Introduction
Lung cancer, a highly malignant tumor, is the
leading cause of cancer-associated mortality in males and females
worldwide (1). The incidence and
mortality rates of non-small cell lung cancer are increasing in
developing countries, including China, due to the increase in the
use of tobacco as well as air pollution (2). According to its pathological pattern,
lung cancer may be divided into small cell lung cancer (SCLC) and
non-small cell lung cancer (NSCLC) (3). NSCLC, which includes adenocarcinoma,
squamous cell carcinoma, adenosquamous cell carcinoma and large
cell carcinoma, accounts for ~80–85% of all lung cancer cases
(4). Despite advances in traditional
treatments, including surgery, supplemented with radiotherapy and
chemotherapy, the prognosis remains poor and the five-year overall
survival rate is extremely low (17.1%) (5). These facts indicate an urgent
requirement to fully understand the molecular mechanisms underlying
the carcinogenesis and progression of NSCLC, and to investigate
novel therapeutic targets to control this malignant disease.
Increasing studies have demonstrated that microRNAs
(miRNAs/miRs) have vital functions in numerous developmental
processes and tumorigenesis, including the progression of NSCLC
(6–8).
miRNAs represent a group of small (~19–25 nucleotides),
non-protein-coding, and endogenous single RNAs, which negatively
regulate gene expression by binding to the 3′ untranslated regions
(3′UTRs) of target genes in an imperfect base pairing manner,
causing mRNA cleavage or translational repression (9,10). To
date, miRNAs have been demonstrated to regulate >30% of all
cellular proteins and serve substantial roles in a wide range of
physiological and pathological processes, including cell growth,
cell cycle, apoptosis, development, migration, invasion, survival
and metastasis (11–13). Accumulating evidence suggests that the
abnormal expression of miRNAs is associated with various types of
human cancer, and may serve as a new therapeutic strategy for
cancers as they act as oncogenes or tumor suppressors in
tumorigenesis and development (14).
In the present study, the potential roles of miR-154
were investigated in NSCLC. The expression levels of miR-154 were
measured in NSCLC tissues and cell lines, and its effects on cell
proliferation, migration and invasion were evaluated. Furthermore,
the direct targets of miR-154 in NSCLC and the underlining
molecular mechanism of its functions were explored. The results are
likely to provide a better understanding of NSCLC carcinogenesis
and progression, and a therapeutic target for patients with
NSCLC.
Materials and methods
Tissue samples, cell lines and cell
transfection
This study was approved by the ethics committee of
XinHua Hospital Affiliated to Shanghai Jiao Tong University School
of Medicine, China. Written informed consent was obtained from all
patients prior to enrollment in the present study. NSCLC tissues
and matched normal lung tissues were collected from 32 patients at
XinHua Hospital Affiliated to Shanghai Jiao Tong University School
of Medicine. None of these NSCLC patients had received chemotherapy
or radiotherapy prior to the surgery. Following collection, all
tissue samples were immediately snap-frozen in liquid nitrogen and
stored at −80°C.
All cell lines were purchased from the American Type
Culture Collection (ATCC; Manassas, VA, USA). The normal human
bronchial epithelial cell line 16HBE and NSCLC cell lines SK-MES-1,
H520, SPC-A1 and A549 were cultured in Dulbecco's modified Eagle's
medium (DMEM; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA,
USA) supplemented with 10% fetal bovine serum (FBS; Gibco), 100
U/ml penicillin and 100 µg/ml streptomycin, and maintained at 37°C
in a 5% CO2 humidified atmosphere.
The miR-154 mimics and negative control (NC) were
obtained from GenePharma (Shanghai, China). pcDNA3.1-BMI-1 or
pcDNA3.1-Ctl were synthesized by Guangzhou RiboBio Co., Ltd.
(Guangzhou, China). For transfection, cells were seeded in six-well
plates and grown to a confluency of 50–60%. Transfections were
performed using a Lipofectamine 2000 kit (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) following the manufacturer's
protocol.
RNA isolation and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was isolated from tissues or cell lines
using TRIzol reagent (Invitrogen) according to the standard
protocol. The concentration and purity of total RNA were determined
by A260/A280 with a NanoDrop ND-2000 spectrophotometer (NanoDrop
Technologies; Thermo Fisher Scientific, Inc., Wilmington, DE, USA).
For miR-154 expression, total RNA was reversed transcribed into
cDNA using a TaqMan microRNA reverse transcription kit (Applied
Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA). qPCR
was performed with a TaqMan microRNA assay kit (Applied
Biosystems). For BMI-1 mRNA expression, the synthesis of cDNA was
performed using an M-MLV First Strand kit (Invitrogen), followed by
qPCR with a SYBR-Green PCR kit (Takara Biotechnology Co., Ltd.,
Dalian, China). U6 and GAPDH mRNA were used as endogenous controls
for miR-154 and BMI-1 mRNA expression levels. All reactions were
performed on an ABI 7500 real-time system (Applied Biosystems).
