Introduction
Gastric cancer is one of the malignant tumor with
the highest mortality rates worldwide (1). Although the incidence and mortality
rates of gastric cancer have declined globally, it remains the
second most common cause of cancer-associated mortality (2). A total of >70% of cases occur in
developing countries and half of cases occur in Eastern Asia,
primarily in China (3,4). Invasion and metastasis are the primary
reasons that lead to gastric cancer-associated mortalities.
Investigating molecular mechanisms of invasion and metastasis is of
great value for the diagnosis, treatment and prevention of gastric
cancer. In the past several decades, a number of molecular
mechanisms have been investigated, but the role of microRNAs
(miRNAs/miRs) in gastric cancer remains to be elucidated.
miRNAs are non-coding RNA molecules containing ~20
nucleotides in length that are major post-transcriptional
regulators. miRNAs regulate numerous cellular processes, including
cellular proliferation, apoptosis, differentiation, invasion and
metastasis (5). miRNAs may undergo
aberrant regulation during carcinogenesis and serve as oncogenes or
tumor suppressors (6). Aberrant
expression of miRNAs is common in various human malignancies and
may modulate cancer-associated genomic regions or fragile sites
(7). miR-28 is an important miRNA,
including miR-28-5p and miR-28-3p. A large body of evidence
indicates that the abnormality of miR-28 expression is associated
with carcinogenesis of breast cancer (8), colorectal cancer (9), B-cell lymphoma (10), glioma and renal cell carcinoma
(11). However, studies concerning
the role of miR-28-5p in human gastric cancer remain to be
performed.
The present study profiled the expression of
miR-28-5p in 91 gastric cancer tissue and adjacent normal mucosal
tissue pairs by reverse transcription-quantitative polymerase chain
reaction (RT-qPCR). In addition, gastric cancer BGC823 and SGC7901
cells were infected with miR-28-5p mimics to evaluate the role of
mir-28-5p in gastric cancer. Low-expression of miR-28-5p in gastric
cancer tissues with poor prognosis was identified, and miR-28-5p
was demonstrated to serve a role in inhibiting gastric cancer
migration and invasion. These results indicate that miR-28-5p may
be a potential biomarker and therapeutic target against gastric
cancer.
Materials and methods
Clinical samples
Fresh samples of 91 pairs of gastric cancer and
adjacent normal mucosal tissues farthest from the tumor (>5 cm)
were obtained from patients who underwent surgical resection for
gastric cancer diagnosed based on the 7th edition of American Joint
Committee on Cancer Tumor Node Metastasis (TNM) staging system
(12) in the First Affiliated
Hospital of China Medical University (Shenyang, China) between
March 2007 and November 2008. The aforementioned samples were
frozen in liquid nitrogen and kept at −80°C instantly until they
were experimentally utilized. The present study was approved by the
Medical Ethics and Human Clinical Trial Committee at the First
Hospital of China Medical University and each patient provided
written informed consent for their inclusion in the present
study.
Cell culture
Human gastric cancer BGC823 and SGC7901 cell lines
obtained from the Department of Cell Biology, China Medical
University were incubated in a humidified incubator at 37°C with 5%
CO2, and cultured in Dulbecco's modified Eagle's medium
(DMEM; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA).
DMEM was supplemented with 10% fetal bovine serum (Gibco; Thermo
Fisher Scientific, Inc.), 100 U/ml penicillin and 100 U/ml
streptomycin according to the supplier's protocol.
Construction of stable cell lines
To construct a stable miR-28-5p-overexpressing cell
line, BGC823 and SGC7901 cells in the logarithmic growth phase in
6-well plates were infected with 50 µl (1×108 TU/ml)
commercial lentiviral packaged pre-miR-28 and the control
lentivirus (Shanghai GeneChem Co., Ltd., Shanghai, China). The
negative control short hairpin RNA sequence was
5′-TTCTCCGAACGTGUCACGT-3′. After 24 h, DMEM containing puromycin
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) was used to culture
and select stable cell lines for two weeks prior to subsequent
phenotypic and functional analyses.
RNA extraction and RT-qPCR
Total RNA was extracted from frozen tissue samples
with TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.). Next, miRNA was purified with the mirVana miRNA
Isolation kit (Ambion; Thermo Fisher Scientific, Inc.) according to
the manufacturer's manual. cDNA was synthesized using the
PrimeScript™ RT Reagent kit (Takara Biotechnology Co.,
Ltd., Dalian, China). qPCR was performed as previously described
(13) to detect the expression level
of miR-28-5p in the 91 pairs of human gastric and adjacent normal
mucosal tissue samples. The expression of miRNAs was calculated
relative to U6 small nuclear RNA. All the quantitation of PCR data
was presented as fold-change and calculated using the
2−ΔΔCq method (14).
