Introduction
Recent data have indicated that breast cancer (BC)
is the most common cancer in females in China in 2015 (1). In the last 2 decades, numerous advances
have occurred in BC diagnosis and treatment; however, BC is a
highly heterogeneous disease, and can exhibit different responses
to existing treatments (2,3). Thus, the molecular characteristics of BC
and the progressive mechanisms underlying it require further
investigation, which may lead to the elucidation of novel,
effective prognostic markers and therapeutic targets.
Long non-coding RNAs (lncRNAs) are
non-protein-coding transcripts of >200 nt (4). Evidence indicates that lncRNAs are
essential participants in the regulation of protein-coding genes,
maintenance of genomic integrity, dosage compensation, genomic
imprinting, mRNA processing, and cell differentiation and
development (5). The aberrant
expression of lncRNAs is associated with multiple diseases,
including cancer, immune diseases and neurological disorders
(6–10). A number of lncRNAs, including
metastasis associated lung adenocarcinoma transcript 1 (MALAT1),
HOX transcript antisense RNA (HOTAIR) and H19, have been revealed
to promote the development of BC (11–14), and a
number of lncRNAs have been demonstrated to be potential prognostic
markers and therapeutic targets of BC (13–18).
BRAF-regulated lncRNA 1 (BANCR) was identified by
Flockhart et al (19) in 2012.
Activating mutations in the BRAF oncogene are present in >70% of
melanomas, 90% of which produce the active mutant
BRAFV600E protein. BANCR was identified as one of the
potentially novel intergenic transcripts on massively parallel cDNA
sequencing (RNA-seq) that genetically analyzed matched normal human
melanocytes with and without BRAFV600E expression.
Further investigations determined that BANCR was a recurrently
overexpressed, previously unannotated 693-bp transcript on
chromosome 9, with a potential functional role in melanoma cell
migration (19). Additionally, BANCR
silencing suppressed melanoma cell migration, which could be
rescued by the addition of the chemokine C-X-C motif chemokine
ligand 11 to the medium (20). Since
its identification, BANCR has been demonstrated to promote the
progression and be associated with the poor prognosis of patients
with multiple types of cancer, including esophageal squamous cell
carcinoma (21), retinoblastoma
(22), lung carcinoma (23), osteosarcoma (24) and gastrointestinal cancer (25); however, the functional role of BANCR
in BC has not yet been elucidated.
The present study aimed to verify the potential role
of BANCR in BC. To do this, the expression level of BANCR in BC
cell lines and clinical samples was detected, and the association
between BANCR expression and BC clinicopathological characteristics
was statistically analyzed. Additionally, the prognostic value of
BANCR expression was investigated. Furthermore, BANCR was
overexpressed or silenced in BC cell lines to assess its function
in motility and proliferation. In addition, the mechanism
underlying BANCR regulating BC metastasis was also evaluated.
Materials and methods
Cell lines and cell culture
All human BC cell lines used in the current study,
including non-invasive BC cell lines (MDA-MB-468, MCF7 and
HCC1569), invasive BC cell lines (BT549 and MDA-MB-231) and normal
human breast epithelial cell line, MCF10A, were obtained from
Institute of Biochemistry and Cell Biology of the Chinese Academy
of Sciences (Shanghai, China). MDA-MB-468, MCF7 and MDA-MB-231
cells were cultured in Dulbecco's modified Eagle's medium (Gibco;
Thermo Fisher Scientific, Inc., Waltham, MA, USA). HCC1569, BT549
and MCF10A cells were cultured in RPMI-1640 medium (Gibco; Thermo
Fisher Scientific, Inc.). The media contained 10% fetal bovine
serum (FBS; Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml
penicillin and 100 mg/ml streptomycin and incubated at 37°C in an
atmosphere containing 5% CO2.
BC cell (MDA-MB-468 and MDA-MB-231) transfection was
performed with Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.), according to the manufacturer
instructions. The number of cells transfected was indicated where
used. Short interfering RNA (shRNA) BANCR silencing vectors
(BANCR-shRNA.1 and BANCR-shRNA.2), a non-targeting shRNA (NC),
pcDNA3.1-Vector and pcDNA3.1-BANCR were purchased from Genewiz,
Inc. (Jiangsu, China). The mass of the plasmids and the shRNAs was
2 µg. After 48 h, the transfected cells were subjected to
subsequent experimentations.
Patient samples
BC tissues and paired non-cancerous tissues of 216
patients treated in The Ningbo No. 2 Hospital (Ningbo, China)
between May, 2010 and October, 2012 were included in the present
study. The following patients were excluded in the present study:
i) Patients who received anticancer treatments prior to surgical
resection; ii) patients who had ≥2 malignances; and iii) patients
who were diagnosed with recurrent BC upon surgery. Final diagnosis
was concluded based on pathological results. All breast samples
were obtained immediately following surgical resection, then frozen
in liquid nitrogen and stored in −80°C freezers until RNA was
extracted. The clinicopathological information of the patients is
presented in Table I. Notably, no
patients with Tumor-Node-Metastasis (TNM) (26) stage IV disease were included in the
present study due to the patients with stage IV disease all
receiving anticancer treatments prior to surgical resection.
Follow-up studies were performed in the outpatient clinic in the
Ningbo No. 2 Hospital (Ningbo, China), where physical examinations,
laboratory analysis and computed tomography were performed if
required. The deadline of follow-up studies was December, 2016.
Overall survival (OS) time was defined as the time from the date of
surgery to mortality or the latest date when censored.
Recurrence-free survival (RFS) time was defined as the interval
between the date of surgery and recurrence. The present study was
approved by the Ethics Committee of the Ningbo No. 2 Hospital.