Cell proliferation assay
Cell proliferation was evaluated using a
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay, according to the manufacturer's protocol. In briefly,
transfected cells were harvested, seeded in 96-well plates at a
concentration of 3,000 cells per well, and cultured for 1, 2, 3 and
4 days. At the indicated time points, cells were incubated with 20
µl MTT solution (5 mg/ml; Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany) for 4 h at 37°C. Then, the MTT solution was removed and
dimethyl sulfoxide was added into each well to dissolve the
formazan crystals. Optical density was determined using a
microplate reader (Pharmacia Biotech, Uppsala, Sweden) at a
wavelength of 490 nm.
Migration and invasion assay
For migration assays, transfected cells were
collected, suspended with FBS-free culture medium, and seeded in
the upper chambers of Transwell plates (BD Biosciences, San Jose,
CA, USA). DMEM containing 20% FBS was added to the lower chamber as
a chemoattractant. The Transwell plates were incubated in 5%
CO2 at 37°C for 24 h. The cells in the upper chamber
were carefully removed with cotton swabs, then the plates were
fixed in 95% methanol and stained with 0.1% crystal violet. The
stained cells were counted in five random fields per Transwell
plate under a microscope, and quantification was performed by
manually counting the stained cells. The invasion assays were
carried out in the same way as migration assays, with the exception
that Matrigel (BD Biosciences) was used in the Transwell
plates.
Bioinformatics methods
TargetScan (http://www.targetscan.org/) and PicTar (http://pictar.mdcberlin.de/) were used to predict the
potential target genes of miR-154.
Luciferase reporter assay
The 3′UTR of BMI-1 containing a putative binding
site (BMI-1-3′UTR WT) or a mutant (BMI-1-3′UTR MUT) cloned into the
psi-CHECK2 vectors were synthesized by GenePharma. HEK293T cells
were seeded in 24-well plates. Following incubation overnight,
BMI-1-3′UTR WT and BMI-1-3′UTR MUT were co-transfected with miR-154
mimics or NC using Lipofectamine 2000 reagent. Forty-eight h after
transfection, cells were collected, and firefly and renilla
luciferase activity was detected using a dual luciferase reporter
assay system (Promega Corporation, Madison, WI, USA) according to
the manufacturer's protocol. Renilla luciferase was used as
an internal control.
Western blot analysis
Proteins were isolated using
radioimmunoprecipitation assay buffer (Sigma-Aldrich) containing 1
mM phenylmethylsulfonyl fluoride (Sigma-Aldrich). Twenty micrograms
of protein were loaded into 10% SDS-PAGE, and transferred to a
polyvinylidene fluoride membrane (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA). The membranes were then blocked for 30 min at
room temperature with 5% skimmed milk in Tris-buffered saline (TBS)
solution containing 0.1% Tween 20 (TBST), and probed with primary
antibodies at 4°C overnight. The primary antibodies used in this
study include mouse anti-human monoclonal BMI-1 antibody
(sc-390443; 1:1,000; Santa Cruz Biotechnology, Inc., Dallas, TX,
USA) and mouse anti-human monoclonal GADPH antibody (sc-365062;
1:1,000; Santa Cruz Biotechnology, Inc.). Subsequently, the
membranes were washed with TBST three times and incubated with
horseradish peroxidase-conjugated secondary antibody (1:5,000;
Santa Cruz Biotechnology, Inc.) for 2 h at room temperature.
Finally, the membranes were washed again with TBST three times and
the proteins were visualized using an enhanced chemiluminescence
detection system (GE Healthcare Life Sciences, Chalfont, UK).
