Introduction
Colorectal carcinoma (CRC), which is prone to
metastasis and recurrence, is a cancer with a high lethality rate
worldwide. More than 50% of patients will develop metastasis and
recurrence (1,2). Multiple factors account for metastasis
and recurrence, including tumor stage, cancer subtypes and cancer
stem cells (CSCs). CSCs are a small population of tumor-initiating
cells that have the ability to self-renew and differentiate in
tumors in vivo. CSCs are involved in various processes
during tumor formation, progression, and angiogenesis and are
considered an important target for novel cancer treatment
strategies (3). Several specific
surface markers are expressed on CSCs, including CD133, CD44 and
aldehyde dehydrogenase 1 (ALDH1) (4).
CD133 (also known as prominin-1) is a 5-transmembrane glycoprotein
of 865 amino acids with a total molecular weight of 120 kDa. CD133
is a CSC marker in colon carcinoma. CD133-positive cells correlate
strongly with poor prognosis and synchronous liver metastasis
(5). Cancer cell populations with
high expression of CD133 are much more aggressive in terms of
metastases compared with those with low expression of CD133,
suggesting that CD133 may be a marker of increased tumorigenesis
ability. CD133-positive cells from clinical biopsy-derived cultures
have been demonstrated to possess multilineage differentiation
potential and are capable of tumor initiation in vivo.
CD133-positive CRC cells are more resistant to chemoradiotherapy,
and CD133 expression is associated with poor prognosis (6,7). However,
the regulation of CD133 expression has not been fully
elucidated.
The present study demonstrated that CD133 expression
was associated with the tumor protein p53 (p53) expression in an
HCT116 p53+/+ cell line. Of note, CD133-negative cells
were detected in the colon cancer cell line HCT116 in a previous
report by Kai et al (8).
Although the p53 status of the HCT116 cell line was not mentioned
in that previous study (8), it was
speculated that an association of CD133 expression with p53 may be
possible. Therefore, the present study focused on CD133 in HCT116
cell lines exhibiting different p53 expression status.
Wild-type p53 functions as a bona fide tumor
suppressor gene that is involved in transcription, DNA replication
and repair, cell cycle arrest, proliferation, apoptosis,
angiogenesis inhibition, and cellular stress responses. Various
types of cancer harbor p53 mutations. In specific, >50% of colon
cancers contain p53 mutations (9,10). p53 is
known to regulate the balance of asymmetric and symmetric divisions
of stem cells. A broken balance of asymmetric and symmetric cell
division leads to either tumor suppression or tumor expansion.
In the present study, CSC subpopulation
distributions, marked by CD133 and CD44 expression, were
significantly different in HCT116 p53+/+ and HCT116
p53−/− cell lines. Therefore, the hypothesis that there
may be a functional interaction between the CSC biomarker CD133 and
p53 was examined.
Materials and methods
Cell culture
The human colon cancer cell lines HCT116
p53+/+ and HCT116 p53−/− were
well-established cell lines (11),
kindly provided by Professor Xingzhi Xu (College of Life Sciences,
Capital Normal University, Beijing, China) and maintained in our
laboratory. All HCT116 cells were cultured in McCoy's 5A medium
(HyClone; GE Healthcare Life Sciences, Logan, UT, USA). The human
embryo kidney epithelial cell line 393T (cat. no. CL00022; Fengh
Bio, Changsha, China) was cultured in Dulbecco's Modified Essential
Medium (HyClone; GE Healthcare Life Sciences), supplemented with
10% fetal bovine serum (HyClone; GE Healthcare Life Sciences), 100
U/ml penicillin and 100 µg/ml streptomycin (both from Invitrogen;
Thermo Fisher Scientific, Inc., Waltham, MA, USA) in a humidified
atmosphere with 5% CO2 and incubated at 37°C.
Flow cytometric analysis
To harvest the cell samples, the cultured cells were
incubated with 0.25% trypsin for 90 sec, collected and washed with
cold PBS. The cell suspension was centrifuged at 300 × g for 10 min
at room temperature, and the supernatant was discarded. The cells
were resuspended in buffer containing PBS (pH 7.2), 0.5% bovine
serum albumin (BSA), and 2 mM EDTA. Next, 10 µl of the CD133/1
(AC133)-fluorescein isothiocyanate (FITC) antibody (clone, 393C3,
cat. no. 130-104-322) and 10 µl of the CD44-phycoerythrin (PE)
antibody (clone DB105, cat. no. 130-098-108) (both from Miltenyi
Biotec, GmbH, Bergisch, Germany) were added, mixed well and
incubated for 10 min in the dark at 4°C. Then, the cells were
washed with 1–2 ml buffer and centrifuged at 300 × g for 10 min at
room temperature. Finally, the cell pellets were resuspended in 1%
triformol for analysis by flow cytometry using the BD FACSDiva
software version 7.0 (BD Biosciences, Franklin Lakes, NJ, USA).