Written informed consent was obtained from each patient, and all
specimens were handled according to accepted ethical standards.
 | Table I.Association between BANCR expression
and clinicopathological characteristics of BC patients. |
Table I.
Association between BANCR expression
and clinicopathological characteristics of BC patients.
|
|
| BANCR expression,
n |
|
|---|
|
|
|
|
|
|---|
| Parameters | Patients, n | Low | High | P-value |
|---|
| Total | 216 | 91 | 125 |
|
| Age, years |
|
|
| 0.335 |
|
<50 | 108 | 49 | 59 |
|
|
≥50 | 108 | 42 | 66 |
|
| Menopausal
status |
|
|
| 0.845 |
|
Pre | 99 | 41 | 58 |
|
|
Post | 117 | 50 | 67 |
|
| Tumor size, cm |
|
|
| 0.008 |
|
<2 | 71 | 39 | 32 |
|
| ≥2 | 145 | 52 | 93 |
|
| Lymph node
status |
|
|
| <0.001 |
|
Negative | 136 | 74 | 62 |
|
|
Positive | 80 | 17 | 63 |
|
| TNM stage |
|
|
| <0.001 |
|
I+II | 138 | 73 | 65 |
|
|
III | 78 | 18 | 60 |
|
| ER status |
|
|
| 0.089 |
|
Negative | 90 | 44 | 46 |
|
|
Positive | 126 | 47 | 79 |
|
| PR status |
|
|
| 0.273 |
|
Negative | 102 | 39 | 63 |
|
|
Positive | 114 | 52 | 62 |
|
| HER2 status |
|
|
| 0.051 |
|
Negative | 121 | 58 | 63 |
|
|
Positive | 95 | 33 | 62 |
|
Total RNA extraction and revere
transcription-quantitative polymerase chain reaction (RT-qPCR)
assay
Total RNA was extracted from clinical tissues or BC
cell lines using TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.), according to manufacturer's protocol.
First-strand cDNA was generated with the extracted total RNA using
the Reverse Transcription System kit (Invitrogen; Thermo Fisher
Scientific, Inc.). The expression level of the target gene was
evaluated by RT-qPCR with the standard SYBR®-Green PCR
kit (Invitrogen; Thermo Fisher Scientific, Inc.) on the ABI 7500
system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The
thermocycling conditions were as follows: initial denaturation at
95°C for 5 min, 40 cycles of denaturation at 95°C for 30 sec,
annealing at 50°C for 30 sec and extension at 72°C for 30 sec.
Melting curve analysis was used to monitor the specificity of the
PCR products and the 2−∆∆Cq method (27) was utilized to evaluate the relative
expression level of the target gene. All experiments were performed
in triplicate with GAPDH serving as the internal control. The
primers used in the present study were: BANCER forward,
5′-ACAGGACTCCATGGCAAACG-3′ and reverse,
5′-ATGAAGAAAGCCTGGTGCAGT-3′; matrix metallopeptidase 2 (MMP2)
forward, 5′-AAGGATGGCAAGTACGGCTT-3′ and reverse,
5′-CGCTGGTACAGCTCTCATACTT-3′; MMP9 forward,
5′-ACCTCGAACTTTGACAGCGAC-3′ and reverse,
5′-GAGGAATGATCTAAGCCCAGC-3′; MMP14 forward,
5′-CGATGTGGTGTTCCAGACAA-3′ and reverse, 5′-TGGATGCAGAAAGTGATTTC-3′;
epithelial cadherin (E-cadherin) forward,
5′-CATTGCCACACATACACTCTCTTCT-3′ and reverse,
5′-CGGTTACCGTGATCAAAATCTC-3′; Vimentin forward,
5′-GGAACAGCATGTCCAAATCG-3′ and reverse, 5′-GCACCTGTCTCCGGTACTCA-3′;
GAPDH forward, 5′-GCACCGTCAAGGCTGAGAAC-3′ and reverse,
5′-GGATCTCGCTCCTGGAAGATG-3′.
Cell proliferation assays
Cell proliferation ability was assessed with MTT and
colony-formation assays. In the MTT assay, transfected BC cells and
corresponding control cells were seeded in 96-well plates. MTT
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) reagent was added
at 0, 24, 48, 72 and 96 h, followed by incubation of the plates at
37°C for another 2 h. Subsequently, dimethyl sulfoxide was used to
solubilize the crystals. Finally, the absorbance was measured at
450 nm using a Multiskan® Spectrum system (Thermo Fisher
Scientific, Inc.).
For the colony formation assay, transfected cells
(1×103 cells per dish) were seeded into a 6-well plate
and incubated at 37°C, and the medium containing 10% FBS was
replaced every 3 days. After 2 weeks, formed colonies were fixed
with pure methanol for 20 min and stained with 0.1% crystal violet
for 20 min at room temperature. Images of the visible colonies were
captured with a light microscope (×10, magnification) and counted
manually.
Cell migration and invasion
assays
For the migration assay, cells (2×104)
were transfected 24 h prior to being suspended in serum-free medium
and then seeded into the upper side of the Transwell chamber (8 µm
pore size) (BD Biosciences, Franklin Lakes, NJ, USA). The lower
side of the chamber was filled with medium containing 10% FBS,
serving as a chemoattractant. After incubation for 24 h, cells
invaded through the membrane were fixed in 4% paraformaldehyde for
30 min and stained with 0.1% crystal violet for another 30 min at
room temperature. For quantitative analysis, images of cells
adhered to the lower surface were captured with a light microscope
(×10) and three random fields were counted. For the invasion assay,
Matrigel (BD Biosciences) was pre-coated in the upper chamber of
the Transwell chamber and the assay was performed as it was in the
migration assay. Three independent experiments were performed for
each experiment.
