Introduction
Lung cancer is the leading cause of
cancer-associated mortality globally (1), with >1 million cases reported
annually (2). Non-small cell lung
cancer (NSCLC) is the dominant histological subtype, accounting for
~85% of lung cancer (3,4). Other types include lung adenocarcinoma
(LUAD), lung squamous cell carcinoma (LUSC) and large cell
carcinoma (5). Due to the nonspecific
symptoms at early stages and poor overall survival rates (6,7),
association studies for NSCLC biomarkers have been investigated
globally (8,9).
The evolution of lung cancer is a complex process
involving the interaction of genetic, epigenetic and environmental
factors (10). As a common epigenetic
modification, DNA methylation serves an important role in human
malignant tumor types, including NSCLC (11). Methylation of the
cytosine-phosphate-guanine (CpG) island affects gene silencing
(12), and provides a novel insight
into lung tumorigenesis and progression. Currently, there are a
number of studies on the potential of DNA methylation biomarkers in
NSCLC, including RASSF1 (13),
CDKN2A (14), MGMT
(14), APC (15), FHIT (16), CDH13 (15) and DAPK (13). Furthermore, a large number of
tumor-specific methylated genes have been identified using
genome-wide CpG island methylation analysis in NSCLC (17). Since aberrant DNA methylation has been
indicated as an early stage event during lung carcinogenesis
(18), it is characterized as dynamic
and reversible (19). DNA methylation
biomarkers may be an ideal tool for early diagnosis and prognosis
due to their non-invasive, high sensitivity, and high specificity
characteristics (20).
MyoD family inhibitor (MDFI) is located in
chromosome 6p21.1, encoding a transcription factor that negatively
regulates myogenic family proteins (21). MDFI has been considered as a
candidate tumor suppressor gene (21,22) and
the domain protein is involved in transcriptional regulation by
affecting the Wnt signaling pathway (23). Additionally, MDFI gene
silencing induced by promoter hypermethylation was observed in
pancreatic cancer (24,25); however, the epigenetic role of
MDFI methylation in NSCLC pathogenesis remains unclear. The
present study aimed to establish the association between the
MDFI promoter methylation and NSCLC.
Materials and methods
Sample collection and data source
Formalin-fixed, paraffin-embedded (FFPE) tumor
tissues and adjacent non-cancerous tissues were collected from 73
male, and 38 female patients with NSCLC at Huzhou First People's
Hospital (Huzhou, Zhejiang, China) between August 2010 and October
2013. The patient age range was 33 to 82 years old (mean,
63.59±10.19 years old). All pathological parameters were defined
according to the World Health Organization guidelines and Union for
International Cancer Control tumor-node-metastasis classifications
(26,27). According to the histological type,
there were 42 patients with LUSC and 69 patients with LUAD. The
adjacent non-cancerous tissues were obtained from ≥5 cm outside the
edge of tumors. All specimens were sliced at 4-µm thickness using a
Leica RM2245 Semi-Automated Rotary Microtome (Leica Microsystems
GmbH, Wetzlar, Germany). Written informed consent form was signed
by all of the participants and the present study was approved by
the Ethics Committee of Huzhou First People's Hospital.
DNA methylation profiles (Illumina Human Methylation
450K, HM450K; Illumina, Inc., San Diego, CA, USA) and clinical
characteristics (age, sex and smoking status) generated from 830
NSCLC tumor tissues and 75 non-tumor tissues were obtained from The
Cancer Genomics Browser of The University of California Santa Cruz
database (https://genome-cancer.ucsc.edu/). The browser contains
data generated from the Cancer Genome Atlas (TCGA) project
(https://cancergenome.nih.gov/).
Therefore, larger samples were used to verify the findings of the
study cases. These samples were then used as a control for the
study data.
DNA extraction and bisulfite
treatment
Genomic DNA from tissues was extracted using the
E.Z.N.A® FFPE DNA kit (Omega Bio-tek, Inc., Norcross,
GA, USA). DNA concentration measurements and bisulfite treatment
were performed as previously described (28).
SYBR® Green-based
quantitative methylation-specific polymerase chain reaction (PCR;
qMSP)
The 20 µl PCR consisted of 20 ng converted DNA, 0.5
µl forward primer (10 µM), 0.5 µl reverse primer (10 µM), 10 µl
LightCycler® 480 SYBR-Green I Master mix (Roche
Diagnostics, Basel, Switzerland) and 8 µl DNAase/RNAase-free water.
