Introduction
Gastric cancer is the second most common cause of
cancer-related mortality worldwide (1). In recent studies, surgical resection
along with chemo-radiation demonstrated significant improvement
compared with surgery alone; however, numerous patients with
gastric cancer have advanced or metastatic diseases at diagnosis
(2–5).
The molecular mechanisms involved in tumor development and
progression remain unclear in gastric cancer (6). A number of studies have reported that
CpG island methylation leads to inactivation and silencing of
respective tumor suppressor genes, including COX-2, APC, HPP1 and
DAPK in gastric cancer (7,8). To improve the prognosis of gastric
cancer patients, a greater understanding of the biological
mechanisms of gastric cancer progression and novel therapeutic
methods is required.
Opioid binding protein/cell adhesion molecule-like
(OPCML), located on 11q25, is a glycosylphosphatidylinositol
(GPI)-anchored cell adhesion-like molecule; it is strongly
associated with cell growth, invasion, and metastasis and
tumorigenesis (9). OPCML is widely
expressed in adult tissues; however, in cancer tissues of various
types, including nasopharyngeal carcinoma, hepatocellular
carcinoma, bladder cancer, ovarian cancer and cervical carcinoma,
its promoter is often methylated and its expression decreased
(9–12). To the best of our knowledge, little is
known regarding the association between OPCML and the occurrence
and development of gastric cancer. Therefore, the aim of the
present study was to investigate the mRNA expression of OPCML and
the degree of CpG island methylation in human gastric cancer cell
lines, and to elucidate the molecular mechanisms that may underlie
the loss of OPCML expression in gastric cancer cell lines.
Materials and methods
Cell lines and tissue
The human gastric cancer SGC7901, MKN74, MKN45, KATO
III, SNU1 cell lines were obtained from RIKEN BioResource Center
(Tsukuba, Japan) and the cell lines AGS, N87 and the immortal
gastric mucosal GES1 cell lines were obtained from the American
Type Culture Collection (Manassas, VA, USA). All cell lines were
maintained in RPMI-1640 medium (HyClone; GE Healthcare Life
Sciences, Logan, UT, USA) with 10% fetal bovine serum in a 5%
CO2 atmosphere at 37°C. In addition, normal gastric
tissue samples were obtained from the gastric antrum (female, 59
years) and corpus gastricum (female, 43 years) by biopsy at the
Affiliated LuoHu Hospital of Shenzhen University in February 2013.
The present study was approved by the Ethics Committee of the
Affiliated LuoHu Hospital of Shenzhen University; written informed
consent was obtained from patients.
Reverse transcription-polymerase chain
reaction (RT-PCR)
The total RNA of all aforementioned cell lines and
normal gastric tissue samples were extracted using the RNA-lyase
Mini kit (Macherey-Nagel GmbH and Co., Düren, Germany), according
to the manufacturer's protocol. For each RNA sample, 1 µg was
reverse-transcribed using the RevertAid First Strand cDNA Synthesis
kit (Fermentas; Thermo Fisher Scientific, Inc., Waltham, MA, USA).
The cDNA was then used to amplify the desired gene with specific
primers using PCR Amplification Reaction kit (Promega Corporation,
Madison, WI, USA). The number of PCR cycles was suitable to each
gene for complete linear amplification. The PCR thermocycling
conditions were as follows: Initial denaturation at 95°C for 5 min,
35 cycles including denaturation at 95°C for 30 sec, annealing at
60°C for 30 sec, elongation at 7°C for 30 sec, and then 72°C for 10
min. PCR primers were designed to amplify a 126-bp cDNA fragment of
the human OPCML gene (forward, 5′-ACACCACTGGGTGGAGAAAG-3′ and
reverse, 5′-AAGGGCAGCTTGCAGTACAT-3′). The mitochondrial ribosomal
protein S12 was used as normalization reference (forward,
5′-GCATTGCTGCTGGAGGTGTAAT-3′ and reverse,
5′-CTGCAACCAACCACTTTACGG-3′). The size of the mitochondrial
ribosomal protein S12 gene was 306-bp. To evaluate the PCR
products, the products were electrophoresed on 1% agarose gel in
Tris base-boric acid-EDTA buffer solution (Sigma-Aldrich; Merck
KGaA; Darmstadt, Germany). To analyze the electrophoresis results,
the samples were stained with 1 mg/ml ethidium bromide solution for
1 min at room temperature, and visualized by an UV transilluminator
apparatus.