Statistical analysis
Statistical analyses were performed with the SPSS
17.0 software (SPSS, Inc., Chicago, IL, USA). Data are presented as
mean ± standard error of the mean. Statistical comparisons of
qualitative data were produced with the χ2 test.
Quantitative data was compared by two-tailed Student's t-test or
analysis of variance test followed by Dunnett's test. The
Kaplan-Meier method was used to estimate survival, and the survival
difference was compared using the log-rank test. Survival data were
evaluated using univariate and multivariate Cox proportional
hazards models, and variables with a value of P<0.05 in
univariate analysis were further analyzed in subsequent
multivariate Cox regression analysis. P<0.05 was considered to
indicate a statistically significant difference.
Results
Significant BANCR overexpression in BC
cell lines and tissues
The expression level of BANCR in BC cell lines was
examined via RT-qPCR to investigated its functional role; the
results of this analysis demonstrated that BANCR was upregulated in
BC cell lines, compared with normal human breast epithelial cell
line MCF10A (Fig. 1A). In addition,
the BANCR expression level in invasive BC cell lines was
significantly higher than that in the noninvasive BC cell lines
(Fig. 1A). The expression of BANCR
was also assessed in 216 BC and paired non-cancerous tissues, the
results of which indicated that BANCR was significantly
overexpressed in BC tissues, compared with paired non-cancerous
tissues (P<0.01; Fig. 1B). BANCR
overexpression in BC cell lines and tissues indicated that it may
serve an oncogenic role in BC.
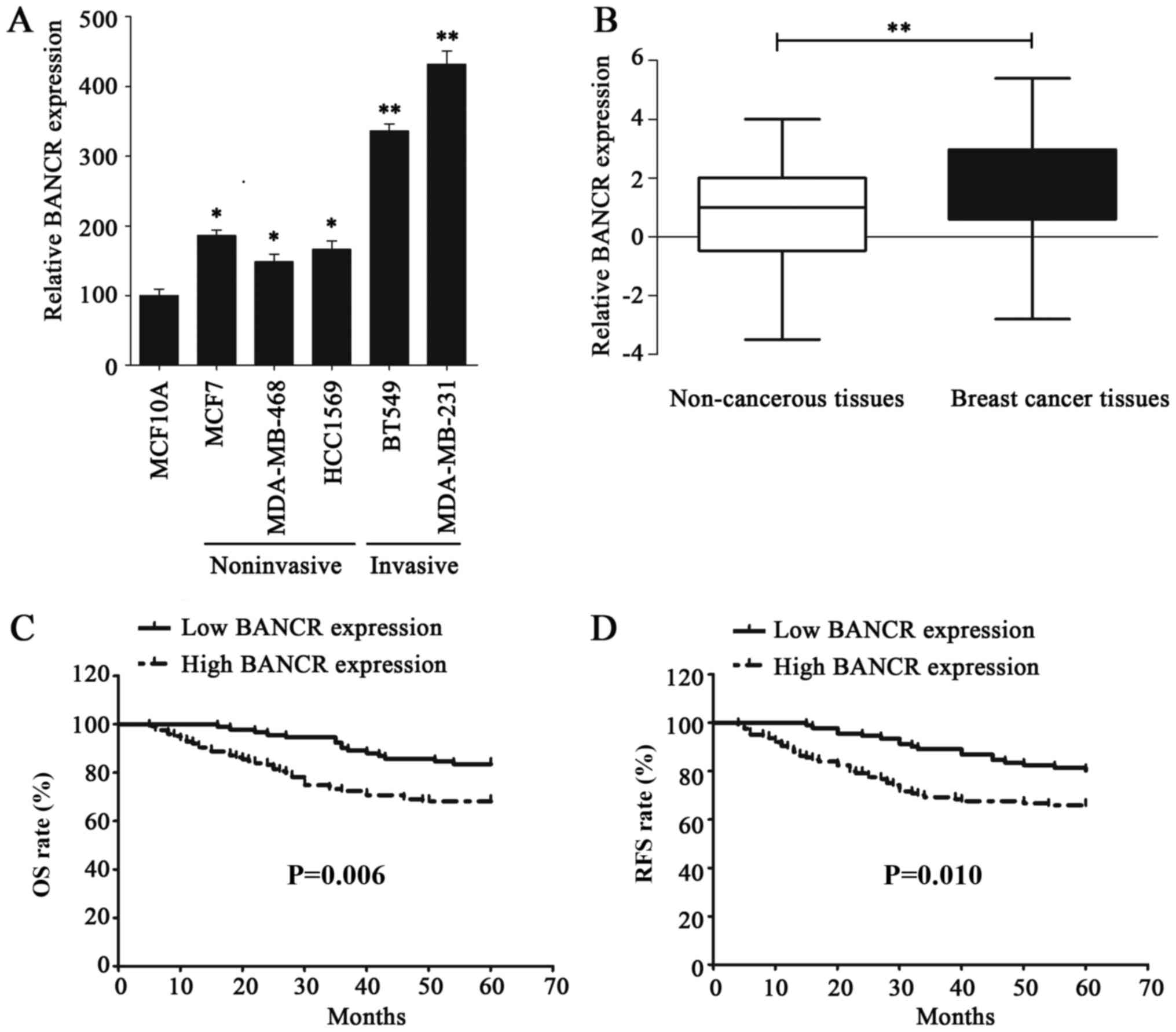 | Figure 1.lncRNA BANCR expression is
upregulated in BC cells and tissues, which is associated with poor
prognosis of patients with BC. (A) The expression of BANCR was
detected via RT-qPCR in normal human breast epithelial cell line,
MCF10A, noninvasive BC cell lines (MDA-MB-468, MCF7 and HCC1569)
and invasive BC cell lines (BT549 and MDA-MB-231). (B) The
expression of BANCR in non-cancerous tissues and BC tissues was
verified by RT-qPCR. (C) The Kaplan-Meier method and log-rank test
were utilized to analyze the difference in OS rate between the low-
and high-BANCR-expression groups. (D) The Kaplan-Meier method and
log-rank test were used to analyze the difference in RFS rate
between the low- and high-BANCR-expression groups. Statistical
significance was determined using Student's t-test between
indicated columns in (A and B), and by log-rank test in (C and D).