All the experiments were performed on the LightCycler 480 system
(Roche Diagnostics) utilizing a 384-well plate platform. The PCR
program was conducted as follows: 95°C for 10 min; followed by 45
cycles at 95°C for 20 sec and 58°C for 20 sec; and a single step of
72°C for 30 sec. Following amplification, melting curve analysis
was performed for PCR product identification that consisted of one
cycle of 95°C for 15 sec, 58°C for 1 min and 95°C for 10 sec (slope
0.11°C/s, acquisition mode: Continuous). The primer sequences of
MDFI were as follows: Forward, 5′-AGAGACGGTGAGGATTGT-3′ and
reverse, 5′-CGACTACTACATTCTTACCTACTT-3′, and the product length was
80 bp. The primer sequences of β-actin were as follows: Forward,
5′-TGGTGATGGAGGAGGTTTAGTAAGT-3′ and reverse,
5′-TGGTGATGGAGGAGGTTTAGTAAGT-3′, and the product length was 133 bp.
Sperm DNA from a healthy individual was methylated with excess SssI
methyltransferase (Thermo Fisher Scientific, Inc., Waltham, MA,
USA) to serve as a positive control. Water without DNA served as a
negative control in each assay. The percentage of methylated
reference (PMR) of the MDFI in each sample was calculated
using the 2−ΔΔCq method, whereby ΔΔCq was calculated as
follows: Sample DNA (Cq target gene-Cq ACTB
control)-fully methylated DNA (Cq target gene-Cq
ACTB control) (29). All
products were confirmed by Sanger sequencing and capillary gel
electrophoresis as previously described (30).
Statistical analysis
Statistical analysis was performed using SPSS
software 18.0 (SPSS Inc., Chicago, IL, USA). Due to the skewed
distribution of methylation data, the non-parametric Wilcoxon
signed-rank test and Mann-Whitney-U test was used to compare the
methylation levels between tumor tissues and non-tumor tissues in
the study cohort, and TCGA dataset. Data were presented as the
median ± interquartile range. Fisher exact test or χ2
test was used to determine the associations between the methylation
status and the clinical characteristics (age, sex, smoking history,
histological types and clinical stage). Kaplan-Meier curve was
implemented to assess the prognostic value of MDFI
methylation in postoperative patients with NSCLC. P<0.05 was
considered to indicate a statistically significant difference.
Results
In order to assess the methylation level of
MDFI promoter region, a fragment (chr6: 41604618-41604697)
amplified with a suitable primer pair was selected. As there was a
high number of CG sites in the promoter region, a specific primer
pair of qMSP method was difficult to design. The primers used were
designed to obtain a single melting curve. Furthermore, this
fragment was located in the region rich in histone modifications
(H3K4me1, H3K4me3 and H3K27Ac) according to human 2009
(GRCh37/hg19) assembly (http://genome.ucsc.edu/), indicating a potential
regulatory mechanism in gene transcription (Fig. 1A). Additionally, the amplified
fragment was demonstrated to match the target sequences by Sanger
sequencing and capillary gel electrophoresis (Fig. 1B).