Statistical analysis
All data are presented as the means ± standard
deviation. SPSS 15.0 (SPSS Inc., Chicago, IL, USA) was used for
statistical analyses. P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-154 is downregulated in NSCLC
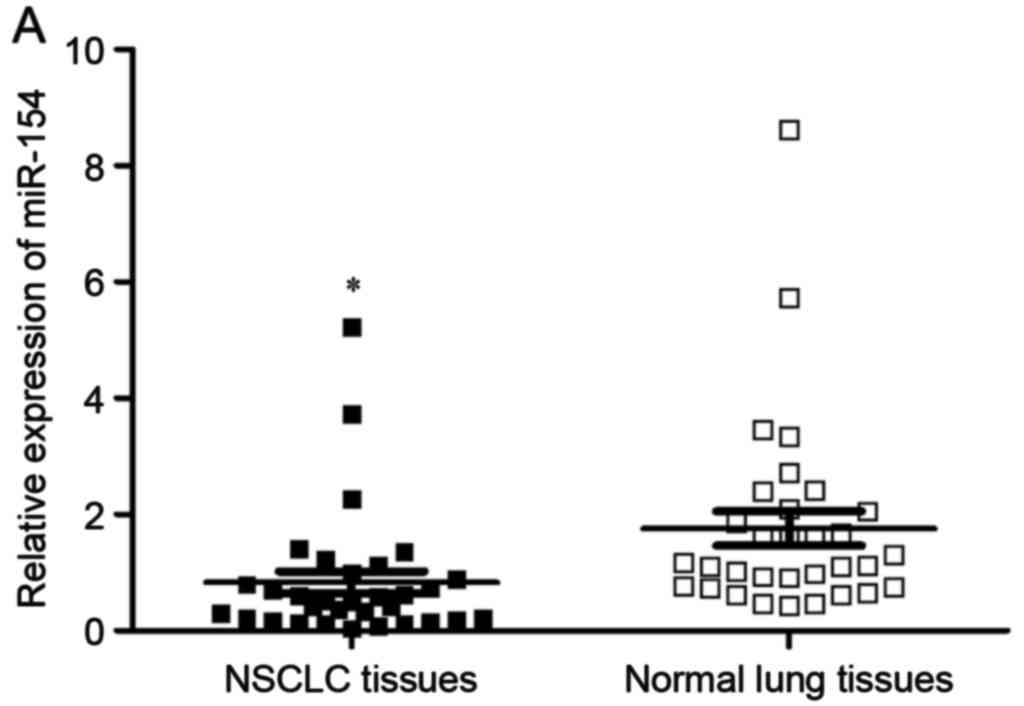
Firstly, RT-qPCR was performed to measure miR-154
expression in NSCLC tissues and matched normal lung tissues. The
results revealed that miR-154 was significantly downregulated in
NSCLC tissues in comparison with matched normal lung tissues
(Fig. 1A, P<0.05). miR-154
expression levels in NSCLC cell lines and normal human bronchial
epithelial cell line 16HBE were also determined using RT-qPCR. As
shown in Fig. 1B, miR-154 expression
levels were decreased in all four NSCLC cell lines compared with
16HBE (P<0.05). Among the four NSCLC cell lines, SPC-A1 and A549
expressed the lowest miR-154 levels and were thus selected for
further analyses.
Effects of miR-154 overexpression on
NSCLC cell proliferation, migration and invasion
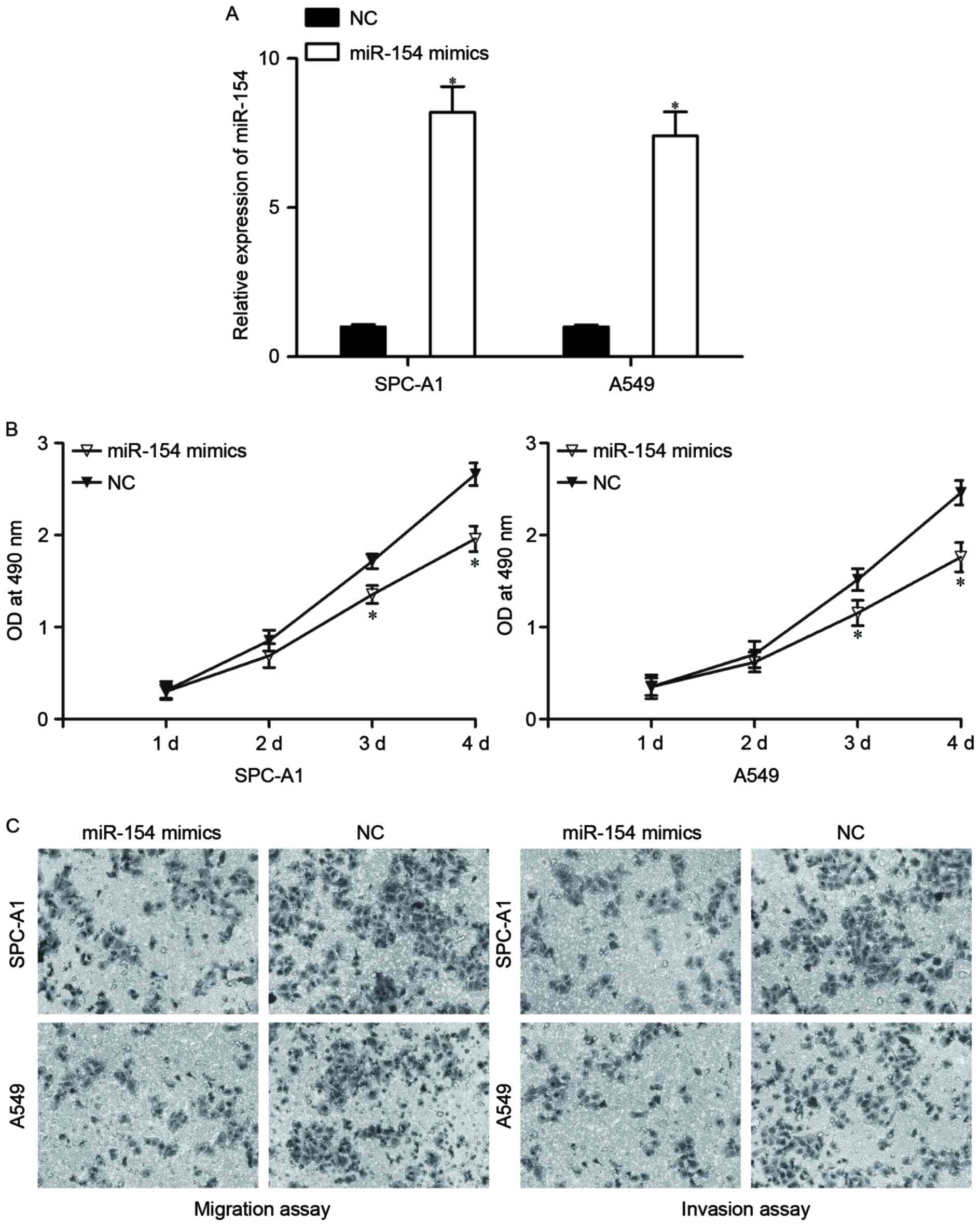
To evaluate the biological roles of miR-154 in
NSCLC, the effects of miR-154 overexpression on NSCLC cell