Sodium bisulfite genomic
sequencing
Genomic DNA from the gastric cancer cell line MKN45
was extracted using the High Pure PCR Template Preparation kit
(Machery-Nagel GmbH), which were selected at random from the
gastric cell lines. A total of 1 µg DNA was then treated using the
CpGenome™ DNA Modification kit (Merck KGaA, Darmstadt,
Germany) according to the manufacturer's protocol. Methylated
primer sequences were as follows: Forward,
5′-CGTTTAGTTTTTCGTGCGTTC-3′ and reverse, 5′-CGAAAACGCGCAACCGACG-3′.
The size of the purpose fragment gene is 129-bp. The unmethylated
primer sequences were as follows: Forward,
5′-TTTGTTTAGTTTTTTGTGTGTTTG-3′ and reverse,
5′-CAAAACAAAAACACACAACAACA-3′. The size of the purpose fragment
gene was 136-bp. The PCR thermocycling conditions were as follows:
98°C for 10 min followed by 40 cycles with denaturation at 97°C for
50 sec, annealing at 60°C for 30 sec, elongation at 72°C for 40
sec, and a final extension step of 72°C for 10 min using PCR
Amplification Reaction kit (Promega Corporation). The methylated
specific primer used to amplify the gene that the promoter was
methylated. Meanwhile the unmethylated specific primer used to
amplify the unmethylated DNA, following methylated and unmethylated
DNA being completely bisulfite modified.
Treatment of cells with
5-aza-2′-deoxycytidine (5-AZA)
The gastric cancer AGS cell line and the immortal
gastric mucosal GES1 cell line were selected at random and seeded
at a density of 1×106 cells on a 60-mm dish. Following a
24 h incubation period in a 5% CO2 atmosphere at 37°C,
cells were treated with 10 µmol/l of 5-AZA (Sigma-Aldrich; Merck
KGaA). The same concentration of DMSO was also used as a control
for nonspecific solvent treatment with these cells. The cells were
extracted 72 h after treatment with 5-AZA using the RNA-lyase Mini
kit (Macherey-Nagel GmbH and Co.), and 1 µg RNA was
reverse-transcribed using the RevertAid First Strand cDNA Synthesis
kit, and then the cDNA was used to amplify OPCML gene using PCR
Amplification Reaction kit, as previously described by Gu et
al (7).
Cells transfected with pcDNA3.1+/OPCML
detected by RT-PCR
The gastric cancer cell lines MKN45, AGS, and 293
were transfected with pcDNA3.1+/OPCML or pcDNA 3.1 vector alone
(Beijing Fungenome Company, Beijing, China), using the transfection
reagent Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). At 48 h post-transfection, the total RNA of
cells was extracted using the RNA lyase Mini kit (Machery-Nagel
GmbH), according to the manufacturer's protocol. For each RNA
sample, 1 µg was reverse-transcribed using a First Strand cDNA
Synthesis kit (Fermentas; Thermo Fisher Scientific, Inc.) according
to the manufacturer's protocol. The PCR primers were designed to
amplify a 828-bp cDNA fragment of the human OPCML gene (forward,
5′-TCCCCAAAGCTATGGACAAC-3′ and reverse, 5′-GCCCATACAATGTGATG-3′).
The conditions on the polymerase chain reaction was set as follows:
First denaturation at 94°C for 5 min, followed by 35 cycles with
denaturation at 94°C for 30 sec, annealing at 60°C for 30 sec,
elongation at 72°C for 30 sec, and a final elongation step of 72°C
for 10 min using PCR Amplification Reaction kit (Promega
Corporation). The mitochondrial ribosomal protein S12 was used as
normalization reference gene (forward 5′-GCATTGCTGCTGGAGGTGTAAT-3′
and reverse, 5′-CTGCAACCAACCACTTTACGG-3′). The size of the
mitochondrial ribosomal protein S12 gene was 306-bp. To evaluate
the PCR products, the products were electrophoresed on 1% agarose
gel in Tris base-boric acid-EDTA buffer solution (Sigma-Aldrich;
Merck KGaA). To analyze the electrophoresis results, the samples
were stained with 1 mg/ml of ethidium bromide solution for 1 min at
room temperature, and visualized by an UV transilluminator
apparatus as aforementioned.