Western blot analysis
Cells were harvested at different times, from 0 to
24 h. After the medium was discarded, cells were washed with
ice-cold PBS twice and trypsinized on ice for 30 min. Cells lysates
were prepared with M-PER™ cell lysis buffer (cat. no.
78501; Thermo Fisher Scientific, Waltham, MA, USA) at 4°C for 30
min, and then centrifuged at 11,500 × g for 15 min at 4°C. The
protein concentration was measured by bicinchoninic acid protein
assay, and A280 absorption was measured using a NanoDrop 2000c/2000
(Thermo Fisher Scientific, Inc.). A total of 40 µg protein extract
was loaded onto a 10% SDS polyacrylamide gel. Following
electrophoresis, proteins were transferred to polyvinylidene
difluoride membranes (Bio-Rad Laboratories, Inc., Hercules, CA,
USA) and incubated with the indicated primary antibodies: Mouse
anti-p53 (cat. no. ab1101, Abcam; Cambridge, UK) or mouse
anti-GAPDH (cat. no. ab9485; Abcam) at a dilution of 1:1,000 at 4°C
overnight. Then, the blots were incubated with a secondary
horseradish peroxidase-conjugated goat anti-mouse antibody
(dilution 1:2,000; cat. no. sc-2005; Santa Cruz Biotechnology,
Inc., Dallas, TX, USA) at room temperature for 1 h. The protein
signal was visualized using an enhanced chemiluminescence detection
kit (Thermo Fisher Scientific, Inc.).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
For detecting the CD133 mRNA expression levels in
HCT116 p53+/+ cells and HCT116 p53−/− cells,
total RNA was isolated with the TRIzol regent and treated with
RNase-free DNase (both from Thermo Fisher Scientific, Inc.), and
reverse transcribed into cDNA using ReverTraAce®qPCR
RTMaster (Toyobo Life Science, Osaka, Japan). qPCR was performed
using a DyNAmo ColorFlash SYBR Green qPCR kit (Thermo Fisher
Scientific, Inc.), according to the manufacturer's instructions.
The primers used were as follows: CD133 forward,
5′-GATTCATACTGGTGGCTGGGTGG-3′ and reverse,
5′-GCAGGTGAAGAGTGCCGTAAGT-3′; and β-actin forward,
5′-TTGAAGGTAGTTTCGTGGAT-3′ and reverse, 5′-ATCACCATTGGCAATGAGCG-3′.
The thermocycling conditions for the qPCR were as follows: 95°C for
2 min, followed by 39 cycles of 95°C for 30 sec and 60°C for 30
sec, and finally 1 min at 68°C. Relative fold changes in mRNA
expression were calculated using the formula 2−ΔΔCq
(12).
CD133 promoter analysis
The CD133 promoter sequence (sequence ID, AY275524;
obtained from http://www.genecopoeia.com/; target gene accession:
NM_006017) was analyzed using a web-based prediction tool
(http://tfbind.hgc.jp/). p53 was identified as one
transcription factor that may interact with the CD133 promoter
sequence.
siRNA and plasmid transfection
p53 siRNA oligonucleotides were from Taihe
Biotechnology Co., Ltd. (Beijing, China) with a sense strand
sequence of: 5′ATGGATCCGTGACACGCTTCCCTGGATTG3′. The
control siRNA (cat. no. B01001) was purchased from GenePharma Co.,
Ltd. (Shanghai, China) and Flag-cmv2-p53 (GenePharma Co., Ltd.,
Shanghai, China) transfections, 1×106 HCT116
p53+/+ or HCT116 p53−/− cells, respectively,
were plated in 60 mm dishes for transfection the following day.
After 24 h, cells were transfected with 660 pmol p53 siRNA or 2 µg
Flag-cmv2-p53 plasmid or its negative control Flag-cmv2 plasmid,
using Lipofectamine 2000 (Thermo Fisher Scientific, Inc.),
following the manufacturer's instructions.
Dual-luciferase reporter assay
To measure the effect of p53 on the CD133 promoter
activity, the Promega dual-luciferase reporter assay system (cat.
no. E1910; Promega Corporation, Madison, WI, USA) was used,
according to the manufacturer's instructions. HCT116
p53+/+ cells were co-transfected with 0.8 µg pGL3-pCD133
reporter vector or pGL3-basic control and 8 ng pRL-CMV vector, and
simultaneously transfected with 660 pmol p53 siRNA or control
siRNA. HCT116 p53−/− cells were co-transfected with 0.8
µg pGL3-pCD133 reporter vector or pGL3-basic control and 8 ng
pRL-CMV vector, and simultaneously transfected with 1 µg
Flag-cmv2-p53 vector or control. Expression of p53 protein and
CD133 promoter activity were analyzed 24 h post-transfection. 293T
cells were transfected exactly as described for the HCT116 cells.
Transfections were performed using Lipofectamine 2000 (Thermo
Fisher Scientific, Inc.), according to manufacturer's instructions.