proliferation, migration and invasion were investigated. miR-154
mimics or NC were transfected into SPC-A1 and A549 cells. Following
transfection for 48 h, RT-qPCR was carried out to assess the
transfection efficiency. As shown in Fig.
2A, miR-154 was significantly elevated by miR-154 mimic
transfection in SPC-A1 and A549 cells (P<0.05).
Cell proliferation assay results revealed that cell
proliferation was notably reduced in SPC-A1 and A549 cells
transfected with miR-154 mimics (Fig.
2B, P<0.05). In addition, migration and invasion assays
revealed that miR-154 decreased the migration and invasion of
SPC-A1 and A549 cells compared with the NC groups (Fig. 2C, P<0.05). Taken together, these
results indicate that miR-154 may act as a tumor suppressor in
NSCLC progression.
BMI-1 is a direct target of miR-154 in
NSCLC
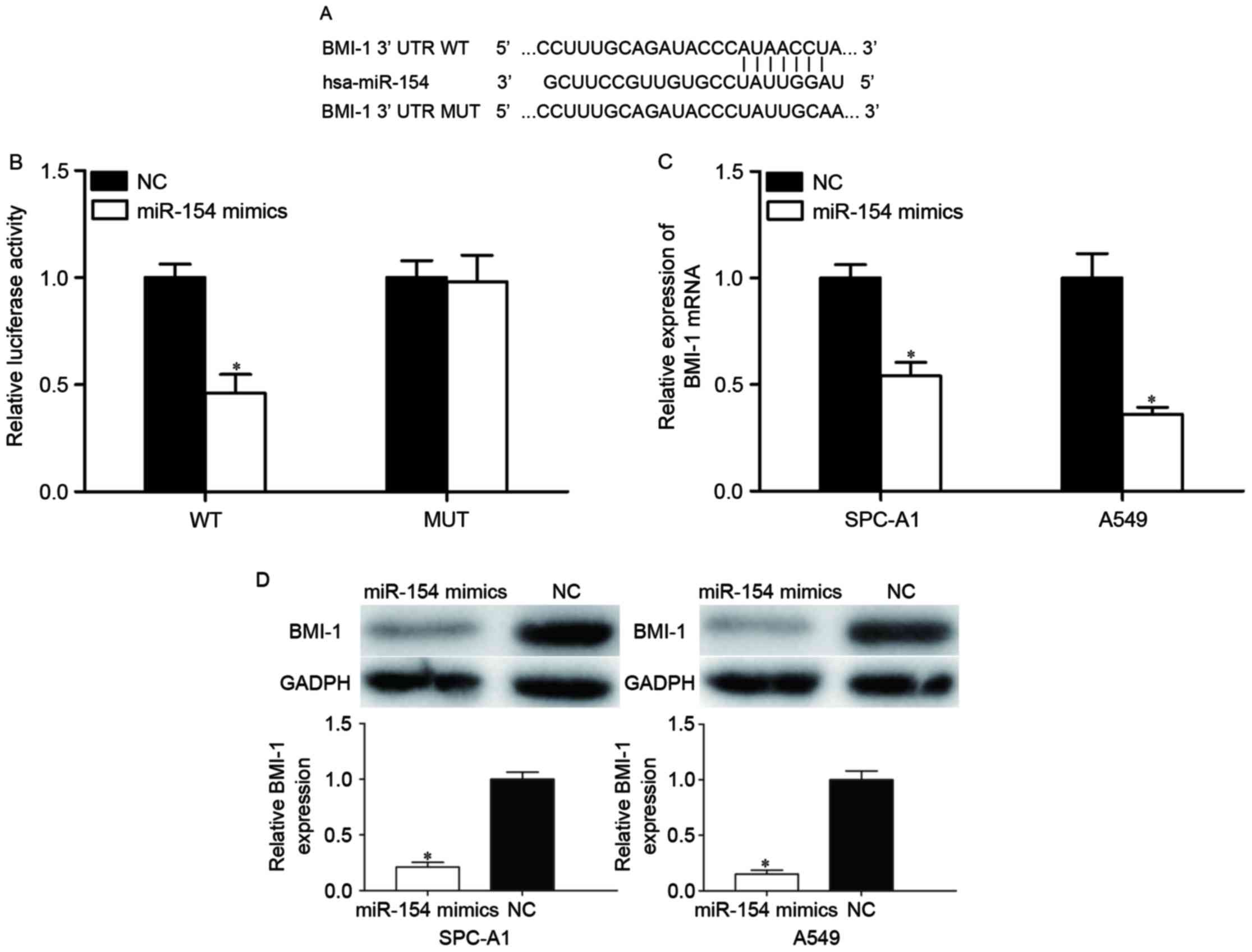
To further reveal the molecular mechanisms
underlying this tumor suppressor role of miR-154, TargetScan and
PicTar were used to predict the potential target genes of miR-154.
Bioinformatics analysis revealed that BMI-1 is a potential target
of miR-154. Subsequently, luciferase reporter assays were adopted
to check whether miR-154 directly targets the 3′UTR of BMI-1.
