Introduction
Breast cancer is the most common cancer in women,
with the fifth highest mortality rate among all types of cancer and
the highest occurrence rate among female cancers globally (1). Furthermore, the mortality rate of breast
cancer in patients aged 25–45 years in Korea is the highest
worldwide (2). Despite the fact that
it is possible to treat hormone receptor-positive breast cancer
with a wide variety of effective regimens in the early stages of
disease and obtain relatively improved survival rates, a
significant number of patients may experience tumor recurrence and
metastasis (3).
The genomic characteristics of a metastatic tumor
are different from those of a primary tumor, owing to the time
interval between recurrence and metastasis, and the occurrence of
the primary tumor. Furthermore, this genomic difference is
intensified after treatment, including chemotherapy (4).
Trastuzumab, a targeted therapeutic agent, markedly
improves progression-free and overall survival rates in patients
with metastatic breast cancer, who have a poor prognosis, in the
short term. However, long-term observation over 30 months
demonstrated similar recurrence and mortality rates in patients
treated with general chemotherapy and targeted therapy (5). Previous reports provided several
hypotheses to explain the resistance to trastuzumab caused by
genomic changes in tumor cells during treatment (6), and other therapies for metastatic cancer
that are resistant to trastuzumab have been reported (7,8).
There is an increasing necessity to monitor the
genomic profiles of tumor cells during cancer onset, recurrence,
and metastasis. However, repeated tumor tissue biopsy is not always
practical. Circulating tumor cells (CTCs) that have shed from a
primary tumor are present in the blood circulation and may cause
tumor metastases (9,10). Liquid biopsy using CTCs is noninvasive
and repeatable; therefore, it is useful for counting tumor cells,
pathological characterization and molecular assays. Furthermore, it
is possible to use a liquid biopsy with CTCs to replace metastatic
tissue biopsy for the prediction of drug sensitivity and
resistance, monitoring of drug responsiveness, and detection of
metastasis (11,12).
Previously, our group developed a novel technology
to enrich and isolate CTCs on the basis of differences in cell size
(13). In the present study, CTCs
were isolated from patients with breast cancer using this method
and a cancer panel analysis of isolated CTCs was performed.
Furthermore, the genetic mutations of CTCs were compared with those
of white blood cells (WBCs) from the same patient in order to
evaluate cancer-specific mutations.
Materials and methods
Cell culture
H358-GFP, MCF7, PC9 and KG-1 cell lines (American
Type Culture Collection, Manassas, VA, USA), were maintained in
RPMI (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
containing 10% fetal bovine serum (Gibco; Thermo Fisher Scientific,
Inc.) and 1% Antibiotic-Antimycotic (Gibco; Thermo Fisher
Scientific, Inc.). Cells were cultured 37°C at 5% CO2 in
an incubator.
Clinical background of patients with
breast cancer
From February 2015 to March 2015, 8 female patients
with breast cancer, with a median age of 45 years (range, 28–48
years), and 4 female healthy volunteers with a median age of 34
years (range, 25–45 years) from the Asan Medical Center (Seoul,
Korea) were included in the present study. A total of 4 patients
(CG 237–240) received neoadjuvant systemic therapy and 4 patients
(CG 242–245) did not receive treatment. Cancer stage was evaluated
on the basis of the seventh American Joint Committee on Cancer
Tumor, Node, and Metastasis Classification (Table I) (14).
All blood samples and medical data used in the present study were
irreversibly anonymized. The present study was approved by the
Institutional Review Board of Asan Medical Center (IRB no.
2013-1048).
 | Table I.Clinical information of patients with
breast cancer, and Whole Genome Amplification results. |
Table I.