Primers used were as follows: miR-28-5p reverse transcription
primer, 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCTCAATAG-3′;
miR-28-5p forward primer, 5′-GCGGAAGGAGCTCACAGTCT-3′; miR-28-5p
reverse primer, 5′-TGGTGTCGTGGAGTCG-3′; U6 forward primer,
5′-CTCGCTTCGGCAGCACA-3′; U6 reverse primer,
5′-AACGCTTCACGAATTTGCGT-3′.
MTT assay
The proliferative rates were assessed by MTT assay.
The stable infected cells and control cells were inoculated in
96-well plates at a density of 2×103 cells/well and
cultured for 1–6 days. At 24, 48, 72, 96, 120 and 144 h, 20 µl MTT
(10 mg/ml) was added to cells and they were incubated for 4 h at
37°C. Next, dimethyl sulfoxide was added to solubilize the formazan
product for 20 min at room temperature. The absorbance was
determined at 490 nm with a spectrophotometer. All experiments were
performed three times in triplicate.
Cell cycle analysis
BGC823 and SGC7901 cells (5×105) were
cultured for 48 h in 6-well plates, and then digested, washed with
cold PBS and fixed in 100% precooled methanol overnight at 4°C.
Cells were washed with cold PBS again, re-suspended in PBS solution
containing 100 µg/ml propidium iodide and 20 µg/ml RNase A (Omega
Bio-Tek, Inc., Norcross, GA, USA), and incubated at 37°C for 30
min. Next, the cells were measured using a flow cytometer and
analyzed with FACSCalibur™ (BD Biosciences, Franklin
Lakes, NJ, USA) and FlowJo software (version 7.6.1; Tree Star,
Inc., Ashland, OR, USA).
Adhesion assay
BGC823 and SGC7901 cells were seeded on 24-well
plates at equal numbers (~2×104), incubated for 20 min
and washed three times with PBS to remove the non-adherent cells.
Next, cells were fixed by 4% paraformaldehyde for 30 min, and
stained with 0.4% Trypan Blue for 20 min at room temperature. Cells
were then counted under a light microscope in 20 independent
symmetrical visual fields at a magnification of ×400.
Wound scratch assay
Equal numbers (~2×105) of stably infected
cells and control cells were incubated in 6-well plates equal
numbers for 24 h until they reached 90–100% confluence. The
capacity of movement on cells was detected by wound-healing assay.
Briefly, a scratch was made across the center of the cell monolayer
in each well using sterile 200-µl pipette tip and the medium was
replaced with fresh medium. The scratch distance between the two
linear regions was imaged and measured at different time points
using an inverted microscope.
Transwell assay
For the cell migration assay, ~1×105
cells were harvested, re-suspended in serum-free DMEM, and added to
the top chamber of 24-well Transwell chambers (Corning
Incorporated, Corning, NY, USA) with a pore size of 8.0 µm; the
lower chamber was filled with 500 µl DMEM with 10% fetal bovine
serum. Cells were fixed with 100% precooled methanol for 10 min and
stained with 0.4% Trypan Blue following incubation at 37°C for 24
h. Cells on the top chambers were removed by a cotton swab. The
number of cells that invaded to the bottom surface was counted
using a light microscope. The invasion assay was performed as
migration assay, with the exception that the cells were seeded onto
the filters of the Transwell chambers with coated Matrigel.
Protein extraction and western blot
analysis
Total protein was extracted from cells using
ice-cold radioimmunoprecipitation assay buffer supplemented with
phenylmethylsulfonyl fluoride and protease inhibitor cocktail
(Sigma-Aldrich; Merck KGaA). ABCA kit with Varioskan multimode
microplates spectrophotometer from Thermo Fisher Scientific Inc.