Putative target sites of miR-154 in 3′-UTR of BMI-1 are presented
in Fig. 3A. As shown in Fig. 3B, miR-154 decreased the luciferase
activity of the BMI-1-3′UTR (P<0.05), whereas BMI-1-3′UTR MUT
blocked this decrease (P>0.05). To further evaluate whether
BMI-1 was modulated by miR-154, miR-154 mimic or NC was transfected
into SPC-A1 and A549 cells, and BMI-1 expression levels were
measured by RT-qPCR and western blot analysis. The results revealed
that BMI-1 was significantly reduced at the mRNA (Fig. 3C, P<0.05) and protein (Fig. 3D, P<0.05) levels in
miR-154-transfected SPC-A1 and A549 cells compared with the NC
groups (P<0.05). These results suggested that miR-154 bound to
the 3′UTR of BMI-1 and thus regulated its expression.
BMI-1 overexpression reverses effects
of miR-154 upregulation in NSCLC cells
As BMI-1 was identified as a direct target of
miR-154, it was then investigated whether BMI-1 had functional
roles in regulating miR-154-induced NSCLC regulation. To do so,
pcDNA3.1-BMI-1 or pcDNA3.1-Ctl were transfected into SPC-A1 and
A549 cells. At 48 h after transfection, the expression levels of
BMI-1 were determined by RT-qPCR (Fig.
4A, P<0.05).