Cells were harvested 24 h post-transfection and plated in 96-well
plates. Then, firefly luciferase and Renilla luciferase
activity were measured.
Statistical analysis
The graphs were plotted and analyzed using t-test or
one-way analysis of variance in GraphPad Prism 6 (GraphPad
Software, Inc., La Jolla, CA, USA). P<0.05 was considered to
indicate a statistically significant difference. All experiments
were repeated at least three independent times. The data are
presented as the mean ± standard deviation (SD).
Results
Distribution of the CD133-positive
subpopulation in HCT116 p53+/+ and HCT116
p53−/− cell lines
After the p53 status was confirmed in both cell
lines (Fig. 1A), flow cytometry was
used to analyze CD133/CD44 expression in HCT116 cells. As
illustrated in Fig. 1B, the
CD133+/CD44+ subpopulation in the HCT116
p53+/+ cell line was 84.84±0.05% of total cells, while
in the HCT116 p53−/− it was 4.13±0.02% of total cells.
The difference in the CD133-positive CSC subpopulations between the
two cell lines was statistically significant (P<0.001; Fig. 1C). However, there was no significant
difference in the % of CD44-positive cell populations between these
two cell lines (Fig. 1C). The CD133
mRNA expression levels, as detected by RT-qPCR (Fig. 1D), and the protein expression levels,
as detected by western blot analysis (Fig. 1E), were also significantly reduced in
HCT116 p53−/− cells compared with HCT116
p53+/+ cells.
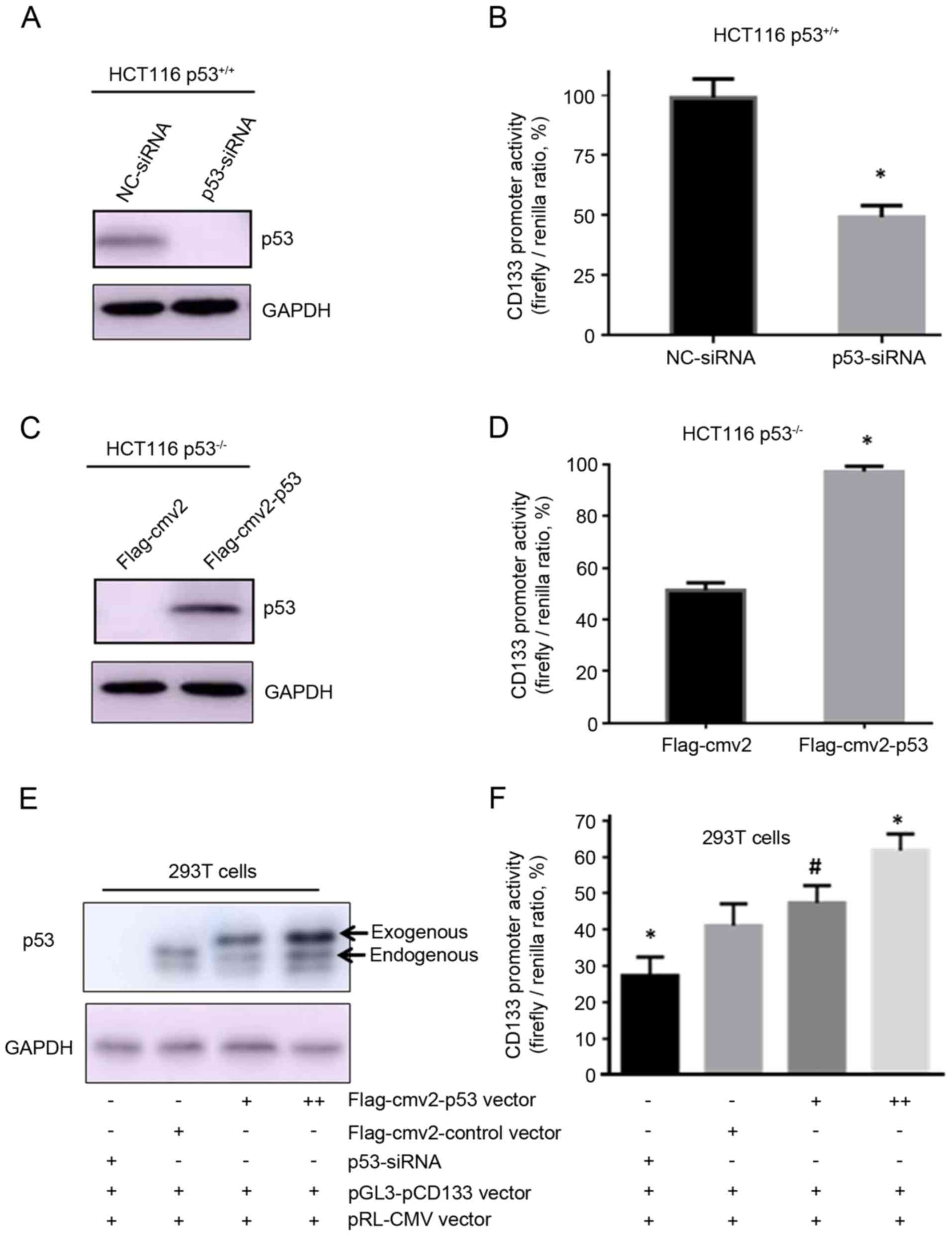
p53 activates CD133 expression
To examine whether p53 could regulate CD133
expression, p53-specific siRNA was used to abrogate p53 expression
in HCT116 p53+/+ cells. The western blot analysis and
qPCR results confirmed that expression of p53 protein (Fig. 2A) and mRNA (Fig. 2B) was significantly reduced in the
HCT116 p53+/+ cells following transfection with the
p53-specific siRNA compared with cells transfected with a negative
control siRNA. p21 protein expression was simultaneously decreased
following siRNA transfection (Fig.