(Waltham, MA, USA) was used to detect the concentration of protein
in the supernatant. A total of 20 µg protein/well was fractionated
by 10% SDS-PAGE and transferred to a polyvinylidene fluoride
membrane. Membranes were blocked at room temperature for 2 h with
5% non-fat dry milk in Tris-buffered saline containing 0.1%
Tween-20 (TBST) and incubated with P-ERK (1:1,000; Cell Signaling
Technology; cat. no. 9101), P-Pak1 (1:1,000; Thermo Fisher
Scientific; cat. no. PA5-37677), P-Pak4 (1:1,000; Cell Signaling
Technology; cat. no. 3241) and P-AKT (1:1,000; Cell Signaling
Technology; cat. no. 4060) at 4°C overnight. The membranes were
washed in TBST and incubated with a horseradish peroxidase
(HRP)-conjugated secondary antibody (Goat anti-rabbit, 1:5,000;
ZSGQ-BIO; cat. no. ZF-0311) for 2 h at room temperature. Bound
antibody complexes were detected and visualized by ECL western
bolting detection kit (Ambion; Thermo Fisher Scientific, Inc.).
Independent verified data set from the
NCBI-Gene Expression Omnibus (GEO) database
The GSE39845 dataset was downloaded from the GEO
database (http://www.ncbi.nlm.nih.gov/geo/, accessed on December
10th 2015). The GSE39845 contained six colorectal cancer samples
(three normal vs. three cancer samples). The platform was based on
the GPL14613 (miRNA-2) Affymetrix Multispecies miRNA-2 Array. Then,
the preprocessing of Microarray Data was conducted by Affy package
in R software (R software, version 3.2.1; Affy package, version
1.56.0; http://www.r-project.org/, accessed on
December 5th 2015), and differentially expressed miRNA was
identified by the limma package in R software (limma package,
version 3.34.6). Only the differentially expressed miRNA with
P<0.05 and |log2 (fold-change)|>1.5 were screened out, as
calculated by Student's t-test. To additionally visualize
differentially expressed miRNA, the heatmap was constructed using
the Pheatmap package in R (Pheatmap package, version 1.0.8;
http://www.r-project.org/).
Statistical analysis
Continuous variables for the summary statistics of
the study are presented as mean ± standard deviation, or as median
value with interquartile ranges. The experimental data were
analyzed by SPSS 16.0 software (SPSS, Inc., Chicago, IL, USA).
Differences were considered statistically significant when
P<0.05. Mann-Whitney test (for two groups) or Kruskal-Wallis
test (for >2 groups) were used to analyze the association
between the expression of miR-28-5p and clinicopathological
features in patients with gastric cancer. Survival curves were
produced using the Kaplan-Meier method. The log-rank test was used
to analyze survival difference. Student's t-test was used to
analyze other parameters of phenotypes of gastric cancer cells.
Results
Clinical significance of miR-28-5p in
gastric cancer tissues
Previous reports have identified that the expression
of miR-28-5p was abnormal in various tumors (9,15–19). The present study used RT-qPCR to
detect the expression level of miR-28-5p in 91 gastric cancer
tissues and corresponding non-tumor adjacent tissues. As presented
in Fig. 1A, the expression level of
miR-28-5p was reduced significantly in gastric cancer tissues
compared with corresponding non-tumor adjacent tissues (P<0.01).
The association between the expression of miR-28-5p and the
clinicopathological factors of gastric cancer was further analyzed
(Table I). No statistical difference
between miR-28-5p expression, and age, gender, location, grosstype,
size, histological type and lymphatic invasion was identified. The
expression level of mir-28-5p in gastric cancer tissues from
patients with TNM stage IIIC and stage IV disease was significantly
lower compared with that in gastric cancer tissues from patients
with TNM stage I, II, IIIA, and IIIB disease (P=0.030). The
miR-28-5p level was significantly reduced in the T4 group compared
with that in T1, T2 and T3 groups (P=0.024). In addition, a
significantly lower expression level of miR-28-5p was observed in
gastric cancer tissues from patients with N3 lymph node
status compared with that in gastric cancer tissues from patients
with N0, N1 and N2 lymph node
status (P=0.035). Kaplan-Meier survival curve analysis illustrated
that the overall survival time was significantly longer in patients
with higher miR-28-5p expression, compared with patients with lower
miR-28-5p expression (P=0.041; Fig.
1B). The dividing standard was that if the median value was
>0.73-fold, the patients were in the higher miR-28-5p expression
group, whereas if the median value was ≤0.73-fold, the patients
were in the lower miR-28-5p expression group. Based on the Limma
package, the published expression dataset GSE39845 of the National
Center for Biotechnology Information Gene Expression Omnibus
database was analyzed and the differentially expressed miRNAs
between colorectal cancer and paired normal tissues were obtained.
The results of the analysis indicated that miR-28-5p expression was
downregulated in colorectal cancer tissues (P<0.001), which
demonstrated that the expression of miR-28-5p was markedly reduced
in colorectal tumors, compared with corresponding normal tissues
(Fig. 1C).
 | Table I.Association between the expression of
miR-28-5p with clinicopathological features in patients with
gastric cancer. |
Table I.