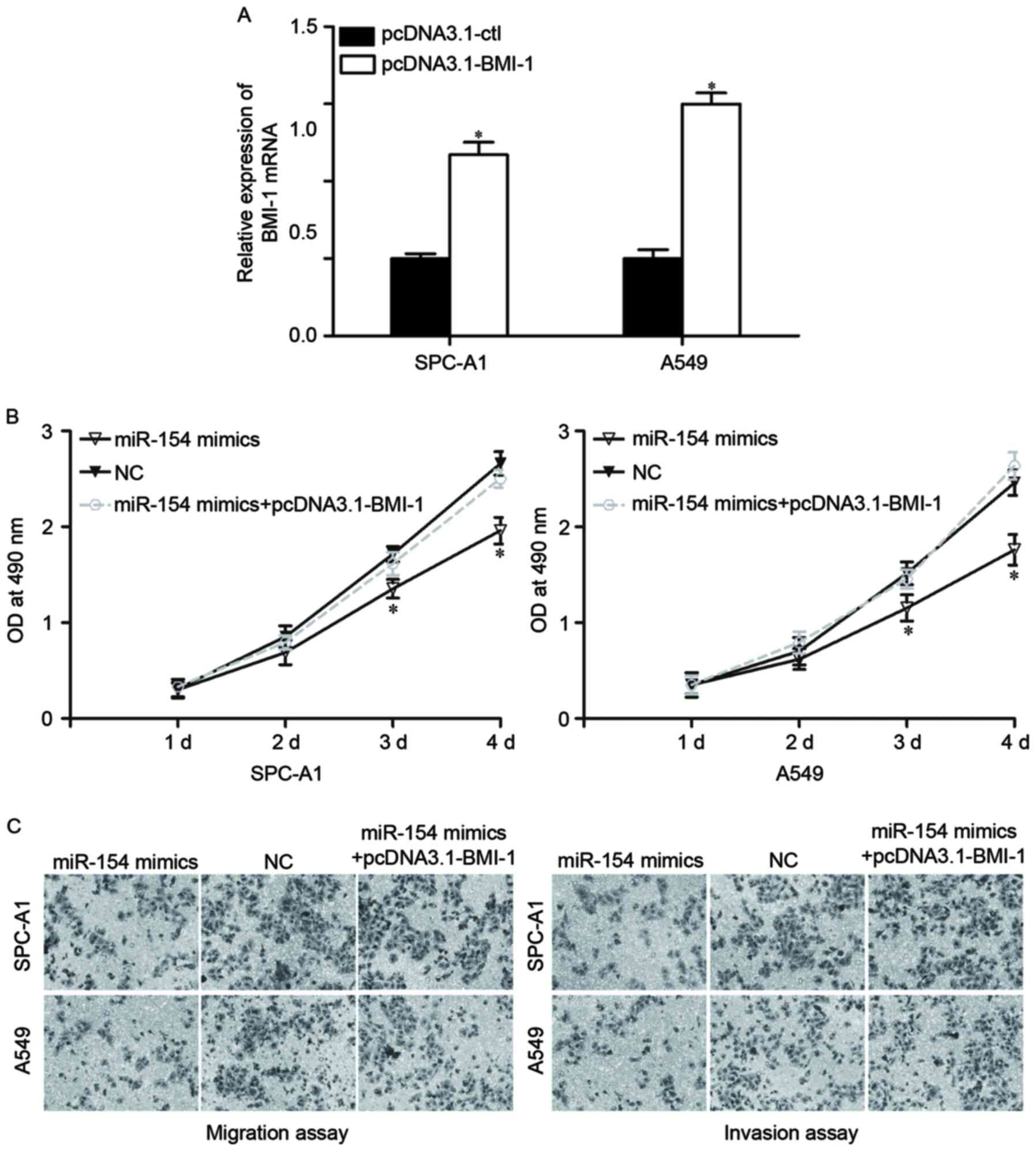 | Figure 4.Overexpression of BMI-1 reverses the
effects induced by miR-154 upregulation in NSCLC cells. (A) SPC-A1
and A549 cells were transfected with pcDNA3.1-BMI-1 or
pcDNA3.1-Ctl. Forty-eight h after transfection, BMI-1 expression at
the mRNA level was measured by RT-qPCR. *P<0.05, compared with
pcDNA3.1-Ctl. Overexpression of BMI-1 reversed the inhibitory
influence on the proliferation (B), migration and invasion (C)
induced by miR-154 overexpression in SPC-A1 and A549 cells.
Migrated and invaded cells were stained and fixed in 95% methanol,
stained with 0.1% crystal violet and imaged at magnification, ×200.
*P<0.05, compared with pcDNA3.1-Ctl. BMI-1, B-cell-specific
Moloney murine leukemia virus insertion site 1; miR, microRNA;
NSCLC, non-small cell lung cancer; RT-qPCR, reverse
transcription-quantitative polymerase chain reaction; OD, optical
density; NC, negative control. |
Next, various functional rescue experiments were
performed. As expected, BMI-1 overexpression mostly reversed the
inhibitory influence of miR-154 on proliferation (Fig. 4B, P<0.05), migration and invasion
(Fig. 4C, P<0.05) in NSCLC cells.
Thus, these data strongly demonstrate that miR-154 acted as a tumor
suppressor in NSCLC, at least in part through negative regulation
of BMI-1.
Discussion
Lung cancer remains the leading cause of
cancer-associated mortality in China and worldwide (15). With frequent local infiltration and
distant metastasis, lung cancer development is a complex process
that involves multiple genes, pathways and steps (16). A great deal of studies have indicated
miRNAs to be critical regulators in the carcinogenesis and
progression of human cancers (17–19).