Colony formation assay
Gastric cancer cell lines AGS were transfected with
pcDNA3.1+/OPCML and pcDNA 3.1 vector, as aforementioned. After 48 h
of transfection, cells were seeded (1×104) on a 60 mm
dish, and selected for 2 weeks in the presence of 400 µg/ml G418
(Invitrogen; thermo Fisher Scientific, Inc.). Surviving colonies
(≥50 cells per colony) were counted following staining with 5%
crystal violet solution for 10 min at room temperature. The data
were obtained from three independent cell cultures and experiments
were repeated three times.
Statistical analysis
Results are presented as the mean ± standard
deviation and were analyzed using SPSS 16.0 (SPSS, Inc., Chicago,
IL, USA) and plotted using Graphpad Prism 6.0 (GraphPad Software,
Inc., La Jolla, CA, USA). Data with two groups were compared using
Student's unpaired t-test. P<0.05 was considered to indicate a
statistically significant difference.
Results
OPCML expression in gastric cancer
cell lines and normal gastric tissue
In the present study, OPCML expression was assessed
using RT-PCR in the gastric cancer AGS, SGC7901, KATO III, MKN74,
N87, SNU and MKN45 cell lines, in the immortal gastric mucosa GES1
cell line and in normal gastric tissue. As Fig. 1 indicates, the expression of the OPCML
gene was reduced in AGS, SGC7901, KATO III, MKN74, N87, SNU, MKN45
and GES1 cell lines, compared with normal gastric tissue (Fig. 1).
OPCML downregulation was mediated by
promoter methylation in gastric cancer cell lines
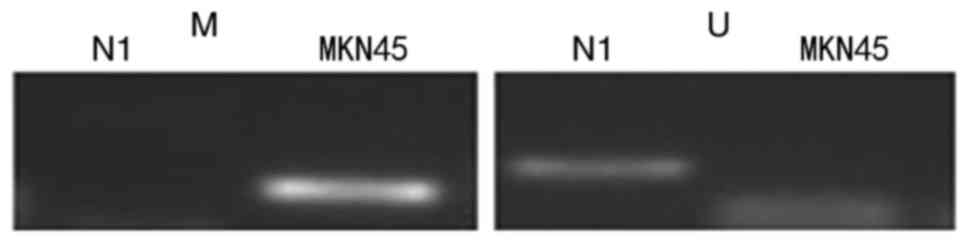
To investigate the role of putative OPCML gene
losses in gastric cells, methylation-specific PCR (MSP) was
performed in MKN45 cells to evaluate the methylation of promoter
CpG islands using specific MSP primers. The data revealed that
hypermethylation of the OPCML promoter commonly occurred in MKN45
cells, and there was no methylation in normal gastric tissues
(Fig. 2). To verify whether CpG
island methylation directly mediates OPCML silencing, the gastric
cancer cell line MKN45 and immortal gastric mucosa cell GES1 were
treated with the demethylating agent 5-AZA. Following this
treatment, OPCML expression was observed to be markedly restored
following drug treatment in MKN45 cells (Fig. 3).