Association between the expression of
miR-28-5p with clinicopathological features in patients with
gastric cancer.
| Parameter | n | miR-28-5p
folda |
P-valueb |
|---|
| Age, years |
|
|
|
|
<65 | 59 | 0.77
(0.29–1.57) | 0.536 |
|
≥65 | 32 | 0.65
(0.30–1.11) |
|
| Gender |
|
|
|
|
Male | 69 | 0.82
(0.34–1.52) | 0.082 |
|
Female | 22 | 0.40
(0.28–0.79) |
|
| Location |
|
|
|
| Upper
third | 4 | 0.23
(0.11–0.38) | 0.131 |
| Middle
third | 12 | 0.62
(0.26–1.01) |
|
| Lower
third | 63 | 0.74
(0.31–1.54) |
|
|
Extensive | 12 | 1.00
(0.42–2.47) |
|
| Gross type |
|
|
|
|
Localized | 11 | 0.37
(0.10–1.54) | 0.247 |
|
Infiltrative | 80 | 0.76
(0.31–1.47) |
|
| Size (max.
diameter), cm |
|
|
|
|
<5 | 42 | 0.76
(0.41–1.65) | 0.324 |
| ≥5 | 49 | 0.67
(0.23–1.40) |
|
| Histological
type |
|
|
|
|
Intestinal | 48 | 0.64
(0.26–1.08) | 0.120 |
|
Diffuse | 43 | 0.84
(0.39–1.95) |
|
| Depth of invasion
(pT) |
|
|
|
|
T1, T2,
T3 | 59 | 0.81
(0.39–1.95) | 0.024 |
| T4 | 32 | 0.50
(0.22–0.94) |
|
| Lymph node status
(pN) |
|
|
|
|
N0, N1,
N2 | 62 | 0.79
(0.37–1.71) | 0.035 |
| N3 | 29 | 0.56
(0.19–0.99) |
|
| Lymph node
metastasis |
|
|
|
| No | 24 | 0.95
(0.43–2.06) | 0.222 |
|
Yes | 67 | 0.71
(0.27–1.12) |
|
| Lymphatic
invasion |
|
|
|
| No | 72 | 0.76
(0.31–1.47) | 0.667 |
|
Yes | 19 | 0.71
(0.20–1.88) |
|
| AJCCTNM stage |
|
|
|
| I, II,
IIIa, IIIb | 79 | 0.79
(0.33–1.57) | 0.030 |
| IIIc,
IV | 12 | 0.44
(0.18–0.80) |
|
miR-28-5p inhibits the migration and
invasion of gastric cancer cells
To investigate the functional significance of
miR-28-5p overexpression in gastric cancer, miR-28-5p-lentivirus
infection was used to establish stable BGC823 and SGC7901 cell
lines overexpressing miR-28-5p. Fig.
2A demonstrated that expression of miR-28-5p was significantly
increased in BGC823 and SGC7901 cell lines following infection with
miR-28-5p-lentivirus. To characterize the functionofmiR-28-5pin
gastric cancer progression, the effect of miR-28-5p expression on