*P<0.05, **P<0.01. BC, breast cancer; RT-qPCR, reverse
transcription-quantitative polymerase chain reaction; BANCR,
BRAF-regulated lncRNA 1; OS, overall survival; RFS, recurrence-free
survival. |
Association between BANCR
overexpression and prognosis of patients with BC
The clinical significance of BANCR expression in BC
was further identified by analyzing the association between BANCR
expression and the clinicopathological characteristics of patients
with BC, who were dichotomized for statistical analysis. All of the
patients were determined to exhibit either low or high BANCR
expression, with the BANCR mean expression level (1.9) serving as
the cutoff value. The patients with BANCR level less than 1.9 were
classified into the low BANCR expression group. Notably, BANCR
overexpression was determined to be significantly associated with a
larger tumor size (P=0.008), lymph node metastasis (P<0.001) and
advanced TNM stage (P<0.001; Table
I). The association between BANCR expression and the prognosis
of patients with BC was calculated using the Kaplan-Meier method
and compared with the log-rank test. As expected, patients with a
high BANCR expression were demonstrated to have a poorer OS rate
(P=0.006) and reduced RFS period (P=0.010), compared with patients
exhibiting low BANCR. (Fig. 1C and
D). Furthermore, univariate analysis indicated that positive
lymph node status [hazard ratio (HR)=2.140, 95% confidence interval
(CI)=1.245–3.650; P=0.005], advanced TNM stage (HR=1.623, 95%
CI=1.115–2.361; P=0.011) and high BANCR expression (HR=1.614, 95%
CI=1.353–1.989; P<0.001) were three risk factors for patients
with BC with a poor OS rate. Additionally, the same three factors
were also identified as risk factors of a reduced RFS, positive
lymph node status (HR=1.999, 95% CI=1.205–3.317; P=0.007), advanced
TNM stage (HR=1.471, 95% CI=1.038–2.085; P=0.030) and high BANCR
expression (HR=1.575, 95% CI=1.317–1.883; P<0.001; Tables II and III). However, further analysis of these
factors using multivariate analysis, only high BANCR expression was
highlighted as an independent risk factor of patients with BC with
a poor OS rate (HR=1.585, 95% CI=1.298–1.935; P<0.001) and early
recurrence (HR=1.532, 95% CI=1.272–1.844; P<0.001; Tables II and III). These data indicated that BANCR
overexpression could promote the clinical progression of BC and
predicts poor prognosis of patients with BC.
 | Table II.Univariate and multivariate Cox
regression analysis of overall survival of BC patients. |
Table II.
Univariate and multivariate Cox
regression analysis of overall survival of BC patients.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Parameters | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Age (≥50 vs. <50
years) | 1.031
(0.604–1.760) | 0.911 |
|
|
| Menopausal status
(pre vs. post) | 1.067
(0.625–1.822) | 0.812 |
|
|
| Tumor size (≥2 vs.
<2 cm) | 1.117
(0.629–1.983) | 0.706 |
|
|
| Lymph node status
(negative vs. positive) | 2.140
(1.254–3.650) | 0.005 | 1.332
(0.639–2.780) | 0.444 |
| TNM (stage I+II vs.
III) | 1.623
(1.115–2.361) | 0.011 | 1.120
(0.665–1.885) | 0.671 |
| ER status (negative
vs. positive) | 0.993
(0.577–1.708) | 0.979 |
|
|
| PR status (negative
vs. positive) | 1.100
(0.643–1.882) | 0.727 |
|
|
| HER2 status
(negative vs. positive) | 1.265
(0.742–2.157) | 0.387 |
|
|
| BANCR expression
(high vs. low) | 1.614
(1.353–1.989) | <0.001 | 1.585
(1.298–1.935) | <0.001 |
 | Table III.Univariate and multivariate Cox
regression analysis of recurrence-free survival of patients with
breast cancer. |
Table III.
Univariate and multivariate Cox
regression analysis of recurrence-free survival of patients with
breast cancer.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Parameters | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Age (≥50 vs. <50
years) | 0.979
(0.589–1.629) | 0.936 |
|
|
| Menopausal status
(pre vs. post) | 1.047
(0.631–1.739) | 0.859 |
|
|
| Tumor size (≥2 vs.
<2 cm) | 1.011
(0.591–1.729) | 0.968 |
|
|
| Lymph node status
(negative vs. positive) | 1.999
(1.205–3.317) | 0.007 | 1.421
(0.703–2.872) | 0.328 |
| TNM (stage I+II vs.