OPCML suppressed gastric cancer colony
formation
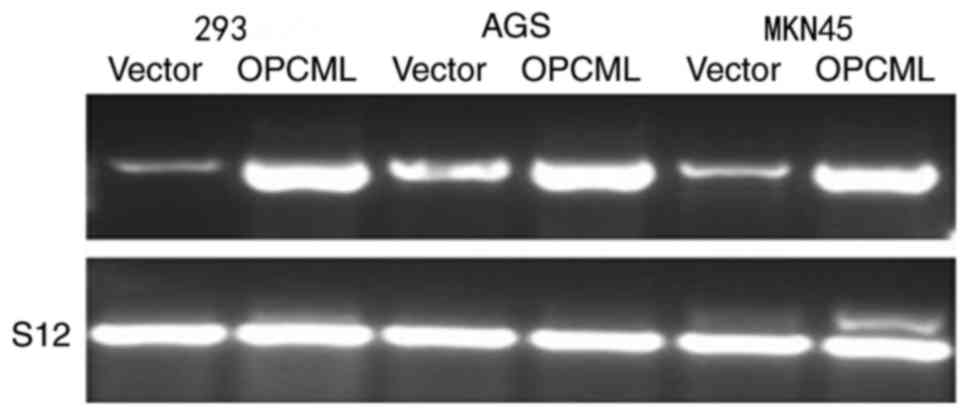
To investigate OPCML gene function further, the
gastric cancer cell MKN45, AGS lines and 293 cells were transfected
with the pcDNA3.1+/OPCML and pcDNA 3.1 vector (Fig. 4). Fewer cells transfected with the
OPCML gene adhered to the culture dish compared with those
transfected with an empty vector (P<0.05; Figs. 5 and 6).
These results indicated that the OPCML gene possesses the ability
to inhibit colony formation in gastric cancer.
Discussion
OPCML belongs to the IgLON family of immunoglobulin
domain-containing glycosylphosphatidylinositol-anchored cell
adhesion molecules, which includes opioid-binding cell adhesion
molecule, neurotbrimin, neuronal growth regulator 1 and limbic
system-associated membrane protein. It has been reported that
IgLONs serve a notable role in cell-cell recognition and adhesion
(13–18). OPCML, which acts as a cell adhesion
molecule, contains several protein-protein interaction domains,
including the three C2-like Ig domains, which are commonly found in
cell surface adhesion molecules and receptor proteins. Through
these domains, OPCML was demonstrated to modulate functions of
growth promotion or inhibition in tumor cells. OPCML was the first
member of the IgLON family identified to possess tumor suppressor
functions in multiple cancer types, which are frequently
epigenetically and genetically silenced at the early stage of
carcinogenesis. The loss of OPCML may reduce heterodimeric complex
formation and cell-cell adhesion, therefore damaging the
corresponding signaling pathways and promoting the progress of
carcinogenesis (13–18).
In the present study, the expression of OPCML in the
SGC7901, KATO III, MKN45, MKN74, SNU1, AGS and N87 cell lines the
immortal gastric mucosal GES1 cell line and normal gastric tissue
were assessed by RT-PCR analysis. OPCML was demonstrated to be
downregulated in SGC7901, KATO III, MKN45, MKN74, SNU1, AGS, N87
and GES1 cell lines, when compared with normal gastric tissue. This
observation was corroborated by Wang et al (6) who also reported that the expression of
OPCML was downregulated in patients with gastric cancer compared
with normal gastric tissue by RT-PCR analysis.
OPCML acts as a tumor suppressor in multiple cancer
types, including nasopharyngeal carcinoma, bladder cancer, ovarian
cancer, cervical carcinoma, esophageal carcinoma and hepatocellular
carcinoma; recent studies have reported that the loss or
downregulation of OPCML expression is associated with OPCML gene
promoter methylation (18–20). However, to the best of our knowledge,
little is known regarding the association between OPCML expression
and promoter methylation in gastric cancer. Therefore, in the
present study, to confirm whether OPCML gene promoter methylation
is the cause of attenuated OPCML expression, MSP analysis was
performed in the MKN45 cell line using the CpGenome™ DNA
Modification kit and hypermethylation of the OPCML promoter was
demonstrated to occur in MKN45.
In the present study the gastric cancer cell line
MKN45 and the immortal gastric mucosal cell line GES1 were treated
with the methylation inhibitor 5-AZA and it was demonstrated that
treatment with 5-AZA was able to restore or upregulate the
expression of OPCML mRNA in these cells. To investigate OPCML gene
function, the gastric cancer cell line AGS was transfected with the
pcDNA3.1+/OPCML and pcDNA 3.1 vector. Ectopic expression of OPCML
in gastric cell lines with endogenous silencing resulted in the
inhibition of cell colony formation, indicating that OPCML acts as
a broad tumor suppressor.