Furthermore, acting as either tumor suppressors or oncogenes,
miRNAs have been demonstrated to be involved in a wide range of
physiological and pathological processes, including cell growth,
cell cycle, apoptosis, migration, invasion and metastasis (11–13).
Therefore, exploring the correlation between NSCLC and miRNAs may
be of benefit in the investigation of therapeutic strategies to
improve the cure and survival rates of this cancer.
The data from the present study revealed that
miR-154 was significantly downregulated in NSCLC tissues and cell
lines, which was consistent with previous findings that expression
levels of miR-154 were lower in several human cancer types and cell
lines, including colorectal cancer (20), NSCLC (21), osteosarcoma (22), hepatocellular carcinoma (23) and prostate cancer (24). This finding indicated that the low
expression levels of miR-154 may contribute to the carcinogenesis
and progression of NSCLC. In addition, functional analysis revealed
that restoration of miR-154 expression in NSCLC cells led to a
significant inhibition of cell proliferation, migration and
invasion. Next, BMI-1 was identified as a direct target gene of
miR-154 in NSCLC via bioinformatics analysis, luciferase reporter
assay, RT-qPCR and western blot analysis. In addition, BMI-1
overexpression reversed the inhibitory influence on NSCLC cells
induced by miR-154, indicating that miR-154 acted as a tumor
suppressor at least in part through the negative regulation of
BMI-1.
The prognostic value of miR-154 has been
investigated in several types of human cancer. For example, Kai
et al reported that, in colorectal cancer, decreased
expression levels of miR-154 were significantly associated with
large tumor size, positive lymph node metastasis and advanced
clinical stage (20). Univariate
analysis revealed that colorectal cancer patients with low miR-154
expression levels had a poorer overall survival rate. In addition,
multivariate analysis identified low miR-154 expression as an
independent predictor of poor survival (20). Pang et al demonstrated that
miR-154 expression was negatively correlated with tumor
differentiation, tumor-node-metastasis (TNM) stage and lymph node
metastasis in hepatocellular carcinoma (23). Lin et al revealed that low
miR-154 expression was significantly correlated with metastasis,
larger tumor size and advanced TNM stage in NSCLC (21). These findings implicated the potential
effects of miR-154 in the prognosis of cancer.
The downregulation of miR-154 in several cancer
types indicates that it may serve a significant role in the
carcinogenesis and progression of cancer. Indeed, miR-154 has been
demonstrated to be involved in several tumor suppressor functions.
Zhou et al observed that miR-154 suppressed osteosarcoma
cell proliferation, colony formation, migration and invasion, as
well as inducing cell cycle arrest at the G1 stage (22). Pang et al reported that
enforced miR-154 expression in hepatocellular carcinoma cells
decreased cell growth and metastasis, and enhanced apoptosis and
cell arrest at the G1 phase in vitro, as well as inhibiting
tumor growth in vivo (23).
Furthermore, miR-154 was noted to serve an essential role in
regulating the growth, colony formation, migration and invasion of
colorectal cancer cells (25). In
prostate cancer, ectopic miR-154 expression inhibited
proliferation, migration and invasion (24,26). These
findings indicated that miR-154 could be investigated as a
therapeutic target for these human cancer types.
With regard to miR-154, several targets have been
determined in previous studies, including Wnt5a in osteosarcoma
(22), Zinc finger E-box-binding
homeobox 2 in hepatocellular carcinoma (23), toll-like receptor 2 in colorectal
cancer (25), and high-mobility group
AT-hook 2 (24) and cyclin D2
(26) in prostate cancer. In the
present study, a novel direct target gene of miR-154, BMI-1, was
identified. Bioinformatics analysis revealed that BMI-1 was one of
the potential target genes of miR-154. Luciferase reporter assays
revealed that luciferase activity was suppressed by cotransfecting
miR-154 mimics and BMI-1-3′UTR WT. However, this inhibition could
be abrogated by cotransfecting miR-154 mimic and BMI-1-3′UTR MUT.
Restoration of miR-154 expression decreased the expression of BMI-1
at the mRNA and protein expression level in NSCLC cells. Finally,
BMI-1 overexpression reversed the inhibitory influence on NSCLC
cells induced by miR-154. These findings indicated that targeting
BMI-1 was involved in the tumor suppressor functions of miR-154 in
NSCLC.
In conclusion, the present study offers evidence
that miR-154 is downregulated in NSCLC and may act as a tumor
suppressor in NSCLC carcinogenesis and progression, partly by
negatively regulating BMI-1. Modulating miR-154 expression
represents a potential strategy for the treatment of NSCLC
patients.