2A). Knockdown of p53 by siRNA resulted in a significant
decrease in CD133-positive cells, from 84.27% in the
control-transfected cells to 5.94% in the p53-siRNA-transfected
HCT116 p53+/+ cells (Fig.
2C). In addition, the CD133 mRNA levels were also decreased
following p53 siRNA knockdown (Fig.
2D). These results suggested that p53 might transcriptionally
regulate CD133 mRNA expression.
Next, the Flag-cmv2-p53 plasmid was transiently
transfected into HCT116 p53−/− cells in order to
overexpress p53 in these cells, while the empty Flag-cmv2 vector
was used as control. Western blot analysis demonstrated increased
levels of p53 and p21 protein at 24 and 48 h following transfection
(Fig. 3A). One-half of the same
transfected sample was then used for flow cytometric detection of
the CSC surface biomarker CD133. The % of CD133-positive cells
increased from 4.19 to 5.13% at 24 h and to 7.25% at 48 h following
p53 overexpression (Fig. 3B).
Although the increase in overall % is relatively small, the change
is statistically significant (Fig.
3C). In addition, the qPCR results indicated that CD133 mRNA
levels were significantly increased in HCT116 p53−/−
cells following p53 overexpression (Fig.
3D).
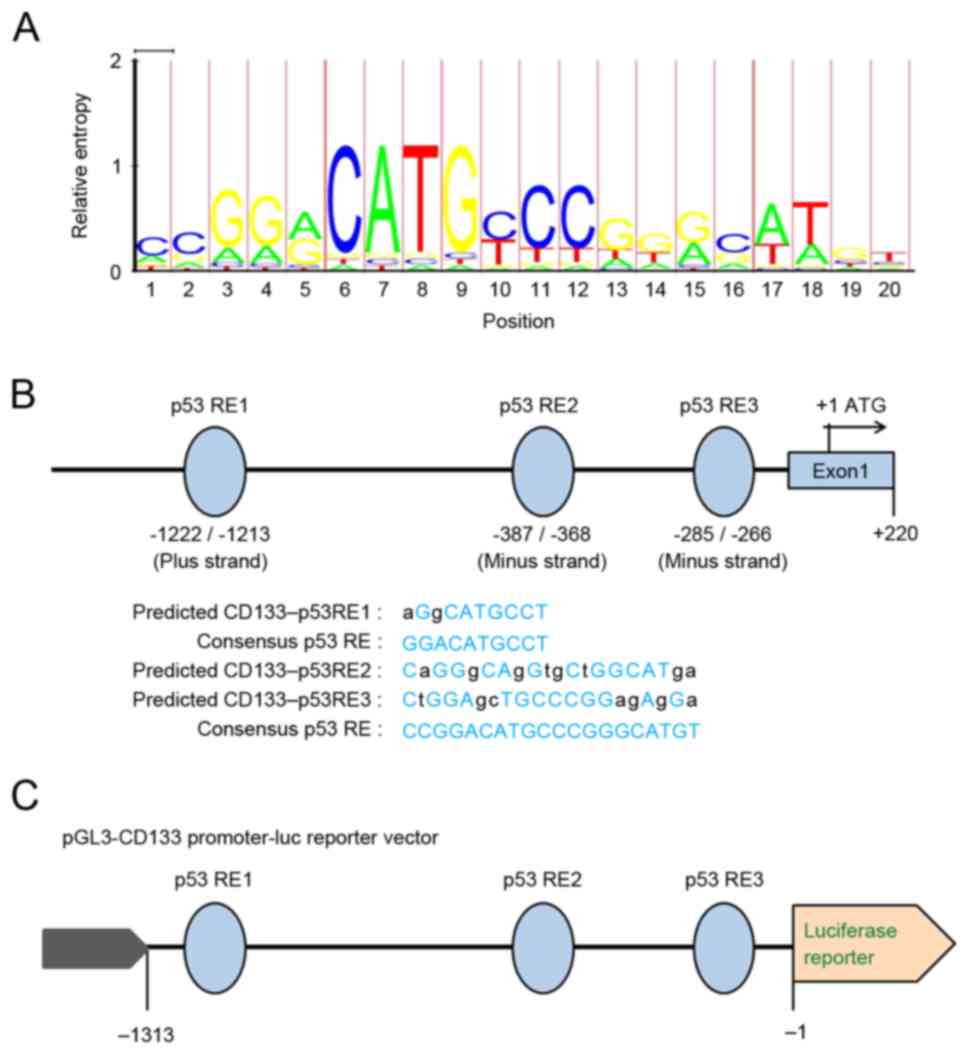
Prediction of p53 response elements in
the CD133 promoter
DNA binding is necessary for p53 transcription
factor activity. p53 modulates the transcription of target genes
mainly through its direct binding to a specific responsive element
(RE), usually within the promoter region of its target genes. The
consensus sequence of the p53 binding site is 5′-RRRCWWGYYY-3′,
where R is a purine, Y is a pyrimidine, and W is either adenine (A)
or (thymine) T (13). Fig. 4A displays the consensus p53 RE
sequence Model logo MA0106 generated using the LogoMat-M software
(http://www.sanger.ac.uk/science/tools/logomat-m)
(14). The Multi-genome Analysis of
Positions and Patterns of Elements of Regulation (MAPPER) Search
Engine (15) was used to identify the
putative p53 binding sites/REs in the promoter of the human
CD133/PROM1 gene. Using the MAPPER Search Engine, the promoter
sequence of the human CD133 gene from the National Center for
Biotechnology Information (NCBI) GenBank genome database (NCBI
reference sequence: NG_011696.1) was queried. Indeed, the sequence
alignment revealed three potential p53 REs at −1222/-1213,
−387/-368 and −285/-266 within the 1.5 kb region of the CD133 gene
promoter (Fig. 4B).
p53 transactivates the CD133
promoter
To investigate whether p53 transactivates the CD133
promoter, the sequence 1,313 bp upstream of the CD133 promoter,
containing these three potential p53 REs, was cloned into a
luciferase reporter plasmid (pGL3-pCD133), thereby producing a
plasmid where the firefly luciferase gene expression is driven by
the CD133 promoter (Fig. 4C). The
cells were co-transfected with the firefly luciferase reporter