DNA methylation is an epigenetic phenomenon that
affects gene expression without altering the DNA sequence (20–24).
Aberrant supermethylation occurs in promoter CpG islands, and is a
mechanism by which tumor suppressor genes are silenced and, in
certain circumstance, may be an important mechanism (20–24). The
present study has demonstrated that OPCML, which acts as a broad
tumor suppressor gene, is silenced in gastric cancer cell lines via
the aberrant supermethylation of promoter CpG islands. This
typically occurred prior to the development of clinical
manifestations in patients and the obtaining of radiographic
evidence, and therefore may provide a novel molecular approach for
the early diagnosis of gastric cancer.
In the present study OPCML gene function in
vivo was not investigated and the OPCML protein expression in
gastric cancer cell lines and normal gastric tissue is unknown.
Future functional studies are required to clarify its role in
signaling pathways, which in turn may result in the identification
of further molecular targets in gastric cancer.
Acknowledgements
Not applicable.
Funding
This study was supported by the National Natural
Science Foundation of China (grant no. 81000887).
Availability of data and materials
The datasets used during the current study are
available from the corresponding author on reasonable request.
Authors' contributions
NZ and XX were responsible for drafting the
manuscript. NZ, YW and XH contributed the experiments. XX, YW, JX
and XH contributed to analysis and interpretation of data. LY and
XX contributed to conducting the study. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the Affiliated LuoHu Hospital of Shenzhen University
and written informed consent was obtained from all patients.
Consent for publication
The patients provided written informed consent for
the publication of any associated data.
Competing interests
The authors declare no competing interests.
References
|
1
|
Kim SJ, Wang YG, Lee HW, Kang HG, La SH,
Choi IJ, Irimura T, Ro JY, Bresalier RS and Chun KH: Up-regulation
of neogenin-1 increases cell proliferation and motility in gastric
cancer. Oncotarget. 5:3386–3398. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wu HH, Lin WC and Tsai KW: Advances in
molecular biomarkers for gastric cancer: miRNAs as emerging novel
cancer markers. Expert Rev Mol Med. 16:e12014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang Y and Wu S: Novel therapy for
advanced gastric cancer. World J Gastrointest Oncol. 7:263–270.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yan S, He F, Luo R, Wu H, Huang M, Huang
C, Li Y and Zhou Z: Decreased expression of BRCA1-associated
protein 1 predicts unfavorable survival in gastric adenocarcinoma.
Tumor Biol. 37:6125–6133. 2016. View Article : Google Scholar
|
|
5
|
Wen R, Gao F, Zhou CJ and Jia YB:
Polymorphisms in mucin genes in the development of gastric cancer.
Word J Gastrointest Oncol. 7:328–337. 2015. View Article : Google Scholar
|
|
6
|
Wang L, Zhu JS, Song MQ, Chen GQ and Chen
JL: Comparison of gene expression profiles between primary tumor
and metastatic lesions in gastric cancer patients using laser
microdissection and cDNA microarray. Word J Gastroenterol.
12:6949–6954. 2006. View Article : Google Scholar
|
|
7
|
Gu P, Xing X, Tänzer M, Röcken C, Weichert
W, Ivanauskas A, Pross M, Peitz U, Malfertheiner P, Schmid RM and
Ebert MP: Frequent loss of TIMP-3 expression in progression of
esophageal and gastric adenocarcinomas. Neoplasia. 10:563–572.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kobayashi K, Inokuchi M, Takagi Y, Otsuki
S, Fujimori Y, Sato Y, Yanaka Y, Higuchi K, Aburatani T, Tomii C,
et al: Prognostic significance of PAK4 expression in gastric
cancer. J Clin Pathol. 69:580–585. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li C, Tang L, Zhao L, Li L, Xiao Q, Luo X,
Peng W, Ren G, Tao Q and Xiang T: OPCML is frequently methylated in
human colorectal cancer and its restored expression reverses EMT
via downregulation of smad signaling. Am J Cancer Res. 5:1635–1648.