References
|
1
|
Molina JR, Yang P, Cassivi SD, Schild SE
and Adjei AA: Non-small cell lung cancer: Epidemiology, risk
factors, treatment, and survivorship. Mayo Clin Proc. 83:584–594.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yang L, Parkin DM, Li L and Chen Y: Time
trends in cancer mortality in China: 1987–1999. Int J Cancer.
106:771–783. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Spira A and Ettinger DS: Multidisciplinary
management of lung cancer. N Engl J Med. 350:379–392. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Goldstraw P, Ball D, Jett JR, Le Chevalier
T, Lim E, Nicholson AG and Shepherd FA: Non-small-cell lung cancer.
Lancet. 378:1727–1740. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
de Cos Sánchez J, González Sojo MA,
Montero MV, Calvo Pérez MC, Vicente MJ and Valle MH: Non-small cell
lung cancer and silent brain metastasis. Survival and prognostic
factors. Lung Cancer. 63:140–145. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chen T, Xu C, Chen J, Ding C, Xu Z, Li C
and Zhao J: MicroRNA-203 inhibits cellular proliferation and
invasion by targeting Bmi1 in non-small cell lung cancer. Oncol
Lett. 9:2639–2646. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hou Y, Zhen J, Xu X, Zhen K, Zhu B, Pan R
and Zhao C: miR-215 functions as a tumor suppressor and directly
targets ZEB2 in human non-small cell lung cancer. Oncol Lett.
10:1985–1992. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li D, Wei Y, Wang D, Gao H and Liu K:
MicroRNA-26b suppresses the metastasis of non-small cell lung
cancer by targeting MIEN1 via NF-kappaB/MMP-9/VEGF pathways.
Biochem Biophys Res Commun. 472:465–470. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Engels BM and Hutvagner G: Principles and
effects of microRNA-mediated post-transcriptional gene regulation.
Oncogene. 25:6163–6169. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Aigner A: MicroRNAs (miRNAs) in cancer
invasion and metastasis: Therapeutic approaches based on
metastasis-related miRNAs. J Mol Med (Berl). 89:445–457. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rottiers V and Näär AM: MicroRNAs in
metabolism and metabolic disorders. Nat Rev Mol Cell Biol.
13:239–250. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cho WC: MicroRNAs: Potential biomarkers
for cancer diagnosis, prognosis and targets for therapy. Int J
Biochem Cell Biol. 42:1273–1281. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li J, Wang Y, Luo J, Fu Z, Ying J, Yu Y
and Yu W: miR-134 inhibits epithelial to mesenchymal transition by
targeting FOXM1 in non-small cell lung cancer cells. FEBS Lett.
586:3761–3765. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Celli BR: Chronic obstructive pulmonary
disease and lung cancer: Common pathogenesis, shared clinical
challenges. Proc Am Thorac Soc. 9:74–79. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jiang YW and Chen LA: microRNAs as tumor
inhibitors, oncogenes, biomarkers for drug efficacy and outcome
predictors in lung cancer (review). Mol Med Rep. 5:890–894. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et
al: A microRNA expression signature of human solid tumors defines
cancer gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mishra PJ and Merlino G: MicroRNA
reexpression as differentiation therapy in cancer. J Clin Invest.
119:2119–2123. 2009.PubMed/NCBI
|
|
20
|
Kai Y, Qiang C, Xinxin P, Miaomiao Z and
Kuailu L: Decreased miR-154 expression and its clinical
significance in human colorectal cancer. World J Surg Oncol.
13:1952015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lin X, Yang Z, Zhang P and Shao G: miR-154
suppresses non-small cell lung cancer growth in vitro and
in vivo. Oncol Rep. 33:3053–3060. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhou H, Zhang M, Yuan H, Zheng W, Meng C
and Zhao D: MicroRNA-154 functions as a tumor suppressor in
osteosarcoma by targeting Wnt5a. Oncol Rep. 35:1851–1858. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Pang X, Huang K, Zhang Q, Zhang Y and Niu
J: miR-154 targeting ZEB2 in hepatocellular carcinoma functions as
a potential tumor suppressor. Oncol Rep. 34:3272–3279. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhu C, Li J, Cheng G, Zhou H, Tao L, Cai
H, Li P, Cao Q, Ju X, Meng X, et al: miR-154 inhibits EMT by
targeting HMGA2 in prostate cancer cells. Mol Cell Biochem.
379:69–75. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xin C, Zhang H and Liu Z: miR-154
suppresses colorectal cancer cell growth and motility by targeting
TLR2. Mol Cell Biochem. 387:271–277. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhu C, Shao P, Bao M, Li P, Zhou H, Cai H,
Cao Q, Tao L, Meng X, Ju X, et al: miR-154 inhibits prostate cancer
cell proliferation by targeting CCND2. Urol Oncol. 32:31.e9–e16.
2014. View Article : Google Scholar
|