pGL3-pCD133 plasmid, the pRL-CMV plasmid (which expresses
Renilla luciferase as the internal control), and other
plasmids as indicated. The empty pGL3-basic vector was transfected
as a control. As presented in Fig. 5A and
B, siRNA-mediated silencing of p53 resulted in a significant
decrease in firefly luciferase activity in p53+/+ HCT116
cells. By contrast, overexpression of 53 by co-transfection of the
p53-expressing vector pCMV2-p53 significantly increased the firefly
luciferase activity in p53−/− HCT116 cells (Fig. 5C and D). These results suggest that
p53 transactivates the CD133 promoter region containing the p53
REs. Similarly, in 293T cells, siRNA-mediated silencing of p53
significantly decreased luciferase activity, while overexpression
of exogenous p53 significantly increased luciferase activity driven
from the CD133 promoter, in a concentration-dependent manner
(Fig. 5E and F). Therefore, the
present results demonstrated that p53 may activate the CD133
promoter.
Discussion
The present study reported the role of p53 in
transcriptionally activating the expression of the cancer stem cell
marker CD133. A significant difference in the ratio of
CD133-positive cell populations was demonstrated between the HCT116
p53+/+ and p53−/− cell lines. Silencing or
overexpression of p53 decreased or restored CD133 expression,
respectively. Informatics analysis predicted that a 1,313 bp
fragment upstream of the CD133 transcriptional start site contained
putative p53 binding REs and a dual-luciferase reporter assay
demonstrated that p53 effectively activated transcription from the
CD133 promoter region.
CD133 has been reported by several studies as a
putative colon cancer stem cell marker (16,17). It
has been demonstrated that CD133-positive cells are more resistant
to radiation and conventional chemotherapy compared with
CD133-negative cells (18).
CD133-positive cells exhibit all the traits of stemness, including
the ability to self-renew, differentiate and form tumors in
immunodeficient mice (19). The
expression of CD133 in different colorectal cancer cell lines
varies. Chen et al (20) and
Yang et al (21) also found
that highly enriched CD133-positive cells existed in the HCT116
cell line and that these cells had characteristics of stem cells.
Their findings are in accordance with the present results, in which
a high population of CD133+/CD44+ cells were
present in the HCT116 p53+/+ cell line, which is poorly
differentiated (22). Multiple
studies have reported that different transcription factors are
involved in the regulation of CD133 expression, including Notch1,
signal transducer and activator of transcription 3, and hypoxia
inducible factor (23–30). DNA methylation and epigenetic factors
are also involved in CD133 expression regulation (31). However, little is known about how
CD133 is regulated in colon cancer.