III) | 1.471
(1.038–2.085) | 0.030 | 0.992
(0.703–2.872) | 0.328 |
| ER status (negative
vs. positive) | 1.024
(0.611–1.717) | 0.927 |
|
|
| PR status (negative
vs. positive) | 0.997
(0.600–1.655) | 0.989 |
|
|
| HER2 status
(negative vs. positive) | 1.268
(0.764–2.103) | 0.358 |
|
|
| BANCR (high vs.
low) | 1.575
(1.317–1.883) | <0.001 | 1.532
(1.272–1.844) | <0.001 |
BANCR promotes BC metastasis and
proliferation
Next, the functional role of BANCR was detected in
BC cell lines. RNA interference was performed in MDA-MB-231 cells,
owing to the relatively high expression level of BANCR; BANCR
ectopic overexpression was conducted in MDA-MB-468 cells, owing to
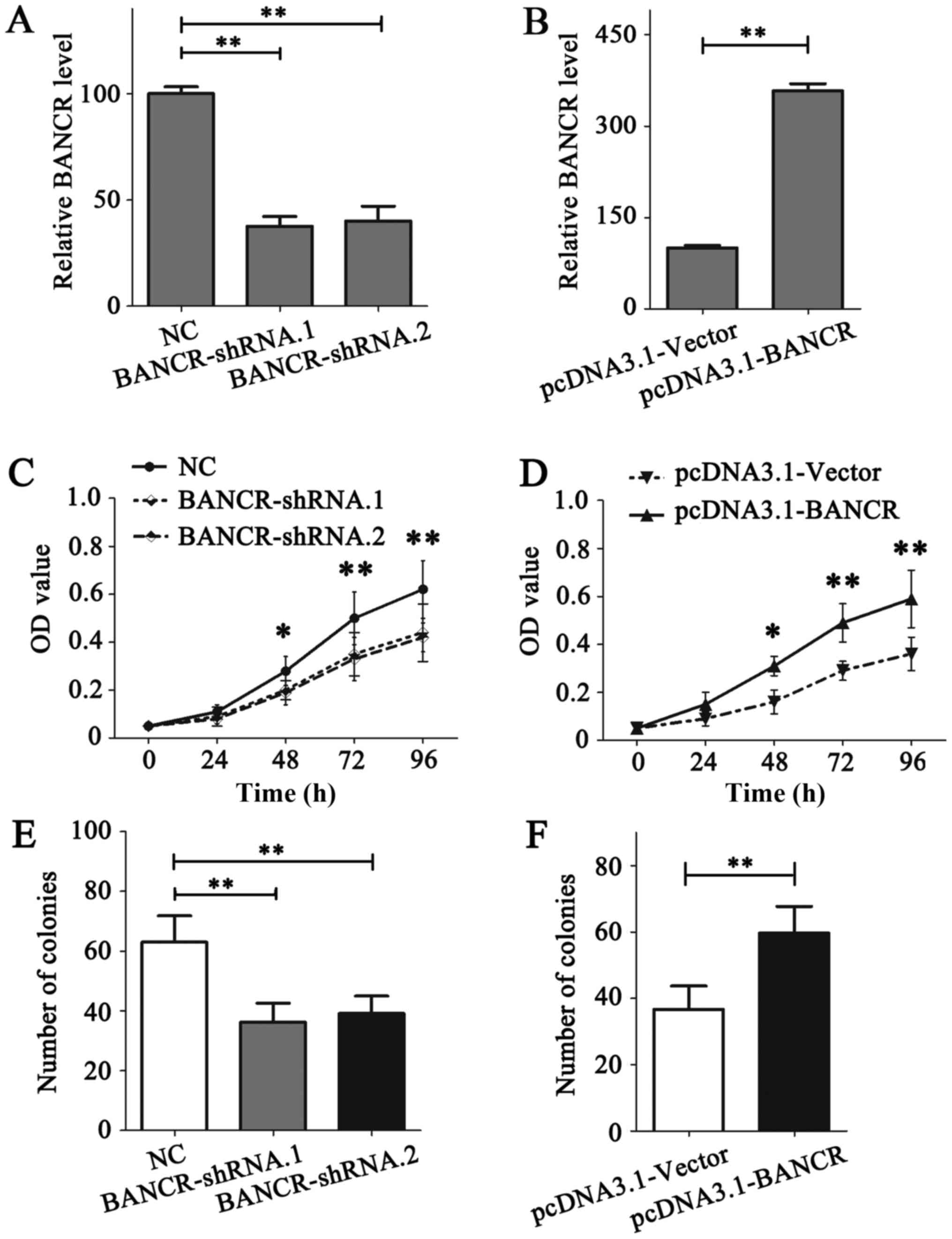
the relatively low expression level of BANCR (Fig. 1A). The expression of BANCR in
MDA-MB-231 and MDA-MB-468 cells was demonstrated to be notably
downregulated or upregulated following transfection, respectively
(Fig. 2A and B).
The role of BANCR on the proliferation of BC cells
was detected using MTT and colony formation assays. The results of
the MTT assay indicated that BANCR deficiency in MDA-MB-231 cells
notably reduced its proliferation ability (P<0.05; Fig. 2C), while BANCR overexpression in
MDA-MB-468 cells promoted the proliferation (P<0.05; Fig. 2D). Similar results were obtained from
the colony formation assay (P<0.05; Fig. 2E and F). Migration and invasion assays
were performed to analyze the association between BANCR expression
levels and the migration and invasion of BC cells. The results
demonstrated that BANCR silencing could significantly inhibit the
migration and invasion of MDA-MB-231 cells (P<0.05; Fig. 3A and B). Contrastingly, BANCR
overexpression significantly facilitated the migration and invasion
of MDA-MB-468 cells (P<0.05; Fig. 3C
and D). Notably, as shown in Fig. 3A
and B, statistical analysis revealed that BANCR interference
left ~50% of invasive cells able to permeate the membrane compared
with the NC group, which may have resulted from the involvement of
a wide range of molecules in cell biology, including protein-coding
genes and microRNAs. lncRNAs function to regulate cell biology.