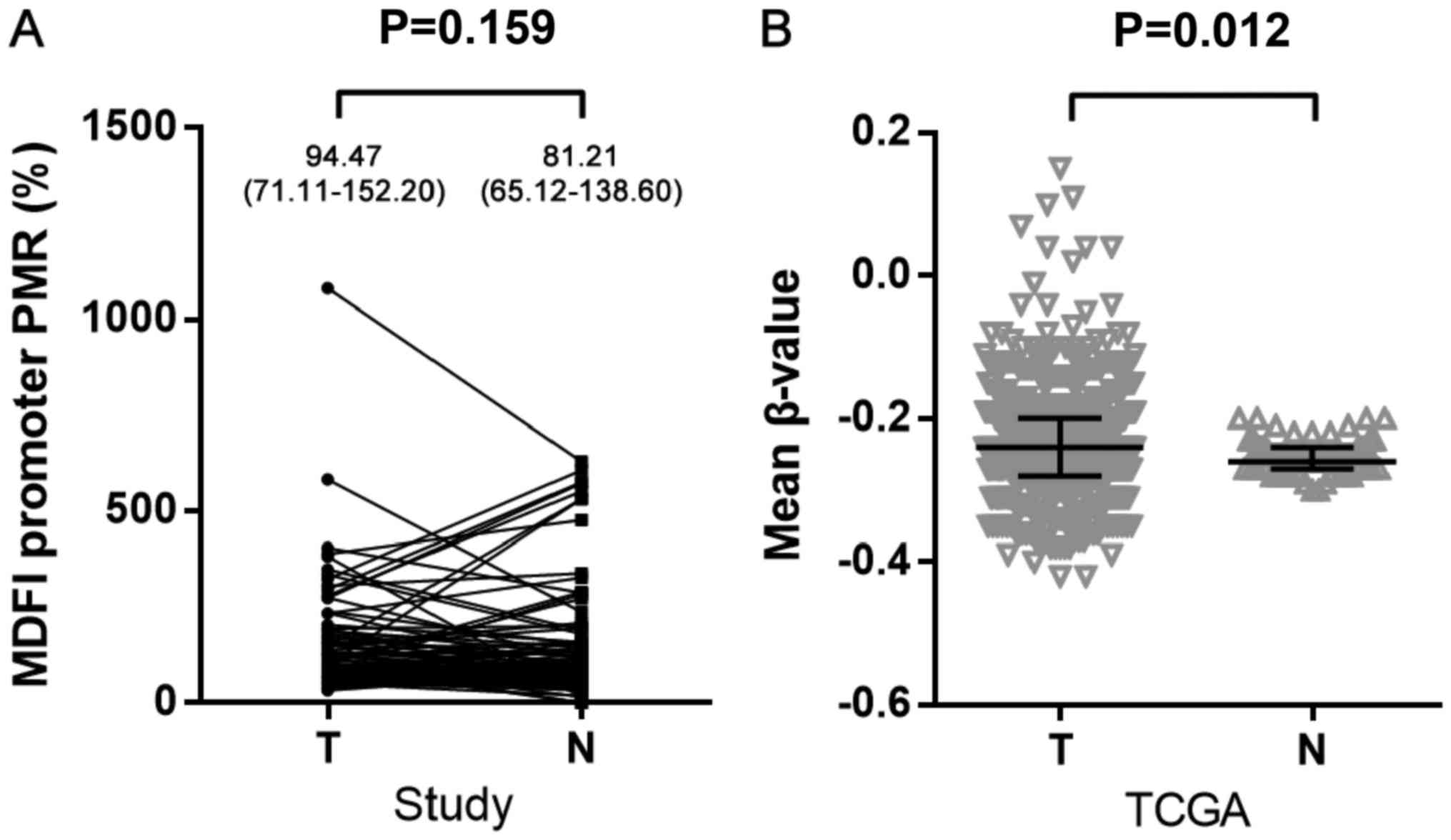
PMR was used to represent the methylation level of
the MDFI promoter. The results indicated that there was no
significant difference in MDFI promoter methylation between
tumor tissues and adjacent non-tumor tissues [median (quartile
range): 94.47% (71.11–152.20%) vs. 81.21% (65.12–138.60%); P=0.159;
Fig. 2A]. Considering the limited
sample size, the TCGA dataset was utilized for further
investigation. There were seven CG probes (cg01520588, cg08625380,
cg14086013, cg06484572, cg02914379, cg17094014 and cg13612207)
located in TSS200 and TSS1500 regions of MDFI. Mean β-value
represented the promoter methylation level. Notably, a
significantly higher mean methylation level was determined in 830
NSCLC tumor tissues compared with 75 non-tumor tissues [median
(quartile range): −0.242 (−0.276–0.200) vs. −0.255 (−0.269–0.239);
P=0.012; Fig. 2B]. Subsequently, sex,
age and smoking history-based comparisons of MDFI promoter
methylation levels between tumor tissues, and paired adjacent
tissues were performed. Significantly increased methylation levels
were observed in tumor tissues compared with paired adjacent
tissues in females [PMR: 90.66% (74.45–153.60%) vs. 75.88%
(48.17–117.53%); P=0.031; Fig. 3A];
however, no significant difference was identified in males
(P=0.832). Furthermore, a significant difference was determined in
younger patients [≤65 years; PMR: 92.98% (70.14–125.37%) vs. 73.74%
(60.79–101.68%); P=0.003; Fig. 3A],
but not in older patients (>65; P=0.327). Additionally,
non-smokers exhibited a significant increase in methylation levels
in tumor tissues compared with non-tumor tissues [PMR: 94.10%
(72.85–153.20%) vs. 75.67% (48.17–115.20%); P=0.014; Fig. 3A], but this association was not
observed in smokers (P=0.832). Following this, the same analysis
process was performed in a TCGA cohort (Fig. 3B). The sample size of tumor
tissues/non-tumor tissues was 481/46 in males, 335/28 in females,
341/32 in those ≤65 years old, 448/42 in those >65 years old,
578/57 in non-smokers and 219/12 in current smokers. The data
demonstrated consistent results with the study cohort whereby
significant differences in the methylation levels between tumor and
non-tumor tissues were only observed in females (P=0.029), and
non-smokers and ex-smokers (P=0.022). No significant difference was
determined in age-based subgroup analysis (all P>0.05).