Surprisingly, the present results demonstrated that
CD133 expression was almost abolished in the HCT116
p53−/− cells. p53 is one of the central regulators
involved in various biological activities, such as transcription,
DNA damage response, metabolism and stem cell maintenance (32). Although several studies have indicated
that p53 may be involved in the regulation of colon cancer stem
cells (33,34), the precise mechanisms are relatively
unclear. The present study demonstrated that wild-type p53
transactivated the promoter of the CD133 gene, indicating that the
CD133 gene may be a novel target of p53 in colorectal cancer.
Although it was previously reported that p53 transcriptionally
suppressed CD133 expression (35),
p53 may function with different transcription factors in the
context of colorectal cancer to maintain the stem cell properties
of HCT116 cells, which may be independent of the tumor suppressor
role of p53 in other types of cancer. The specificity and
robustness of CD133 as a colorectal CSC marker may also need
further investigation (36,37). The combinatorial use of other markers,
such as CD44 and ALDH1, in colorectal cancers may better clarify
CSC features and how p53 may affect stem cell characteristics of
colorectal cancer (38).
It is well-acknowledged that p53 transactivates its
target genes through binding to a consensus RE (39–44).
However, the mechanism by which p53 elicits cell-type specific
responses is not fully understood. A previous study has examined
the different binding modes of p53 between HCT116 and IMR90 cells
(45). It will be interesting to
identify other binding sites of p53 in addition to the 1,313 bp
fragment upstream of the CD133 promoter. In the present findings,
overexpression of p53 in HCT116 p53−/− cells increased
the % of CD133-positive cells only moderately, suggesting that
other epigenetic machinery may also be involved in the
transactivation of CD133 expression by p53 (31,46).
In summary, the present study identified CD133 as a
target gene of p53 in colorectal cancer cells. The clinical
significance of CD133 in other types of cancer cells is already
well established (47–52). Large-scale detection of CD133
expression in clinical colorectal cancer specimens may further
evaluate the prognostic value of CD133. The identification of
positive regulation of CD133 by p53 may provide new clues for the
study of CSC in specific colorectal cancer subtypes.
Acknowledgements
This study was supported by the National Key Basic
Research Program (973 Program) of MOST, China (grant no.
2015CB910601), and the National Natural Science Foundation of China
(grant no. U1432248).
References
|
1
|
Cunningham D, Atkin W, Lenz HJ, Lynch HT,
Minsky B, Nordlinger B and Starling N: Colorectal cancer. Lancet.
375:1030–1047. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kerr D: Clinical development of gene
therapy for colorectal cancer. Nat Rev Cancer. 3:615–622. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dalerba P, Dylla SJ, Park IK, Liu R, Wang
X, Cho RW, Hoey T, Gurney A, Huang EH, Simeone DM, et al:
Phenotypic characterization of human colorectal cancer stem cells.
Proc Natl Acad Sci USA. 104:10158–10163. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Puglisi MA, Tesori V, Lattanzi W,
Gasbarrini GB and Gasbarrini A: Colon cancer stem cells:
Controversies and perspectives. World J Gastroenterol.
19:2997–3006. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Horst D, Scheel SK, Liebmann S, Neumann J,
Maatz S, Kirchner T and Jung A: The cancer stem cell marker CD133
has high prognostic impact but unknown functional relevance for the
metastasis of human colon cancer. J Pathol. 219:427–434. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hongo K, Kazama S, Sunami E, Tsuno NH,
Takahashi K, Nagawa H and Kitayama J: Immunohistochemical detection
of CD133 is associated with tumor regression grade after
chemoradiotherapy in rectal cancer. Med oncol. 29:2849–2857. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ying X, Wu J, Meng X, Zuo Y, Xia Q, Chen
J, Feng Y, Liu R, Li L and Huang W: AC133 expression associated
with poor prognosis in stage II colorectal cancer. Med Oncol.
30:3562013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kai K, Nagano O, Sugihara E, Arima Y,
Sampetrean O, Ishimoto T, Nakanishi M, Ueno NT, Iwase H and Saya H:
Maintenance of HCT116 colon cancer cell line conforms to a
stochastic model but not a cancer stem cell model. Cancer Sci.
100:2275–2282. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li XL, Zhou J, Chen ZR and Chng WJ: P53
mutations in colorectal cancer-molecular pathogenesis and
pharmacological reactivation. World J Gastroenterol. 21:84–93.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Naccarati A, Polakova V, Pardini B,
Vodickova L, Hemminki K, Kumar R and Vodicka P: Mutations and
polymorphisms in TP53 gene-an overview on the role in colorectal
cancer. Mutagenesis. 27:211–218. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hall EH, Schoenbach KH and Beebe SJ:
Nanosecond pulsed electric fields induce apoptosis in p53-wildtype
and p53-null HCT116 colon carcinoma cells. Apoptosis. 12:1721–1731.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang B, Xiao Z and Ren EC: Redefining the
p53 response element. Proc Natl Acad Sci USA. 106:14373–14378.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Schuster-Bockler B, Schultz J and Rahmann
S: HMM Logos for visualization of protein families. BMC
Bioinformatics. 5:72004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Marinescu VD, Kohane IS and Riva A:
MAPPER: A search engine for the computational identification of
putative transcription factor binding sites in multiple genomes.