Taken together, these data indicate that BANCR could promote the
proliferation and metastasis of BC cells.
BANCR promotes BC metastasis through
regulating MMP2/9 and epithelial-mesenchymal transition (EMT)
To investigate the mechanisms underlying the
BANCR-dependent promotion of BC metastasis, the expression of
invasive markers, including MMP2, MMP9 and MMP14, were quantified
by RT-qPCR (Fig. 4). Expression of
MMP2/9 were significantly suppressed in BANCR-knockdown MDA-MB-231
cells, whereas MDA-MB-468 cells overexpressing BANCR exhibited
significantly higher MMP2/9 expression (Fig. 4A, B, F and G). The expression level of
MMP14 was relatively stable despite the overexpression or knockdown
of BANCR (Fig. 4C and H). EMT is a
fundamental process in cancer cell metastasis (28). The expression of the epithelial marker
E-cadherin and the mesenchymal marker vimentin was thus analyzed
using RT-qPCR. BANCR upregulation increased the expression of
vimentin and reduced the expression of E-cadherin. Contrastingly,
the vimentin expression level was decreased and E-cadherin
expression level was upregulated following BANCR silencing
(Fig. 4D, E, I and J).
 | Figure 4.BANCR promotes the metastasis of
breast cancer cells by regulating MMP2/9 and the
epithelial-mesenchymal transition. The expression of (A) MMP2, (B)
MMP9, (C) MMP14, (D) Vimentin and (E) E-cadherin was analyzed in
MDA-MB-231 cells following BANCR knockdown. The expression of (F)
MMP2, (G) MMP9, (H) MMP14, (I) Vimentin and (J) E-cadherin was
analyzed in MDA-MB-468 cells following BANCR overexpression.
Statistical significance was determined using Student's t-test
between indicated columns in (A-J). **P<0.01. NC, negative
control; BNCR, BRAF-regulated lncRNA 1; MMP, matrix
metallopeptidase; E-cadherin, epithelial cadherin. |
Discussion
lncRNAs are regarded as pivotal regulators in the
development and progression of cancer (8,29). A
number of lncRNAs have been revealed as pivotal regulators and
associated with clinical outcomes in patients with BC through
various regulating pathways (17,30). A
number of lncRNAs, including HOTAIR, lncRNA activated by TGF-β,
breast cancer anti-estrogen resistance 4, urothelial
cancer-associated 1 and growth arrest specific 5, have been
demonstrated to be associated with anticancer drug resistance
(31). The lncRNAs 91H (32), lncRNA inhibiting metastasis (33) and eosinophil granule ontogeny
transcript (16) have been indicated
to promote an aggressive BC phenotype. BC is a highly heterogeneous
disease, and lncRNAs also display specific expression patterns in
different subtypes of BC (11,34). For
example, HOTAIR only provided a prognostic insight in patients with
ER-negative BC (34), whereas MALAT1
overexpression was associated with poor RFS in tamoxifen treated
patients with ER-positive BC, and may serve as a potential
biomarker to predict endocrine treatment sensitivity (11). Each patient's lncRNA expression
signature may provide a novel method for individualized anticancer
treatments. Several lncRNAs have already been highlighted as
prognostic markers and therapeutic targets in BC (30). However, the significance of BANCR has
not yet been identified.
Upregulation of BANCR has been observed in multiple
cancer types, including gastrointestinal cancer (25), lung cancer (35), esophageal squamous cell carcinoma
(21), papillary thyroid carcinoma
(36), endometrial cancer (37), osteosarcoma (24) and retinoblastoma (22). In demonstrating the functional role of
BANCR in BC, the present study first observed that BANCR was
overexpressed in BC cell lines and clinical tissues. It has been
reported that BANCR overexpression accelerates the progression of
various cancer types (21,23,24,35–41).
The present study determined that high BANCR expression was
associated with a larger tumor size, lymph node metastasis and
advanced TNM stages in patients with BC (P<0.05). However,
further research is required to investigate the association between
BANCR expression and distant metastasis. Additionally, high BANCR
expression predicted a poor OS rate (HR=1.585, 95% CI=1.298–1.935;
P<0.001) and early recurrence (HR=1.532, 95% CI=1.272–1.844;
P<0.001) in patients with BC. BANCR upregulation was also
identified as an independent risk factor of poor OS and RFS rates
for patients with BC. In vitro assays demonstrated that
BANCR could facilitate the migration, invasion and proliferation of
BC cells. Mechanistically, the expression of invasive markers, MMP2
and MMP9 was positively associated with BANCR expression. BANCR was
also demonstrated to accelerate EMT in BC cells. However, as a
limitation of the present study, the aforementioned observations
were only obtained in the in vitro setting, meaning that
further research is required to verify the association between
BANCR and markers of invasion in patient samples. To the best of
our knowledge, the present study is the first to demonstrate the
oncogenetic role of BANCR in BC and to indicate that BANCR could
serve as an effective prognostic marker and therapeutic target in
BC.
The detailed mechanisms by which BANCR regulates
cancer biology have been investigated in a number of studies
(37–39,41–43). BANCR
actively functions as a regulator of EMT during non-small cell lung
cancer and colorectal cancer metastasis (42,43). The
mitogen-activated protein kinase signaling pathway has been
implicated as the target of BANCR in promoting the proliferation of
melanoma and endometrial cancer (37,38).