The sample was defined as ‘hypermethylation’ if the
PMR value was higher in tumor tissue compared with adjacent
non-tumor tissues; otherwise, it was termed ‘hypomethylation’. As
in Table I depicts, the
hypermethylation percentage, used to determine the probability of a
hypermethylation event, among 111 patients with NSCLC was 58.56%
(65/111). The association analysis with clinical variables
indicated that MDFI was more significantly frequently
hypermethylated in the tumors of female patients compared with male
patients (73.68% vs. 50.68%; P=0.020). MDFI was also
significantly more frequently hypermethylated in younger patients
compared with older patients (67.74% vs. 46.94%; P=0.027); and in
patients without a history of smoking compared with current smokers
(70.00% vs. 49.18%; P=0.027).
 | Table I.Association between MyoD family
inhibitor promoter methylation and clinical characteristics in
patients with non-small cell lung cancer. |
Table I.
Association between MyoD family
inhibitor promoter methylation and clinical characteristics in
patients with non-small cell lung cancer.
| Variables | No. | Hypermethylation, n
(%) | Hypomethylation, n
(%) | P-value |
|---|
| Total | 111 | 65 (58.56) | 46 (40.54) |
|
| Sex |
|
|
| 0.020a |
|
Male | 73 | 37 (50.68) | 36 (49.32) |
|
|
Female | 38 | 28 (73.68) | 10 (26.32) |
|
| Age, years |
|
|
| 0.027a |
|
≤65 | 62 | 42 (67.74) | 20 (32.26) |
|
|
>65 | 49 | 23 (46.94) | 26 (53.06) |
|
| Smoking
history |
|
|
| 0.027a |
|
Non-smoker | 50 | 35 (70.00) | 15 (30.00) |
|
|
Smoker | 61 | 30 (49.18) | 31 (50.82) |
|
| Histological
type |
|
|
| 0.068 |
|
LUSC | 42 | 20 (47.62) | 22 (52.38) |
|
|
LUAD | 69 | 45 (65.22) | 24 (34.78) |
|
| Clinical stage |
|
|
| 0.824 |
|
I+II | 88 | 52 (59.09) | 36 (40.91) |
|
|
III+IV | 23 | 13 (56.52) | 10 (43.48) |
|
The next focus was on the prognostic value of
aberrant MDFI promoter methylation status on predicting the
outcomes of postoperative patients with NSCLC. Mortality occurred
in 11/111 patients with NSCLC; however, the Kaplan-Meier survival
curve indicated that there was no significant association between
MDFI hypermethylation and the overall survival of patients
with NSCLC (P=0.344; Fig. 4A). No
significant association was determined in the subgroup analysis of
sex, age and smoking behavior, even in females (P=0.979; Fig. 4B), the younger population (P=0.709;
Fig. 4C) and non-smokers (P=0.837;
Fig. 4D).
In order to investigate the potential epigenetic
role in regulating gene expression, the gene expression changes of
four lung cancer cell lines (A549, H1299, Hotz and U1752) was
further analyzed with different regimens of 5-aza-2′-deoxycytidine
(5-AZA) treatment from the Gene Expression Omnibus (GEO) database.
As demonstrated in Fig. 5, the
expression of MDFI was increased significantly in the H1299
cell line with the increasing doses (0.3 and 3.0 µM) of 5-AZA for
48 h (GSE29077; P=0.027). Following 5-AZA treatment in the Hotz
cell line, the expression of MDFI was also increased,
compared with the cell line without treatment (GSE14315;
P=0.006).