Clinical information of patients with
breast cancer, and Whole Genome Amplification results.
| Patient ID | Age, years | AJCC TNM stage | EpCAM+
cells, na | Cell type | DNA amount, µg | Purity of DNA
(A260/A280) | On target,
%b |
|---|
| CG 237 | 41 | IIA | 20 | CTC | 35.97 | 1.8 | 97.83 |
|
|
|
|
| WBC | 33.22 | 1.84 | 98.89 |
| CG 238 | 45 | IIIA | 1 | CTC | 30.69 | 1.88 | 98.93 |
|
|
|
|
| WBC | 32.78 | 1.89 | 98.91 |
| CG 239 | 48 | IIA | 7 | CTC | 31.35 | 1.89 | 99.17 |
|
|
|
|
| WBC | 33.55 | 1.91 | 98.54 |
| CG 240 | 47 | IIB | 23 | CTC | 30.58 | 1.84 | 98.81 |
|
|
|
|
| WBC | 34.1 | 1.87 | 99.13 |
| CG 242 | 48 | IIIA | 2 | CTC | 33.44 | 1.88 | 98.67 |
|
|
|
|
| WBC | 30.69 | 1.92 | 98.74 |
| CG 243 | 30 | IIA | 6 | CTC | 31.02 | 1.91 | 98.21 |
|
|
|
|
| WBC | 31.9 | 1.86 | 98.64 |
| CG 244 | 45 | IIA | 0 | CTC | 31.02 | 1.88 | 98.87 |
|
|
|
|
| WBC | 34.43 | 1.85 | 98.42 |
| CG 245 | 28 | IIA | 1 | CTC | 35.09 | 1.86 | 98.19 |
|
|
|
|
| WBC | 33.55 | 1.88 | 98.19 |
Blood collection and CTC
enrichment
Blood from each patient (10 ml) was collected in BD
Vacutainer acid citrate dextrose-solution A tubes and processed
within 4 h. The blood samples were divided into two groups as
follows: One for immunofluorescent staining, and the other for the
cancer panel analysis of CTCs. The samples were processed using the
same procedure, with a CTC isolation kit (cat. no. CIKW10; Cytogen,
Inc.) used according to the protocol. Briefly, blood samples were
incubated for 20 min with an antibody cocktail against WBCs and red
blood cells, and then mixed with preactivation buffer prior to
density gradient centrifugation (400 × g for 30 min at 25°C). A
cell suspension containing CTCs was collected and gradually diluted
with dilution buffer. The diluted cell suspensions were then
filtered through a high-density microporous (HDM) chip as
previously described (13). Cells on
the HDM chip were retrieved and transferred to a microtube. For
immunofluorescent staining, isolated cells were fixed in 4%
paraformaldehyde for 5 min at room temperature. For cancer panel
analysis, isolated cells were pelleted and kept at −80°C until
further processing. From the same patient, 500 µl blood was layered
onto a density gradient medium (Ficoll-Paque™ PLUS; GE
Healthcare Life Sciences, Little Chalfont, UK) and centrifuged (400
× g for 30 min at 25°C). From the peripheral blood mononuclear cell
layer, 100 WBCs were isolated as a negative control for cancer
panel analysis. In addition, different numbers (5, 10, 20 and 100)
of MCF7 cells were spiked into 1 ml blood from healthy volunteers,
isolated using the same CTC isolation procedure, and used as a
positive control for evaluation of the Cytogen protocol.
Immunofluorescent staining
Cells on slides were permeabilized with 0.2%
Triton-X 100 in PBS for 10 min, and quenched with 0.3% hydrogen
peroxide for 1 h. Cells were then blocked with 1% bovine serum
albumin (Hyclone; GE Healthcare Life Sciences, Logan, UT, USA) in
PBS for 30 min, and incubated with primary antibodies followed by
secondary antibodies for 1 h each at room temperature. The primary
antibodies were as follows: Mouse anti-epithelial cell adhesion
molecule (EpCAM; dilution 1:200; cat. no. #2929; Cell Signaling
Technology, Inc., Danvers, MA, USA) and rabbit anti-CD45 (dilution
1:10; cat. no. SC-25590; Santa Cruz Biotechnology, Inc., Dallas,
TX, USA). EpCAM signals were amplified using the Tyramide Signal
Amplification system (Thermo Fisher Scientific, Inc.) according to
the manufacturer's protocol. The secondary antibody for CD45 was
Alexa 594-conjugated goat anti-rabbit immunoglobulin G (H+L)
(dilution 1:200; cat. no. A11012; Invitrogen; Thermo Fisher
Scientific, Inc.). The slides were mounted with Fluoroshield with
DAPI (Immunobioscience Corporation, Mukilteo, WA, USA). Stained
cells were observed and photographed 3 fields using a fluorescence
microscope (Eclipse Ti; Nikon Corporation, Tokyo, Japan) at a
magnification of ×400.
