,
Tarrell R, Karaus M, McGuffog L, Pharaoh PD, Ponder BA, Dunning AM,
Peock S, et al: Common variants associated with breast cancer in
genome-wide association studies are modifiers of breast cancer risk
in BRCA1 and BRCA2 mutation carriers. Hum Mol Genet. 19:2886–2897.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Coon TA, Glasser JR, Mallampalli RK and
Chen BB: Novel E3 ligase component FBXL7 ubiquitinates and degrades
Aurora A, causing mitotic arrest. Cell Cycle. 11:721–729. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Liu Y, Lear T, Iannone O, Shiva S, Corey
C, Rajbhandari S, Jerome J, Chen BB and Mallampalli RK: The
proapoptotic F-box protein Fbxl7 regulates mitochondrial function
by mediating the ubiquitylation and proteasomal degradation of
survivin. J Biol Chem. 290:11843–11852. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Kamran M, Long ZJ, Xu D, Lv SS, Liu B,
Wang CL, Xu J, Lam EW and Liu Q: Aurora kinase A regulates Survivin
stability through targeting FBXL7 in gastric cancer drug resistance
and prognosis. Oncogenesis. 6:e2982017. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Lehman NL, Tibshirani R, Hsu JY, Natkunam
Y, Harris BT, West RB, Masek MA, Montgomery K, van de Rijn M and
Jackson PK: Oncogenic regulators and substrates of the anaphase
promoting complex/cyclosome are frequently overexpressed in
malignant tumors. Am J Pathol. 170:1793–1805. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Gütgemann I, Lehman NL, Jackson PK and
Longacre TA: Emi1 protein accumulation implicates misregulation of
the anaphase promoting complex/cyclosome pathway in ovarian clear
cell carcinoma. Mod Pathol. 21:445–454. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Shimizu N, Nakajima N, Tsunematsu T, Ogawa
I, Kawai H, Hirayama R, Fujimori A, Yamada A, Okayasu R, Ishimaru
N, et al: Selective enhancing effect of early mitotic inhibitor 1
(Emi1) depletion on the sensitivity of doxorubicin or X-ray
treatment in human cancer cells. J Biol Chem. 288:17238–17252.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Zhang YW, Brognard J, Coughlin C, You Z,
Dolled-Filhart M, Aslanian A, Manning G, Abraham RT and Hunter T:
The F box protein Fbx6 regulates Chk1 stability and cellular
sensitivity to replication stress. Mol Cell. 35:442–453. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Katayama K, Noguchi K and Sugimoto Y:
FBXO15 regulates P-glycoprotein/ABCB1 expression through the
ubiquitin-proteasome pathway in cancer cells. Cancer Sci.
104:694–702. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Ravindranath AK, Kaur S, Wernyj RP,
Kumaran MN, Miletti-Gonzalez KE, Chan R, Lim E, Madura K and
Rodriguez-Rodriguez L: CD44 promotes multi-drug resistance by
protecting P-glycoprotein from FBXO21-mediated ubiquitination.
Oncotarget. 6:26308–26321. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Wu B, Liu ZY, Cui J, Yang XM, Jing L, Zhou
Y, Chen ZN and Jiang JL: F-box protein FBXO22 mediates
polyubiquitination and degradation of CD147 to reverse cisplatin
resistance of tumor cells. Int J Mol Sci. 18:pii: E212. 2017.
View Article : Google Scholar
|
|
102
|
Hanai J, Cao P, Tanksale P, Imamura S,
Koshimizu E, Zhao J, Kishi S, Yamashita M, Phillips PS, Sukhatme VP
and Lecker SH: The muscle-specific ubiquitin ligase atrogin-1/MAFbx
mediates statin-induced muscle toxicity. J Clin Invest.
117:3940–3951. 2007.PubMed/NCBI
|
|
103
|
Tanaka N, Kosaka T, Miyazaki Y, Mikami S,
Niwa N, Otsuka Y, Minamishima YA, Mizuno R, Kikuchi E, Miyajima A,
et al: Acquired platinum resistance involves epithelial to
mesenchymal transition through ubiquitin ligase FBXO32
dysregulation. JCI Insight. 1:e836542016. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Tan J, Yang X, Zhuang L, Jiang X, Chen W,
Lee PL, Karuturi RK, Tan PB, Liu ET and Yu Q: Pharmacologic
disruption of Polycomb-repressive complex 2-mediated gene
repression selectively induces apoptosis in cancer cells. Genes
Dev. 21:1050–1063. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Lorenzi F, Babaei-Jadidi R, Sheard J,
Spencer-Dene B and Nateri AS: Fbxw7-associated drug resistance is
reversed by induction of terminal differentiation in murine
intestinal organoid culture. Mol Ther Methods Clin Dev.
3:160242016. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Zhang X, Kong Y, Xu X, Xing H, Zhang Y,
Han F, Li W, Yang Q, Zeng J, Jia J and Lui Z: F-box protein FBXO31
is down-regulated in gastric cancer and negatively regulated by
miR-17 and miR-20a. Oncotarget. 5:6178–6190. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Huang HL, Zheng WL, Zhao R, Zhang B and Ma
WL: FBXO31 is down-regulated and may function as a tumor suppressor
in hepatocellular carcinoma. Oncol Rep. 24:715–720. 2010.PubMed/NCBI
|
|
108
|
Liu J, Lv L, Gong J, Tan Y, Zhu Y, Dai Y,
Pan X, Huen MS, Li B, Tsao SW, et al: Overexpression of F-box only
protein 31 predicts poor prognosis and deregulates p38α- and
JNK-mediated apoptosis in esophageal squamous cell carcinoma. Int J
Cancer. 142:145–155. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Kogo R, Mimori K, Tanaka F, Komune S and
Mori M: FBXO31 determines poor prognosis in esophageal squamous
cell carcinoma. Int J Oncol. 39:155–159. 2011.PubMed/NCBI
|
|
110
|
Guerrero-Garcia TA, Mogollon RJ and
Castillo JJ: Bortezomib in plasmablastic lymphoma: A glimpse of
hope for a hard-to-treat disease. Leuk Res. 62:12–16. 2017.
View Article : Google Scholar : PubMed/NCBI
|