Discussion
The aim of the present study was to determine
whether DNA methylation of MDFI promoter was associated with
NSCLC risk. The results demonstrated that the association between
MDFI promoter hypermethylation and NSCLC was specific to
younger patients with younger age, females, and non-smokers.
Age is a crucial factor in carcinogenesis (31). Numerous studies have reported that
aging is associated with highly reproducible changes in DNA
methylation at specific sites in the genome (32,33).
Age-associated hypermethylation is enriched close to CpG islands,
whereas hypomethylation occurs outside of CpG islands (34,35). DNA
methylation may be one of the important mechanisms by which aging
predisposes to numerous age-associated diseases, including cancer
(36). In the present study,
MDFI promoter hypermethylation occurrence was determined in
the younger population, which provided as a potential age-specific
biomarker of NSCLC.
In female patients with NSCLC, MDFI
hypermethylation occurred more frequently in the tumor groups
compared with the non-tumor groups, in the study and TCGA cohorts,
which indicated MDFI hypermethylation may be an effective
marker for female patients with NSCLC. Previous studies have
reported that sexual hormones influence neoplastic diseases by
altering the DNA methylation levels (37,38).
Estrogen induces decreased thymic autoimmune regulator
(AIRE) expression by increasing the number of methylation
sites within the AIRE promoter (39). MDFI has been considered as a
potential estrogen-associated gene in lung tumorigenesis (40); however, further investigation is
required to verify the hypothesis of the present study.
Cigarette smoking, the top risk factor, is
attributed to >80% cases of lung cancer cases (41). A previous study identified a group of
aberrantly-methylated smoking-associated genes in patients with
NSCLC (42). Notably, breakdown
analysis by smoking history indicated that a significant
association with MDFI hypermethylation existed in
non-smoking patients, but not in smoking patients. Furthermore,
this association existed in ex-smokers, but not in the current
smokers. Although it was unclear why MDFI promoter
methylation was predominantly associated with less tobacco
exposure, it was considered that the methylation was caused by
carcinogens other than those contained in tobacco smoke. A previous
study indicated that the infections caused by human papilloma virus
was an influencing factor of lung cancer in female non-smokers
(43); therefore, further analysis of
carcinogenesis and the progression of NSCLC in non-smokers should
be performed in the future.
Similar to other cancer types, NSCLC is influenced
by regional hypermethylation of promoters of common
cancer-associated genes (10).
Significant differences in MDFI methylation have been
observed between pancreatic tumor tissues and normal controls
(24,25); furthermore, MDFI methylation
has been considered as a promising diagnostic marker in pancreatic
cancer (24). Furthermore, compared
with non-tumor tissues, the MDFI promoter was most
frequently hypermethylated in colorectal cancer tissues (44), indicating a tumor suppressor effect of
MDFI in human cancer types. In the present study, an
increased trend in MDFI methylation level was determined in
patients with NSCLC, although the result was not statistically
significant. Using the TCGA database with an increased sample size
demonstrated an elevated methylation level in NSCLC tumor tissues.
Considering the divergence caused by sample size, different
ethnicities and different detection methods, a more comprehensive
study is required.
Aberrant methylation of promoter regions is
generally associated with gene transcriptional dysfunction through
different underlying mechanisms, including the direct inhibition of
transcription factor binding, and the recruitment of methyl-binding
domain proteins (MBD1, MBD2 and MeCP2) and their associated
complexes (45). Currently, there are
a limited number of studies on the underlying epigenetic mechanism
of the MDFI gene in NSCLC. The MDFI gene is commonly
downregulated in invasive hepatic cellular cancer cells and is a
repressor of myogenic helix-loop-helix class transportation factors
(46). Furthermore, Pan et al
(46) determined an increasing Wnt
reporter gene activity in the canonical Wnt signaling pathway by
knocking down endogenous MDFI expression, which indicated
the MDFI gene as a tumor suppressor gene (21). As poor prognosis of NSCLC has been
reported to be associated with aberrant methylation through Wnt
signaling, including WNT inhibitory factor 1 (47) and secreted frizzled related protein 3
(48), a similar role of MDFI
promoter methylation may participate in Wnt signaling pathway
regulation for different types of cancer with aggressive
phenotypes. The evidence for MDFI promoter methylation as a
regulatory mechanism of gene expression in NSCLC is notable and
should be further explored. Additionally, the GEO analysis in the
present study demonstrated gene expression changes in partial lung
cancer cells following demethylation, indicating that other
epigenetic mechanisms, including histone modifications and
non-coding RNA, may exert this interaction in NSCLC. Therefore,
this complex network of gene activity establishment and maintenance
requires further research to be understood.