BMC Bioinformatics. 6:792005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ricci-Vitiani L, Lombardi DG, Pilozzi E,
Biffoni M, Todaro M, Peschle C and De Maria R: Identification and
expansion of human colon-cancer-initiating cells. Nature.
445:111–115. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
O'Brien CA, Pollett A, Gallinger S and
Dick JE: A human colon cancer cell capable of initiating tumour
growth in immunodeficient mice. Nature. 445:106–110. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bao S, Wu Q, McLendon RE, Hao Y, Shi Q,
Hjelmeland AB, Dewhirst MW, Bigner DD and Rich JN: Glioma stem
cells promote radioresistance by preferential activation of the DNA
damage response. Nature. 444:756–760. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wu Y and Wu PY: CD133 as a marker for
cancer stem cells: Progresses and concerns. Stem Cells Dev.
18:1127–1134. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen KL, Pan F, Jiang H, Chen JF, Pei L,
Xie FW and Liang HJ: Highly enriched CD133(+)CD44(+) stem-like
cells with CD133(+)CD44 (high) metastatic subset in HCT116 colon
cancer cells. Clin Exp Metastasis. 28:751–763. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yang ZL, Zheng Q, Yan J, Pan Y and Wang
ZG: Upregulated CD133 expression in tumorigenesis of colon cancer
cells. World J Gastroenterol. 17:932–937. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chantret I, Barbat A, Dussaulx E, Brattain
MG and Zweibaum A: Epithelial polarity, villin expression and
enterocytic differentiation of cultured human colon carcinoma
cells: A survey of twenty cell lines. Cancer Res. 48:1936–1942.
1988.PubMed/NCBI
|
|
23
|
Konishi H, Asano N, Imatani A, Kimura O,
Kondo Y, Jin X, Kanno T, Hatta W, Ara N, Asanuma K, et al: Notch1
directly induced CD133 expression in human diffuse type gastric
cancers. Oncotarget. 30:56598–56607. 2016.
|
|
24
|
Matsumoto K, Arao T, Tanaka K, Kaneda H,
Kudo K, Fujita Y, Tamura D, Aomatsu K, Tamura T, Yamada Y, et al:
mTOR signal and hypoxia-inducible factor-1 alpha regulate CD133
expression in cancer cells. Cancer Res. 69:7160–7164. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mathieu J, Zhang Z, Zhou W, Wang AJ,
Heddleston JM, Pinna CM, Hubaud A, Stadler B, Choi M, Bar M, et al:
HIF induces human embryonic stem cell markers in cancer cells.
Cancer Res. 71:4640–4652. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Won C, Kim BH, Yi EH, Choi KJ, Kim EK,
Jeong JM, Lee JH, Jang JJ, Yoon JH, Jeong WI, et al: Signal
transducer and activator of transcription 3-mediated CD133
up-regulation contributes to promotion of hepatocellular carcinoma.
Hepatology. 62:1160–1173. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Mak AB, Nixon AM and Moffat J: The mixed
lineage leukemia (MLL) fusion-associated gene AF4 promotes CD133
transcription. Cancer Res. 72:1929–1934. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Iida H, Suzuki M, Goitsuka R and Ueno H:
Hypoxia induces CD133 expression in human lung cancer cells by
up-regulation of OCT3/4 and SOX2. Int J Oncol. 40:71–79.
2012.PubMed/NCBI
|
|
29
|
Zhang L, Li H, Ge C, Li M, Zhao FY, Hou
HL, Zhu MX, Tian H, Zhang LX, Chen TY, et al: Inhibitory effects of
transcription factor Ikaros on the expression of liver cancer stem
cell marker CD133 in hepatocellular carcinoma. Oncotarget.
5:10621–10635. 2014.PubMed/NCBI
|
|
30
|
Ohnishi S, Maehara O, Nakagawa K, Kameya
A, Otaki K, Fujita H, Higashi R, Takagi K, Asaka M, Sakamoto N, et
al: hypoxia-inducible factors activate CD133 promoter through ETS
family transcription factors. PLoS One. 8:e662552013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Min KJ, So KA, Ouh YT, Hong JH and Lee JK:
The effects of DNA methylation and epigenetic factors on the
expression of CD133 in ovarian cancers. J Ovarian Res. 5:282012.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Vogelstein B, Lane D and Levine AJ:
Surfing the p53 network. Nature. 408:307–310. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Puca F, Colamaio M, Federico A, Gemei M,
Tosti N, Bastos AU, Del Vecchio L, Pece S, Battista S and Fusco A:
HMGA1 silencing restores normal stem cell characteristics in colon
cancer stem cells by increasing p53 levels. Oncotarget.