Whole genome amplification
The cell pellets that were kept at −80°C were
amplified using the REPLI-g Single Cell kit (Qiagen GmbH, Hilden,
Germany) according to the manufacturer's protocol. Briefly, the
cell pellets were mixed with a denaturation buffer (included in the
kit) and incubated at 65°C for 10 min. Following the addition of a
stop solution (included in the kit), the denatured DNA samples were
mixed with REPLI-g sc DNA polymerase and a reaction buffer
(included in the kit) and incubated at 30°C for 8 h and then at
65°C for 3 min.
Ion AmpliSeq comprehensive cancer
panel (CCP) analysis
Genomic mutations were analyzed using the Ion
AmpliSeq CCP (Thermo Fisher Scientific, Inc.), which is a
next-generation sequencing assay that provides all-exon coverage of
409 oncogenes and tumor suppressor genes. The Ion AmpliSeq CCP was
designed to target all exons of key tumor suppressor genes and
oncogenes most frequently cited and most frequently mutated.
Briefly, genomic DNA was amplified using the Ion AmpliSeq Cancer
Panel and the amplicons were purified using Agencourt AM-Pure XP
(Beckman Coulter, Inc., Brea, CA, USA). This was followed by end
repairing and ligation with Ion Xpress barcode adapters (Thermo
Fisher Scientific, Inc.). The median fragment size and
concentration of the final library were detected using a
BioAnalyzer instrument with a high sensitivity chip (Agilent
Technologies, Inc., Santa Clara, CA, USA). The library was diluted
to 10 pM by low TE buffer included in the kit; and the library (5
µl) was used for emulsion PCR reactions using the Ion PI™ Hi-Q™ OT2
200 kit (Invitrogen; Thermo Fisher Scientific, Inc.). The following
thermocycling conditions were used: 80°C for 3 min, followed by 18
cycles of 99°C for 20 sec, 58°C for 30 sec, 72°C for 1 min, 99°C
for 20 sec, 56°C for 30 sec and 70°C for 1 min, and 10 cycles of
99°C for 20 sec and 58°C for extended durations from 3–20 min. The
emulsion PCR product was enriched using Dynabeads MyOne
Streptavidin C1 beads (Invitrogen; Thermo Fisher Scientific, Inc.).
The final enriched Ion spheres were mixed with a sequencing primer
(included in the kit) and polymerase (included in the kit) and
loaded onto a total of five chips of Ion 316™ Chip kit. Base
calling was generated by Torrent Suite 3.0 software (Thermo Fisher
Scientific, Inc.), using tmap-f3 on the Ion Torrent server for
further analysis. Bam and FASTQ alignment files were generated on
the basis of the base calling result and were used to report the
variant calling, including single nucleotide polymorphisms and
insertions/deletions.
Catalogue of somatic mutations in
cancer (COSMIC) database
COSMIC is an online database of somatically acquired
mutations in human cancer. It is the most comprehensive resource
for exploring the impact of somatic mutations in human cancer
(15).
Statistical analysis
Correlation analysis was performed by simple linear
regression analysis. The equation used was ‘Y=0.6179× + 5.6398’,
and was calculated using Microsoft Excel 2010 (Microsoft
Corporation, Redmond, WA, USA).