the proliferation of gastric cancer cells was detected using an MTT
assay and flow cytometry analysis. No statistically significant
difference was identified in the proliferative ability of cells
overexpressing miR-28-5p compared with the control cells (Fig. 2B). Similarly, cell cycle analysis also
revealed that there were no significant differences between the
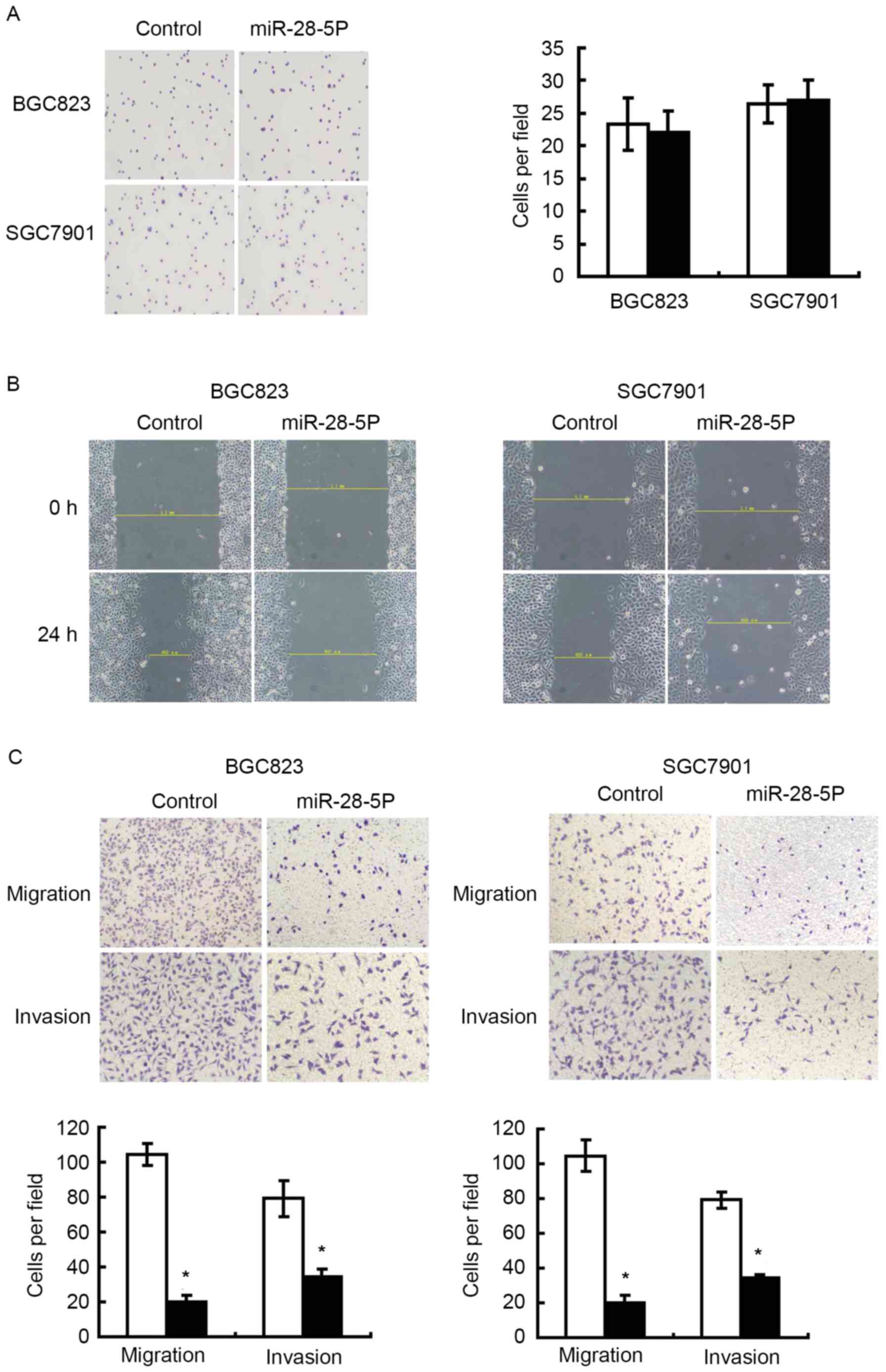
cells overexpressing miR-28-5p and control cells (Fig. 2C and D). A cell adhesion assay was
performed to assess whether miR-28-5p expression affected cell
adhesion capability of gastric cancer cells, and revealed that
upregulation of miR-28-5p exhibited no effect on cell adhesion
(Fig. 3A). Wound-scratch and cell
migration assays were performed to evaluate whether miR-28-5p was
able to affect the migration of gastric cancer cells. Fig. 3B and C revealed that there was a
significant inhibition in migration in cells infected with
miR-28-5p mimics compared with cells infected with negative
control. In Fig. 3C, an invasion
assay revealed miR-28-5p inhibited the invasion ability of gastric
cancer cells.
Overexpressing miR-28-5p inhibits the
phosphorylation of RAC serine/threonine-protein kinase (AKT) in
gastric cancer cells
Previous studies have validated thatP21-activated
kinase-1 (Pak1) (20), P21-activated
kinase-4 (Pak4) (21),
extracellular-signal regulated kinase (ERK) (22) and AKT (23) are involved in invasion and metastasis
of gastric cancer. Western blotting detected the phosphorylation
level of Pak1, Pak4, ERK and AKT protein in BGC823, and SGC7901
cells overexpressing miR-28-5p. Only the phosphorylation level of
AKT protein revealed a marked reduction in BGC823 and SGC7901 cells
overexpressing miR-28-5p, compared with cells that were infected
with negative control (Fig. 4). These
results revealed that overexpression miR-28-5p inhibited the
phosphorylation of AKT protein in gastric cancer cells.