In conclusion, hypermethylation of MDFI
promoter may contribute to the risk of NSCLC in females,
non-smokers and the younger population. The evidence for
MDFI promoter methylation as a regulatory mechanism of gene
expression is notable and should be further explored.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the
National Natural Science Foundation of China (grant no. 81371469),
K. C. Wong Magna Fund in Ningbo University and Ningbo Social and
Science Development Fund (grant nos. 2012C50006 and
2015C50012).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
SD and HM contributed to the conception, design and
final approval of the submitted version. XC, HH and BL contributed
to the interpretation of data and completion of figures and tables.
XY, CZ and JZ contributed to performing the experiments and
analyzing the data. GZ analyzed the data, revised and approved the
manuscript. XC and HH contributed to writing the paper. All the
authors have read and approved the final manuscript.
Ethics approval and consent to
participate
A written informed consent form was signed by all
participants prior to their inclusion within the study. The study
was approved by the Ethics Committee of Huzhou First People's
Hospital.
Consent for publication
All patients have provided written informed consent
for the publication of any associated data and accompanying
images.
Competing interest
The authors declare that they have no competing
interests.
References
|
1
|
Han Y, Shi K, Zhou SJ, Yu DP and Liu ZD:
The clinicopathological significance of hMLH1 hypermethylation in
non-small-cell lung cancer: A meta-analysis and literature review.
Onco Targets Ther. 9:5081–5090. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Drzewiecka H, Gałęcki B,
Jarmołowska-Jurczyszyn D, Kluk A, Dyszkiewicz W and Jagodziński PP:
Decreased expression of connective tissue growth factor in
non-small cell lung cancer is associated with clinicopathological
variables and can be restored by epigenetic modifiers. J Cancer Res
Clin Oncol. 142:1927–1946. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Devarakonda S, Morgensztern D and Govindan
R: Genomic alterations in lung adenocarcinoma. Lancet Oncol.
16:e342–e351. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shackelford RE, Vora M, Mayhall K and
Cotelingam J: ALK-rearrangements and testing methods in non-small
cell lung cancer: A review. Genes Cancer. 5:1–14. 2014.PubMed/NCBI
|
|
5
|
Travis WD: Pathology of lung cancer. Clin
Chest Med. 32:669–692. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Broodman I, VanDuijn MM, Stingl C, Dekker
LJ, Germenis AE, de Koning HJ, van Klaveren RJ, Aerts JG, Lindemans
J and Luider TM: Survivin autoantibodies are not elevated in lung
cancer when assayed controlling for specificity and smoking status.
Cancer Immunol Res. 4:165–172. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tsai MF, Wang CC and Chen JJ: Tumour
suppressor HLJ1: A potential diagnostic, preventive and therapeutic
target in non-small cell lung cancer. World J Clin Oncol.
5:865–873. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pérez-Ramírez C, Cañadas-Garre M, Robles
AI, Molina MÁ, Faus-Dáder MJ and Calleja-Hernández MA: Liquid
biopsy in early stage lung cancer. Transl Lung Cancer Res.
5:517–524. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shien K, Papadimitrakopoulou VA and
Wistuba II: Predictive biomarkers of response to PD-1/PD-L1 immune
checkpoint inhibitors in non-small cell lung cancer. Lung Cancer.
99:79–87. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ansari J, Shackelford RE and El-Osta H:
Epigenetics in non-small cell lung cancer: From basics to
therapeutics. Transl Lung Cancer Res. 5:155–171. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Song X, Shi K, Zhou SJ, Yu DP, Liu Z and
Han Y: Clinicopathological significance and a potential drugtarget
of RARβ in non-small-cell lung carcinoma: A meta-analysis and a
systematic review. Drug Des Devel Ther. 10:1345–1354.