Results
Mutations are detectable in purified
MCF7 cells
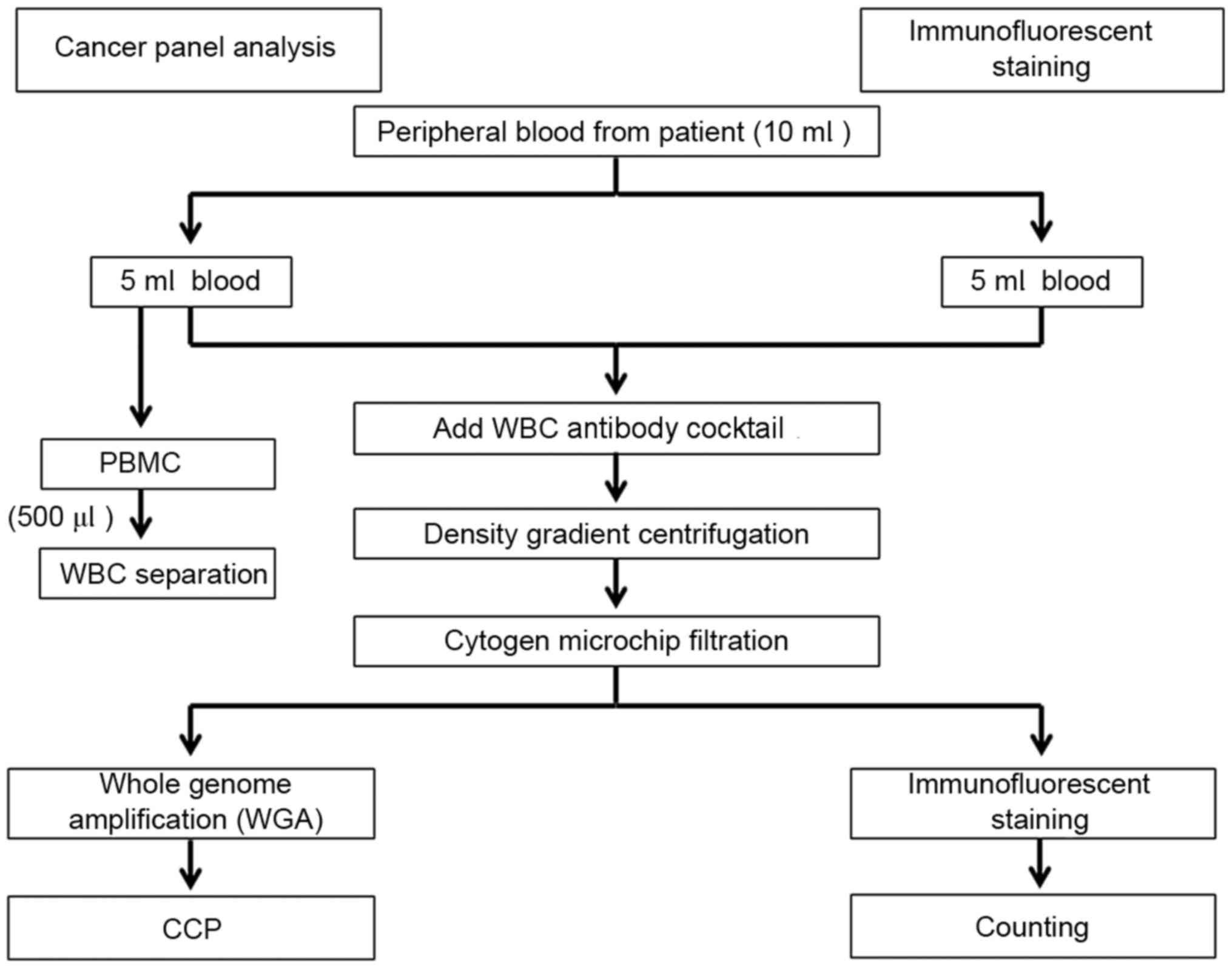
Fig. 1 depicts the
methodologies employed by the present study. Immunofluorescent
cells were counted to determine the number of EpCAM-positive cells.
The purity of MCF7 cells increased with the number of cells
(Fig. 2A). The recovery rate of our
method using H358-GFP cell lines was 84% (data not shown).
Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit α
(PIK3CA) mutation, which is a known mutation in MCF7, was detected
in MCF7 cells isolated through the Cytogen protocol. The
frequencies of mutation were increased when purity was high;
however even in samples with low purity, mutations were detected
(Fig. 2B). Therefore, R2
(between the purity and frequency of PIK3CA mut. in cancer panel
results) of Fig. 2B indicates that
the panel analysis was reliable.
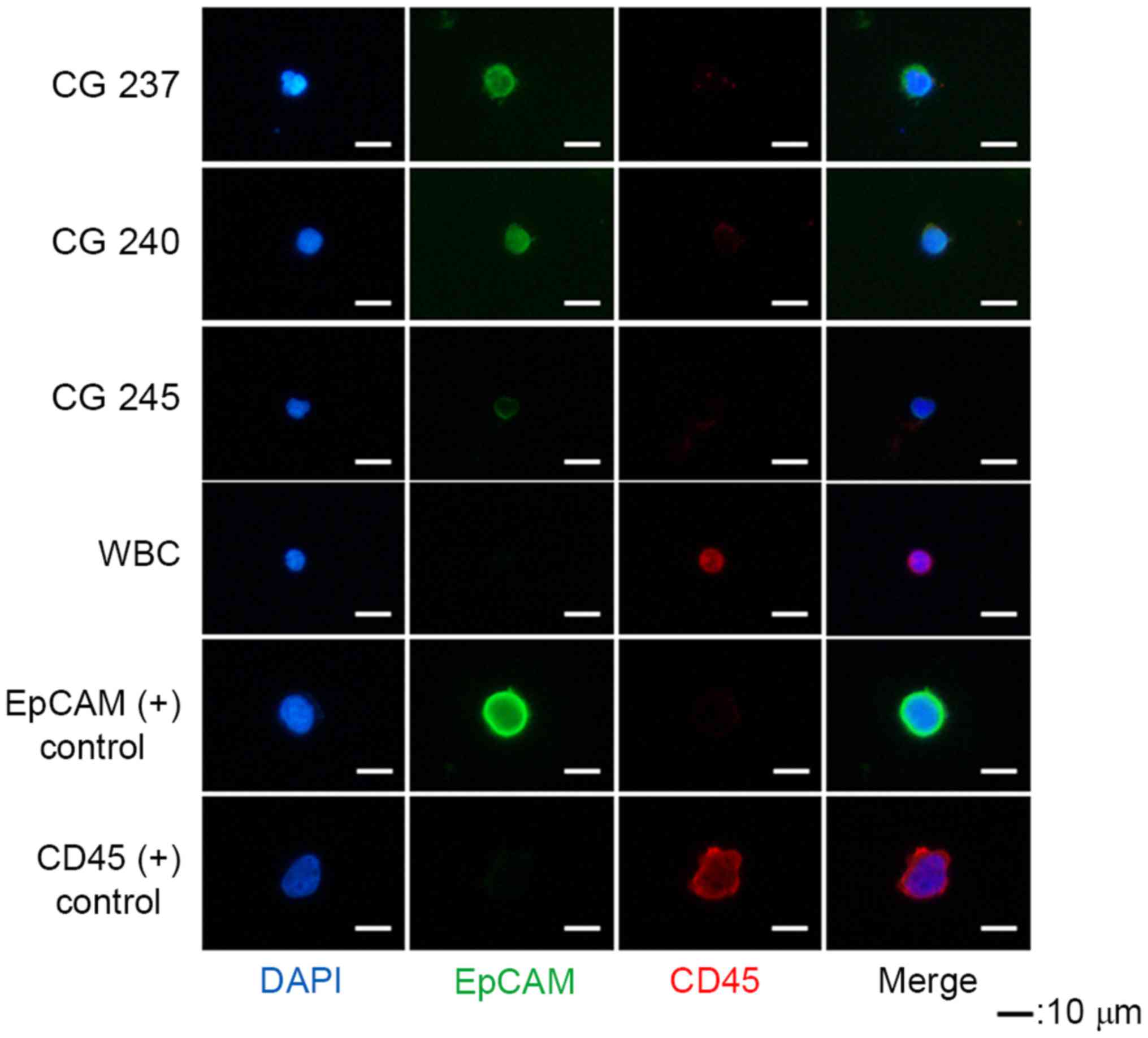
Successful isolation of CTCs
CTCs were defined as EpCAM+ and
CD45− cells (Fig. 3).