Discussion
Gastric cancer is the second-leading cause of
cancer-associated mortality in the world (24). A large proportion of gastric cancer
patients, particularly in China, are diagnosed at advanced stage
with metastasis, and therefore are unable to undergo curative
resection (25,26). It has been documented that patients
with advanced stage gastric cancer have a poor prognosis, with a
reduced 5-year survival rate (27).
Therefore, metastasis is a key event in determining the prognosis
of gastric cancer. miRNAs possess essential roles in angiogenesis
and cancer metastasis (28). The
Snail-regulated miR-375 inhibited the migration and invasion of
gastric cancer cells partially by targeting Janus kinase 2 (JAK2)
(29). Liu et al (30) reported that miR-10b promoted the
invasion of gastric cells by activating RhoC-AKT signaling, through
targeting homeobox protein Hox-D10. Mir-28-5p has been reported to
be associated with development and progression of a number of
tumors, including hepatocellular carcinoma, colorectal cancer and
renal cell carcinoma (9,17,31,32).
However, the role of miR-28-5p in gastric cancer remains unknown.
The present study revealed the significant downregulation of
miR-28-5p in gastric cancer tissues. The overall survival time was
significantly longer in patients with higher miR-28-5p expression
compared with those with low expression. Detailed analysis of
miR-28-5p expression and the clinicopathological characteristics of
gastric cancer revealed significant association between the
expression level of miR-28-5p, and depth of invasion, lymph node
status and TNM stage. The data indicated that the expression of
miR-28-5pwas clinically relevant and maybe a potent biomarker for
the prognosis of gastric cancer.
The present study investigated the biological
function of miR-28-5p in gastric cancer in vitro. MTT assay
and flow cytometry analysis revealed that overexpression of
miR-28-5p in human gastric cell line BGC823 and SGC7901 did not
affect cellular proliferation, and cell cycle progression. To
further characterize the role of miR-28-5p in invasion and
metastasis of gastric cancer, cell adhesion, wound healing,
migration, and invasion assays were performed. Overexpression of
miR-28-5p inhibited the migratory and invasive ability of gastric
cancer cells, but not the adhesion ability (Fig. 3). These data suggested that miR-28-5p
might be a tumor suppressor gene, which inhibited the invasion and
metastasis of gastric cancer.
ERK, Pak1, Pak4 and AKT have been reported to be
involved in the invasion and metastasis of gastric cancer. ERK
signaling pathway can be activated by transforming growth factor-β
to regulate the invasion and metastasis of gastric cancer (33). Pak1 activates ERK and c-Jun N-terminal
kinase (JNK) to induce the metastasis of gastric cancer (34). Pak4 kinase mediates the
phosphorylation of stathmin 2 to promote the invasive potential of
gastric cancer cells (21). AKT has
three isoforms, AKT1, AKT2 and AKT3, and serves an essential role
in the regulation of diverse cellular functions, including cell
growth, proliferation, glucose metabolism, survival, genome
stability, transcription and neovascularization (35). AKT signaling regulates the
epithelial-mesenchymal transition, affecting the migration and
invasion of circulating gastric cancer cells (36). Western blot analysis was used to
profile the phosphorylation level of ERK, Pak1, Pak4 and AKT in
BGC823, and SGC7901 cells overexpressing miR-28-5p. The results
revealed that overexpression of miR-28-5p markedly inhibited the
phosphorylation level of AKT, but not ERK, Pak1 and Pak4, in
gastric cancer cells. Overexpression of miR-28-5p reduced the
migratory and invasive capacity of gastric cancer cells, and the
possible mechanism may involve the inhibition of the activation of
the AKT signaling pathway via miR-28-5p.
In summary, to the best of our knowledge, the
present study demonstrated that miR-28-5p expression was
significantly downregulated in gastric cancer and associated with
poor gastric cancer patient prognosis for the first time.
miR-28-5p, as a tumor suppressor, inhibited gastric cancer cell
invasion and metastasis by repressing the phosphorylation level of
AKT. miR-28-5p may be a potential biomarker for prognosis of
gastric cancer and a therapeutic target for the treatment of
advanced gastric cancer.
Acknowledgements
Not applicable.
Funding
This study was supported by grants from the National
Natural Science Foundation of China (grant no. 81001092) and
Natural Science Foundation of Liaoning (grant no. 2013021097). The
funding body had no role in the design the study and collection,
analysis, and interpretation of data and in writing the
manuscript.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
FX, ZC and FL conceived and designed the study. FX,
ZC and HH performed the experiments. PW and BG performed the
statistical and bioinformatics analyses. YX performed statistical
analyses. FX and ZC wrote the manuscript. YX and FL revised the
final manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Our study was approved by the Medical Ethics and
Human Clinical Trial Committee at the First Hospital of China
Medical University and the written informed consent of each patient
included in our study was also provided.