Additionally, Zhang et al (39) revealed that BANCR could promote
gastric cancer cell proliferation by regulating nuclear factor-κB1,
whereas p21 was indicated to be the target gene of BANCR, affecting
the proliferation of colorectal cancer cells (41). Notably, the mechanisms underlying
BANCR functioning vary in different cancer types, which requires
further study.
In conclusion, the present study demonstrated that
BANCR was overexpressed in BC, and this expression was
significantly associated with the clinical progression of BC.
Furthermore, high BANCR expression was indicated to be an
independent risk factor for patients with BC with poor OS and RFS
rates. In addition, the functional role of BANCR promoting BC
metastasis and proliferation was demonstrated in BC cells. These
observations indicated that BANCR may serve as a promising
prognostic marker and therapeutic target in BC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
Not applicable.
Authors' contributions
JJ collected the clinical specimens and conducted
RT-qPCR, performed in vitro studies and wrote the
manuscript. SHS, XJL and LS performed RT-qPCR and statistical
analysis. QDG and CL collected the clinical specimens and wrote the
manuscript. WZ designed the study.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the Ningbo No. 2 Hospital.
Consent for publication
Written informed consent was obtained from each
patient.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cancer Genome Atlas Network: Comprehensive
molecular portraits of human breast tumours. Nature. 490:61–70.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ciriello G, Gatza ML, Beck AH, Wilkerson
MD, Rhie SK, Pastore A, Zhang H, McLellan M, Yau C, Kandoth C, et
al: Comprehensive molecular portraits of invasive lobular breast
cancer. Cell. 163:506–519. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Boon RA, Jaé N, Holdt L and Dimmeler S:
Long noncoding RNAs: From clinical genetics to therapeutic targets?
J Am Coll Cardiol. 67:1214–1226. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bhan A and Mandal SS: Long noncoding RNAs:
Emerging stars in gene regulation, epigenetics and human disease.
Chem Med Chem. 9:1932–1956. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ulitsky I and Bartel DP: lincRNAs:
Genomics, evolution, and mechanisms. Cell. 154:26–46. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Isin M and Dalay N: LncRNAs and neoplasia.
Clin Chim Acta. 444:280–288. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li X, Wu Z, Fu X and Han W: lncRNAs:
Insights into their function and mechanics in underlying disorders.
Mutat Res Rev Mutat Res. 762:1–21. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Maass PG, Luft FC and Bähring S: Long
non-coding RNA in health and disease. J Mol Med (Berl). 92:337–346.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Huang NS, Chi YY, Xue JY, Liu MY, Huang S,
Mo M, Zhou SL and Wu J: Long non-coding RNA metastasis associated
in lung adenocarcinoma transcript 1 (MALAT1) interacts with
estrogen receptor and predicted poor survival in breast cancer.
Oncotarget. 7:37957–37965. 2016.PubMed/NCBI
|
|
12
|
Si X, Zang R, Zhang E, Liu Y, Shi X, Zhang
E, Shao L, Li A, Yang N, Han X, et al: LncRNA H19 confers
chemoresistance in ERα-positive breast cancer through epigenetic
silencing of the pro-apoptotic gene BIK. Oncotarget. 7:81452–81462.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xu S, Sui S, Zhang J, Bai N, Shi Q, Zhang
G, Gao S, You Z, Zhan C, Liu F and Pang D: Downregulation of long
noncoding RNA MALAT1 induces epithelial-to-mesenchymal transition
via the PI3K-AKT pathway in breast cancer. Int J Clin Exp Pathol.
8:4881–4891. 2015.PubMed/NCBI
|
|
14
|
Milevskiy MJ, Al-Ejeh F, Saunus JM,
Northwood KS, Bailey PJ, Betts JA, McCart Reed AE, Nephew KP, Stone
A, Gee JM, et al: Long-range regulators of the lncRNA HOTAIR
enhance its prognostic potential in breast cancer. Hum Mol Genet.
25:3269–3283. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Deng LL, Chi YY, Liu L, Huang NS, Wang L
and Wu J: LINC00978 predicts poor prognosis in breast cancer
patients. Sci Rep. 6:379362016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xu SP, Zhang JF, Sui SY, Bai NX, Gao S,
Zhang GW, Shi QY, You ZL, Zhan C and Pang D: Downregulation of the
long noncoding RNA EGOT correlates with malignant status and poor
prognosis in breast cancer. Tumor Biol. 36:9807–9812. 2015.
View Article : Google Scholar
|
|
17
|
Mendell JT: Targeting a long noncoding RNA
in breast cancer. N Engl J Med. 374:2287–2289. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yang Y, Qian J, Xiang Y, Chen Y and Qu J:
The prognostic value of long noncoding RNA HOTTIP on clinical
outcomes in breast cancer. Oncotarget. 8:6833–6844. 2017.PubMed/NCBI
|
|
19
|
Flockhart RJ, Webster DE, Qu K,
Mascarenhas N, Kovalski J, Kretz M and Khavari PA: BRAFV600E
remodels the melanocyte transcriptome and induces BANCR to regulate
melanoma cell migration. Genome Res. 22:1006–1014. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
McCarthy N: Epigenetics. Going places with
BANCR. Nat Rev Cancer. 12:4512012. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu Z, Yang T, Xu Z and Cao X:
Upregulation of the long non-coding RNA BANCR correlates with tumor
progression and poor prognosis in esophageal squamous cell
carcinoma. Biomed Pharmacother. 82:406–412. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Su S, Gao J, Wang T, Wang J, Li H and Wang
Z: Long non-coding RNA BANCR regulates growth and metastasis and is
associated with poor prognosis in retinoblastoma. Tumor Biol.