2016.PubMed/NCBI
|
|
12
|
Zhang YA, Ma X, Sathe A, Fujimoto J,
Wistuba I, Lam S, Yatabe Y, Wang YW, Stastny V, Gao B, et al:
Validation of SCT methylation as a hallmark biomarker for lung
cancers. J Thorac Oncol. 11:346–360. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fischer JR, Ohnmacht U, Rieger N, Zemaitis
M, Stoffregen C, Manegold C and Lahm H: Prognostic significance of
RASSF1A promoter methylation on survival of non-small cell lung
cancer patients treated with gemcitabine. Lung Cancer. 56:115–123.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zöchbauer-Müller S, Fong KM, Virmani AK,
Geradts J, Gazdar AF and Minna JD: Aberrant promoter methylation of
multiple genes in non-small cell lung cancers. Cancer Res.
61:249–255. 2001.PubMed/NCBI
|
|
15
|
Kim DS, Cha SI, Lee JH, Lee YM, Choi JE,
Kim MJ, Lim JS, Lee EB, Kim CH, Park TI, et al: Aberrant DNA
methylation profiles of non-small cell lung cancers in a Korean
population. Lung Cancer. 58:1–6. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tomizawa Y, Iijima H, Nomoto T, Iwasaki Y,
Otani Y, Tsuchiya S, Saito R, Dobashi K, Nakajima T and Mori M:
Clinicopathological significance of aberrant methylation of
RARbeta2 at 3p24, RASSF1A at 3p21.3, and FHIT at 3p14.2 in patients
with non-small cell lung cancer. Lung Cancer. 46:305–312. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Heller G, Babinsky VN, Ziegler B,
Weinzierl M, Noll C, Altenberger C, Müllauer L, Dekan G, Grin Y,
Lang G, et al: Genome-wide CpG island methylation analyses in
non-small cell lung cancer patients. Carcinogenesis. 34:513–521.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Di Paolo A, Del Re M, Petrini I, Altavilla
G and Danesi R: Recent advances in epigenomics in NSCLC: Real-time
detection and therapeutic implications. Epigenomics. 8:1151–1167.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Egger G, Liang G, Aparicio A and Jones PA:
Epigenetics in human disease and prospects for epigenetic therapy.
Nature. 429:457–463. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mehta A, Dobersch S, Romero-Olmedo AJ and
Barreto G: Epigenetics in lung cancer diagnosis and therapy. Cancer
Metastasis Rev. 34:229–241. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kusano S, Shiimura Y and Eizuru Y: I-mfa
domain proteins specifically interact with SERTA domain proteins
and repress their transactivating functions. Biochimie.
93:1555–1564. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kusano S and Eizuru Y: Human I-mfa domain
proteins specifically interact with KSHV LANA and affect its
regulation of Wnt signaling-dependent transcription. Biochem
Biophys Res Commun. 396:608–613. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Snider L, Thirlwell H, Miller JR, Moon RT,
Groudine M and Tapscott SJ: Inhibition of Tcf3 binding by I-mfa
domain proteins. Mol Cell Biol. 21:1866–1873. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kisiel JB, Yab TC, Taylor WR, Chari ST,
Petersen GM, Mahoney DW and Ahlquist DA: Stool DNA testing for the
detection of pancreatic cancer: Assessment of methylation marker
candidates. Cancer. 118:2623–2631. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Omura N, Li CP, Li A, Hong SM, Walter K,
Jimeno A, Hidalgo M and Goggins M: Genome-wide profiling of
methylated promoters in pancreatic adenocarcinoma. Cancer Biol
Ther. 7:1146–1156. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Edge SB and Compton CC: The american joint
committee on cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Travis WD: The 2015 WHO classification of
lung tumors. Pathologe. 35 Suppl 2:S1882014. View Article : Google Scholar
|
|
28
|
Chen X, Yang Y, Liu J, Li B, Xu Y, Li C,
Xu Q, Liu G, Chen Y, Ying J and Duan S: NDRG4 hypermethylation is a
potential biomarker for diagnosis and prognosis of gastric cancer
in Chinese population. Oncotarget. 8:8105–8119. 2017.PubMed/NCBI
|
|
29
|
Kristensen LS, Mikeska T, Krypuy M and
Dobrovic A: Sensitive melting analysis after real time-methylation
specific PCR (SMART-MSP): High-throughput and probe-free
quantitative DNA methylation detection. Nucleic Acids Res.