Consent for publication
All patients have provided the written informed
consent for the publication for the associated data.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
|
miRNA/miR
|
microRNA
|
|
Pak1
|
p21-activated kinase-1
|
|
Pak4
|
p21-activated kinase-4
|
|
ERK
|
extracellular-signal regulated
kinase
|
|
JAK
|
Janus kinase
|
|
JNK
|
c-Jun N-terminal kinase
|
References
|
1
|
Arnold M, Moore SP, Hassler S,
Ellison-Loschmann L, Forman D and Bray F: The burden of stomach
cancer in indigenous populations: A systematic review and global
assessment. Gut. 63:64–71. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yang L: Incidence and mortality of gastric
cancer in China. World J Gastroenterol. 12:17–20. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fock KM and Ang TL: Epidemiology of
Helicobacter pylori infection and gastric cancer in Asia. J
Gastroenterol Hepatol. 25:479–486. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhao X, Li X and Yuan H: microRNAs in
gastric cancer invasion and metastasis. Front Biosci (Landmark Ed).
18:803–810. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ventura A and Jacks T: MicroRNAs and
cancer: Short RNAs go a long way. Cell. 136:586–591. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wagner S, Ngezahayo A, Escobar Murua H and
Nolte I: Role of miRNA let-7 and its major targets in prostate
cancer. Biomed Res Int. 2014:3763262014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yang M, Yao Y, Eades G, Zhang Y and Zhou
Q: miR-28 regulates Nrf2 expression through a Keap1-independent
mechanism. Breast Cancer Res Treat. 129:983–991. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Almeida MI, Nicoloso MS, Zeng L, Ivan C,
Spizzo R, Gafà R, Xiao L, Zhang X, Vannini I, Fanini F, et al:
Strand-specific miR-28-5p and miR-28-3p have distinct effects in
colorectal cancer cells. Gastroenterology. 142(886–896):
e92012.
|
|
10
|
Schneider C, Setty M, Holmes AB, Maute RL,
Leslie CS, Mussolin L, Rosolen A, Dalla-Favera R and Basso K:
MicroRNA 28 controls cell proliferation and is down-regulated in
B-cell lymphomas. Proc Natl Acad Sci USA. 111:8185–8190. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Malzkorn B, Wolter M, Liesenberg F,
Grzendowski M, Stühler K, Meyer HE and Reifenberger G:
Identification and functional characterization of microRNAs
involved in the malignant progression of gliomas. Brain Pathol.
20:539–550. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Edge SB, Byrd DR, Compton CC, Fritz AG,
Greene FL and Trotti A: AJCC Cancer Staging Manual. 7th edition.
Springer-Verlag; New York: pp. 347–377. 2010
|
|
13
|
Cheng Z, Liu F, Wang G, Li Y, Zhang H and
Li F: miR-133 is a key negative regulator of CDC42-PAK pathway in
gastric cancer. Cell Signal. 26:2667–2673. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li Z, Gu X, Fang Y, Xiang J and Chen Z:
microRNA expression profiles in human colorectal cancers with brain
metastases. Oncol Lett. 3:346–350. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li LL, Qu LL, Fu HJ, Zheng XF, Tang CH, Li
XY, Chen J, Wang WX, Yang SX, Wang L, et al: Circulating microRNAs
as novel biomarkers of ALK-positive nonsmall cell lung cancer and
predictors of response to crizotinib therapy. Oncotarget.
8:45399–45414. 2017.PubMed/NCBI
|
|
17
|
Zhou SL, Hu ZQ, Zhou ZJ, Dai Z, Wang Z,
Cao Y, Fan J, Huang XW and Zhou J: miR-28-5p-IL-34-macrophage
feedback loop modulates hepatocellular carcinoma metastasis.
Hepatology. 63:1560–1575. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lim EL, Trinh DL, Scott DW, Chu A,
Krzywinski M, Zhao Y, Robertson AG, Mungall AJ, Schein J, Boyle M,
et al: Comprehensive miRNA sequence analysis reveals survival
differences in diffuse large B-cell lymphoma patients. Genome Biol.