5:3234–3245. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zeilstra J, Joosten SP, Vermeulen L,
Koster J, Medema JP, Versteeg R, Spaargaren M and Pals ST: CD44
expression in intestinal epithelium and colorectal cancer is
independent of p53 status. PLoS One. 8:e728492013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Park EK, Lee JC, Park JW, Bang SY, Yi SA,
Kim BK, Park JH, Kwon SH, You JS, Nam SW, et al: Transcriptional
repression of cancer stem cell marker CD133 by tumor suppressor
p53. Cell Death Dis. 6:e19642015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Shmelkov SV, Butler JM, Hooper AT, Hormigo
A, Kushner J, Milde T, St Clair R, Baljevic M, White I, Jin DK, et
al: CD133 expression is not restricted to stem cells and both
CD133+ and CD133− metastatic colon cancer
cells initiate tumors. J Clin Invest. 118:2111–2120.
2008.PubMed/NCBI
|
|
37
|
Kawamoto H, Yuasa T, Kubota Y, Seita M,
Sasamoto H, Shahid JM, Hayashi T, Nakahara H, Hassan R, Iwamuro M,
et al: Characteristics of CD133(+) human colon cancer SW620 cells.
Cell Transplant. 19:857–864. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Rassouli FB, Matin MM and Saeinasab M:
Cancer stem cells in human digestive tract malignancies. Tumour
Biol. 37:7–21. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Smeenk L, van Heeringen SJ, Koeppel M, van
Driel MA, Bartels SJ, Akkers RC, Denissov S, Stunnenberg HG and
Lohrum M: Characterization of genome-wide p53-binding sites upon
stress response. Nucleic Acids Res. 36:3639–3654. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hoh J, Jin S, Parrado T, Edington J,
Levine AJ and Ott J: The p53MH algorithm and its application in
detecting p53-responsive genes. Proc Natl Acad Sci USA.
99:8467–8472. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Gowrisankar S and Jegga AG: Regression
based predictor for p53 transactivation. BMC Bioinformatics.
10:2152009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ma B, Pan Y, Zheng J, Levine AJ and
Nussinov R: Sequence analysis of p53 response-elements suggests
multiple binding modes of the p53 tetramer to DNA targets. Nucleic
Acids Res. 35:2986–3001. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Veprintsev DB and Fersht AR: Algorithm for
prediction of tumour suppressor p53 affinity for binding sites in
DNA. Nucleic Acids Res. 36:1589–1598. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Tebaldi T, Zaccara S, Alessandrini F,
Bisio A, Ciribilli Y and Inga A: Whole-genome cartography of p53
response elements ranked on transactivation potential. BMC
Genomics. 16:4642015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Botcheva K and McCorkle SR: Cell context
dependent p53 genome-wide binding patterns and enrichment at
repeats. PLoS One. 9:e1134922014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Williams K, Christensen J, Rappsilber J,
Nielsen AL, Johansen JV and Helin K: The histone lysine demethylase
JMJD3/KDM6B is recruited to p53 bound promoters and enhancer
elements in a p53 dependent manner. PLoS One. 9:e965452014.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhao Q, Zhou H, Liu Q, Cao Y, Wang G, Hu
A, Ruan L, Wang S, Bo Q, Chen W, et al: Prognostic value of the
expression of cancer stem cell-related markers CD133 and CD44 in
hepatocellular carcinoma: From patients to patient-derived tumor
xenograft models. Oncotarget. 7:47431–47443. 2016.PubMed/NCBI
|
|
48
|
Thanan R, Pairojkul C, Pinlaor S,
Khuntikeo N, Wongkham C, Sripa B, Ma N, Vaeteewoottacharn K,
Furukawa A, Kobayashi H, et al: Inflammation-related DNA damage and
expression of CD133 and Oct3/4 in cholangiocarcinoma patients with
poor prognosis. Free Radic Biol Med. 65:1464–1472. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Le H, Zeng F, Xu L, Liu X and Huang Y: The
role of CD133 expression in the carcinogenesis and prognosis of
patients with lung cancer. Mol Med Rep. 8:1511–1518. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhuang HW, Mo TT, Hou WJ, Xiong GX, Zhu
XL, Fu QL and Wen WP: Biological characteristics of CD133(+) cells
in nasopharyngeal carcinoma. Oncol Rep. 30:57–63. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Wen L, Chen XZ, Yang K, Chen ZX, Zhang B,
Chen JP, Zhou ZG, Mo XM and Hu JK: Prognostic value of cancer stem
cell marker CD133 expression in gastric cancer: A systematic
review. PLoS One. 8:e591542013. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Kim K, Ro JY, Kim S and Cho YM: Expression
of stem-cell markers OCT-4 and CD133: Important prognostic factors
in papillary renal cell carcinoma. Human Pathol. 43:2109–2116.
2012. View Article : Google Scholar
|