36:7205–7211. 2015. View Article : Google Scholar
|
|
23
|
Jiang W, Zhang D, Xu B, Wu Z, Liu S, Zhang
L, Tian Y, Han X and Tian D: Long non-coding RNA BANCR promotes
proliferation and migration of lung carcinoma via MAPK pathways.
Biomed Pharmacother. 69:90–95. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Peng ZQ, Lu RB, Xiao DM and Xiao ZM:
Increased expression of the lncRNA BANCR and its prognostic
significance in human osteosarcoma. Genet Mol Res. 15:2016.
View Article : Google Scholar
|
|
25
|
Fan YH, Ye MH, Wu L, Wu MJ, Lu SG and Zhu
XG: BRAF-activated lncRNA predicts gastrointestinal cancer patient
prognosis: A meta-analysis. Oncotarget. 8:6295–6303.
2017.PubMed/NCBI
|
|
26
|
Amin MB, Edge SB, Greene FL, Byrd DR,
Brookland RK, Washington MK, Gershenwald JE, Compton CC, Hess KR,
Sullivan DC, et al: American Joint Committee on Cancer (AJCC): AJCC
Cancer Staging Manual. 8th edition. Springer; New York, NY:
2017
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tsai JH and Yang J: Epithelial-mesenchymal
plasticity in carcinoma metastasis. Genes Dev. 27:2192–2206. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tsai MC, Spitale RC and Chang HY: Long
intergenic noncoding RNAs: New links in cancer progression. Cancer
Res. 71:3–7. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu H, Li J, Koirala P, Ding X, Chen B,
Wang Y, Wang Z, Wang C, Zhang X and Mo YY: Long non-coding RNAs as
prognostic markers in human breast cancer. Oncotarget.
7:20584–20596. 2016.PubMed/NCBI
|
|
31
|
Chen QN, Wei CC, Wang ZX and Sun M: Long
non-coding RNAs in anti-cancer drug resistance. Oncotarget.
8:1925–1936. 2017.PubMed/NCBI
|
|
32
|
Vennin C, Spruyt N, Robin YM, Chassat T,
Le Bourhis X and Adriaenssens E: The long non-coding RNA 91H
increases aggressive phenotype of breast cancer cells and
up-regulates H19/IGF2 expression through epigenetic modifications.
Cancer Lett. 385:198–206. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sas-Chen A, Aure MR, Leibovich L, Carvalho
S, Enuka Y, Körner C, Polycarpou-Schwarz M, Lavi S, Nevo N,
Kuznetsov Y, et al: LIMT is a novel metastasis inhibiting lncRNA
suppressed by EGF and downregulated in aggressive breast cancer.
EMBO Mol Med. 8:1052–1064. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gökmen-Polar Y, Vladislav IT, Neelamraju
Y, Janga SC and Badve S: Prognostic impact of HOTAIR expression is
restricted to ER-negative breast cancers. Sci Rep. 5:87652015.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chen JX, Chen M, Zheng YD, Wang SY and
Shen ZP: Up-regulation of BRAF activated non-coding RNA is
associated with radiation therapy for lung cancer. Biomed
Pharmacother. 71:79–83. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zheng H, Wang M, Jiang L, Chu H, Hu J,
Ning J, Li B, Wang D and Xu J: BRAF-activated long noncoding RNA
modulates papillary thyroid carcinoma cell proliferation through
regulating thyroid stimulating hormone receptor. Cancer Res Treat.
48:698–707. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang D, Wang D, Wang N, Long Z and Ren X:
Long non-coding RNA BANCR promotes endometrial cancer cell
proliferation and invasion by regulating MMP2 and MMP1 via ERK/MAPK
signaling pathway. Cell Physiol Biochem. 40:644–656. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li R, Zhang L, Jia L, Duan Y, Li Y, Bao L
and Sha N: Long non-coding RNA BANCR promotes proliferation in
malignant melanoma by regulating MAPK pathway activation. PLoS One.
9:e1008932014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang ZX, Liu ZQ, Jiang B, Lu XY, Ning XF,
Yuan CT and Wang AL: BRAF activated non-coding RNA (BANCR)
promoting gastric cancer cells proliferation via regulation of
NF-κB1. Biochem Biophys Res Commun. 465:225–231. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li L, Zhang L, Zhang Y and Zhou F:
Increased expression of LncRNA BANCR is associated with clinical
progression and poor prognosis in gastric cancer. Biomed
Pharmacother. 72:109–112. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Scaria V, Shi Y, Liu Y, Jie D, Yun T, Li
W, Yan L, Wang K and Feng J: Downregulated long noncoding RNA BANCR
promotes the proliferation of colorectal cancer cells via
downregualtion of p21 expression. PloS One. 10:e01226792015.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Guo Q, Zhao Y, Chen J, Hu J, Wang S, Zhang
D and Sun Y: BRAF-activated long non-coding RNA contributes to
colorectal cancer migration by inducing epithelial-mesenchymal
transition. Oncol Lett. 8:869–887. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Sun M, Liu XH, Wang KM, Nie FQ, Kong R,
Yang JS, Xia R, Xu TP, Jin FY, Liu ZJ, et al: Downregulation of
BRAF activated non-coding RNA is associated with poor prognosis for
non-small cell lung cancer and promotes metastasis by affecting
epithelial-mesenchymal transition. Mol Cancer. 13:682014.
View Article : Google Scholar : PubMed/NCBI
|