36:e422008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xia Y, Hong Q, Chen X, Ye H, Fang L, Zhou
A, Gao Y, Jiang D and Duan S: APC2 and CYP1B1 methylation changes
in the bone marrow of acute myeloid leukemia patients during
chemotherapy. Exp Ther Med. 12:3047–3052. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chen K, Song F, He M, Li H, Qian B, Zhang
W, Wei Q and Hao X: Trends in head and neck cancer incidence in
Tianjin, China, between 1981 and 2002. Head Neck. 31:175–182. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Rakyan VK, Down TA, Maslau S, Andrew T,
Yang TP, Beyan H, Whittaker P, McCann OT, Finer S, Valdes AM, et
al: Human aging-associated DNA hypermethylation occurs
preferentially at bivalent chromatin domains. Genome Res.
20:434–439. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Shennib H and Nguyen D: Bronchoalveolar
lavage in lung transplantation. Ann Thorac Surg. 51:335–340. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Florath I, Butterbach K, Müller H,
Bewerunge-Hudler M and Brenner H: Cross-sectional and longitudinal
changes in DNA methylation with age: An epigenome-wide analysis
revealing over 60 novel age-associated CpG sites. Hum Mol Genet.
23:1186–1201. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Johansson A, Enroth S and Gyllensten U:
Continuous aging of the human DNA methylome throughout the human
lifespan. PLoS One. 8:e673782013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ben-Avraham D: Epigenetics of aging. Adv
Exp Med Biol. 847:179–191. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Bredfeldt TG, Greathouse KL, Safe SH, Hung
MC, Bedford MT and Walker CL: Xenoestrogen-induced regulation of
EZH2 and histone methylation via estrogen receptor signaling to
PI3K/AKT. Mol Endocrinol. 24:993–1006. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kulig E, Landefeld TD and Lloyd RV: The
effects of estrogen on prolactin gene methylation in normal and
neoplastic rat pituitary tissues. Am J Pathol. 140:207–214.
1992.PubMed/NCBI
|
|
39
|
Dragin N, Le Panse R and Berrih-Aknin S:
Autoimmune disease predisposition: Aire « protects » men. Med Sci
(Paris). 33:169–175. 2017.(In French). View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Pedraza-Alva G, Zingg JM, Donda A and
Pérez-Martínez L: Estrogen receptor regulates MyoD gene expression
by preventing AP-1-mediated repression. Biochem Biophys Res Commun.
389:360–365. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Xie D, Lan L, Huang K, Chen L, Xu C, Wang
R, Shi Y, Wu X, Wang L, Liu Y and Lu B: Association of p53/p21
expression and cigarette smoking with tumor progression and poor
prognosis in non-small cell lung cancer patients. Oncol Rep.
32:2517–2526. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Huang T, Chen X, Hong Q, Deng Z, Ma H, Xin
Y, Fang Y, Ye H, Wang R, Zhang C, et al: Meta-analyses of gene
methylation and smoking behavior in non-small cell lung cancer
patients. Sci Rep. 5:88972015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Cheng YW, Chiou HL, Sheu GT, Hsieh LL,
Chen JT, Chen CY, Su JM and Lee H: The association of human
papillomavirus 16/18 infection with lung cancer among nonsmoking
Taiwanese women. Cancer Res. 61:2799–2803. 2001.PubMed/NCBI
|
|
44
|
Lin PC, Lin JK, Lin CH, Lin HH, Yang SH,
Jiang JK, Chen WS, Chou CC, Tsai SF and Chang SC: Clinical
relevance of plasma DNA methylation in colorectal cancer patients
identified by using a genome-wide high-resolution array. Ann Surg
Oncol. 22 Suppl 3:S1419–S1427. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Lopez-Serra L and Esteller M: Proteins
that bind methylated DNA and human cancer: Reading the wrong words.
Br J Cancer. 98:1881–1885. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Pan W, Jia Y, Huang T, Wang J, Tao D, Gan
X and Li L: Beta-catenin relieves I-mfa-mediated suppression of
LEF-1 in mammalian cells. J Cell Sci. 119:4850–4856. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Xie J, Zhang Y, Hu X, Lv R, Xiao D, Jiang
L and Bao Q: Norcantharidin inhibits Wnt signal pathway via
promoter demethylation of WIF-1 in human non-small cell lung
cancer. Med Oncol. 32:1452015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Schlensog M, Magnus L, Heide T,
Eschenbruch J, Steib F, Tator M, Kloten V, Rose M, Noetzel E, Gaisa
NT, et al: Epigenetic loss of putative tumor suppressor SFRP3
correlates with poor prognosis of lung adenocarcinoma patients.
Epigenetics. Sep 13–2016.(Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|