16:182015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Rizzo M, Berti G, Russo F, Evangelista M,
Pellegrini M and Rainaldi G: The miRNA pull out assay as a method
to validate the miR-28-5p targets identified in other tumor
contexts in prostate cancer. Int J Genomics. 2017:52148062017.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang G, Zhang Q, Song Y, Wang X, Guo Q,
Zhang J, Li J, Han Y, Miao Z and Li F: PAK1 regulates
RUFY3-mediated gastric cancer cell migration and invasion. Cell
Death Dis. 6:e16822015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Guo Q, Su N, Zhang J, Li X, Miao Z, Wang
G, Cheng M, Xu H, Cao L and Li F: PAK4 kinase-mediated SCG10
phosphorylation involved in gastric cancer metastasis. Oncogene.
33:3277–3287. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fukui H, Zhang X, Sun C, Hara K, Kikuchi
S, Yamasaki T, Kondo T, Tomita T, Oshima T, Watari J, et al: IL-22
produced by cancer-associated fibroblasts promotes gastric cancer
cell invasion via STAT3 and ERK signaling. Br J Cancer.
111:763–771. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sasaki T and Kuniyasu H: Significance of
AKT in gastric cancer (Review). Int J Oncol. 45:2187–2192. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Danaei G, Vander Hoorn S, Lopez AD, Murray
CJ and Ezzati M: Comparative Risk Assessment collaborating group
(Cancers): Causes of cancer in the world: Comparative risk
assessment of nine behavioural and environmental risk factors.
Lancet. 366:1784–1793. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shen L, Shan YS, Hu HM, Price TJ, Sirohi
B, Yeh KH, Yang YH, Sano T, Yang HK, Zhang X, et al: Management of
gastric cancer in Asia: Resource-stratified guidelines. Lancet
Oncol. 14:e535–e547. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Takahashi T, Saikawa Y and Kitagawa Y:
Gastric cancer: Current status of diagnosis and treatment. Cancers
(Basel). 5:48–63. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hochwald SN, Kim S, Klimstra DS, Brennan
MF and Karpeh MS: Analysis of 154 actual five-year survivors of
gastric cancer. J Gastrointest Surg. 4:520–525. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhong J, Chen Y and Wang LJ: Emerging
molecular basis of hematogenous metastasis in gastric cancer. World
J Gastroenterol. 22:2434–2440. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xu Y, Jin J, Liu Y, Huang Z, Deng Y, You
T, Zhou T, Si J and Zhuo W: Snail-regulated miR-375 inhibits
migration and invasion of gastric cancer cells by targeting JAK2.
PLoS One. 9:e995162014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu Z, Zhu J, Cao H, Ren H and Fang X:
miR-10b promotes cell invasion through RhoC-AKT signaling pathway
by targeting HOXD10 in gastric cancer. Int J Oncol. 40:1553–1560.
2012.PubMed/NCBI
|
|
31
|
Shi X and Teng F: Down-regulated miR-28-5p
in human hepatocellular carcinoma correlated with tumor
proliferation and migration by targeting insulin-like growth
factor-1 (IGF-1). Mol Cell Biochem. 408:283–293. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang C, Hu J, Lu M, Gu H, Zhou X, Chen X,
Zen K, Zhang CY, Zhang T, Ge J, et al: A panel of five serum miRNAs
as a potential diagnostic tool for early-stage renal cell
carcinoma. Sci Rep. 5:76102015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Fu H, Hu Z, Wen J, Wang K and Liu Y:
TGF-beta promotes invasion and metastasis of gastric cancer cells
by increasing fascin1 expression via ERK and JNK signal pathways.
Acta Biochim Biophys Sin (Shanghai). 41:648–656. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li LH, Luo Q, Zheng MH, Pan C, Wu GY, Lu
YZ, Feng B, Chen XH and Liu BY: P21-activated protein kinase 1 is
overexpressed in gastric cancer and induces cancer metastasis.
Oncol Rep. 27:1435–1442. 2012.PubMed/NCBI
|
|
35
|
Bellacosa A, Kumar CC, Di Cristofano A and
Testa JR: Activation of AKT kinases in cancer: Implications for
therapeutic targeting. Adv Cancer Res. 94:29–86. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yuan D, Xia H, Zhang Y, Chen L, Leng W,
Chen T, Chen Q, Tang Q, Mo X, Liu M and Bi F: P-Akt/miR-200
signaling regulates epithelial-mesenchymal transition, migration
and invasion in circulating gastric tumor cells. Int J Oncol.
45:2430–2438. 2014. View Article : Google Scholar : PubMed/NCBI
|