2015.PubMed/NCBI
|
|
10
|
Sanz R, Ferraro GB and Fournier AE: IgLON
cell adhesion molecules are shed from the cell surface of cortical
neurons to promote neuronal growth. J Biol Chem. 290:4330–4342.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Minhas HM, Pescosolido MF, Schwede M,
Piasecka J, Gaitanis J, Tantravahi U and Morrow EM: An unbalanced
translocation involving loss of 10q26.2 and gain of 11q25 in a
pedigree with autism spectrum disorder and cerebellar juvenile
pilocytic astrocytoma. Am J Med Genet A. 161A:787–791. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhou F, Tao G, Chen X, Xie W, Liu M and
Cao X: Methylation of OPCML promoter in ovarian cancer tissues
predicts poor patient survival. Clin Chem Lab Med. 52:735–742.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhou F, Ma M, Tao G, Chen X, Xie W, Wang Y
and Cao X: Detection of circulating methylated opioid binding
protein/cell adhesion molecule-like gene as a biomarker for ovarian
carcinoma. Clin Lab. 60:759–765. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wu SY and Sood AK: New roles opined for
OPCML. Cancer Discov. 2:115–116. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
McKie AB, Vaughan S, Zanini E, Okon IS,
Louis L, de Sousa C, Greene MI, Wang Q, Agarwal R, Shaposhnikov D,
et al: The OPCML tumor suppressor functions as a cell surface
repressor-adaptor, negatively regulating receptor tyrosine kinases
in epithelial ovarian cancer. Cancer Discov. 2:156–171. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Amoenpisutt R, Proungvitaya S,
Jearanaikoon P and Limpaiboon T: DNA methylation level of OPCML and
SFRP1: A potential diagnostic biomarker of cholangiocarcinoma.
Tumor Biol. 36:4973–4978. 2015. View Article : Google Scholar
|
|
17
|
Rein BJ, Gupta S, Dada R, Safi J, Michener
C and Agarwal A: Potential markers for detection and monitoring of
ovarian cancer. J Oncol. 2011:4759832011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen W, Xiang J, Chen DF, Ni BB, Chen H,
Fan XJ, Wang PN, Song SX, Fang LK, Xiao HY, et al: Screening for
differentially methylated genes among human colorectal cancer
tissues and normal mucosa by microarray chip. Mol Biol Rep.
40:3457–3464. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Reed JE, Dunn JR, du Plessis DG, Shaw EJ,
Reeves P, Gee AL, Warnke PC, Sellar GC, Moss DJ and Walker C:
Expression of cellular adhesion molecule ‘OPCML’ is down-regulated
in gliomas and other brain tumours. Neuropathol Appl Neurobiol.
33:77–85. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Amornpisutt R, Sriraksa R and Limpaiboon
T: Validation of methylation-sensitive high resolution melting for
the detection of DNA methylation in cholangiocarcinoma. Clin
Biochem. 45:1092–1094. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cui Y, Ying Y, van Hasselt A, Ng KM, Yu J,
Zhang Q, Jin J, Liu D, Rhim JS, Rha SY, et al: OPCML is a broad
tumor suppressor for multiple carcinomas and lymphomas with
frequently epigenetic inactivation. PLoS One. 3:e29902008.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang B, Yu L, Yang GZ, Luo X and Huang L:
Applicaion of multiplex nested methylated specific PCR in early
diagnosis of epithelial ovarian cancer. Asian Pac J Cancer Prev.
16:3003–3007. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ye F, Zhang SF, Xie X and Lu WG: OPCML
gene promoter methylation and gene expression in tumor and stroma
cells of invasive cervical carcinoma. Cancer Invest. 26:569–574.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang Q, Hu G, Yang Q, Dong R, Xie X, Ma
D, Shen K and Kong B: A multiplex methylation-specific PCR assay
for the detection of early-stage ovarian cancer using cell-free
serum DNA. Gynecol Oncol. 130:132–139. 2013. View Article : Google Scholar : PubMed/NCBI
|