EpCAM+ cells were detected in 7/8 patients, and the
average number of EpCAM+ cells was 8.6 (1–23; Table I). PC9 (EpCAM+) and KG-1
(CD45+) cell lines were used as positive controls during
immunostaining. CTCs isolated for cancer panel analysis were
amplified using whole genome amplification, and the average DNA
amount was 32.7 µg with high purity
(A260/A280 above 1.80). On target, the
percentage of reads mapped to any targeted region relative to all
reads mapped to the reference was 98.6% (range 97.8–99.2%; Table I).
Cancer gene panel analysis
COSMIC was used to confirm the CCP results. When
mutations were in the WBCs analyzed as negative controls, they were
considered germ line mutations. These mutations in CTCs were
excluded from the analysis. CTC-specific mutations were validated
by comparing mutations between CTCs and WBCs (Table II). CTC-specific mutations had a
detection rate of 62.5%, and these were enhancer of zeste polycomb
repressive complex 2 subunit (EZH2), notch 1 (NOTCH1), AT-rich
interaction domain 1A, serine/threonine kinase 11, fms related
tyrosine kinase 3, MYCN proto-oncogene, bHLH transcription factor,
APC, WNT signaling pathway regulator, and phosphatase and tensin
homolog (PTEN).
 | Table II.Comprehensive cancer panel analysis
results. |
Table II.
Comprehensive cancer panel analysis
results.
|
| WBC | CTC | CTC specific |
|---|
|
|
|
|
|
|---|
| Patient ID | Gene | AA mutation | Gene | AA mutation | Gene | AA mutation |
|---|
| CG 237 | MSH2 | Unknown |
|
|
|
|
|
| ARID1A | p.D1850fs*4 |
|
|
|
|
|
| HNF1A | p.G292fs*25 |
|
|
|
|
|
|
|
| EZH2 | p.D730fs*1 | EZH2 | p.D730fs*1 |
|
|
|
| NOTCH1 | p.D1698D | NOTCH1 | p.D1698D |
| CG 238 | PDGFRA | p.V824V | PDGFRA | p.V824V |
|
|
|
| NOTCH1 | p.D1698D | NOTCH1 | p.D1698D |
|
|
|
| RET | p.T278N | RET | p.T278N |
|
|
|
| NF2 | p.N371N | NF2 | p.N371N |
|
|
|
| MSH2 | Unknown |
|
|
|
|
|
| PTCH1 | p.C727fs*11 |
|
|
|
|
| CG 239 | NOTCH1 | p.D1698D |
|
|
|
|
| CG 240 | MSH2 | Unknown | MSH2 | Unknown |
|
|
|
| PDGFRA | p.V824V | PDGFRA | p.V824V |
|
|
|
| FLT3 | p.L561L | FLT3 | p.L561L |
|
|
|
| STK11 | p.T32T | STK11 | p.T32T |
|
|
|
| SMARCB1 | p.P383fs*4 |
|
|
|
|
|
| SMARCB1 | p.P383fs |
|
|
|
|
|
|
|
| NOTCH1 | p.A2463fs*14 | NOTCH1 | p.A2463fs*14 |
|
|
|
| NOTCH1 | p.D1698D | NOTCH1 | p.D1698D |
| CG 242 | PDGFRA | p.V824V | PDGFRA | p.V824V |
|
|
|
| FLT3 | p.L561L | FLT3 | p.L561L |
|
|
|
| MSH2 | Unknown |
|
|
|
|
|
| EZH2 | p.D730fs*1 |
|
|
|
|
|
| NOTCH1 | p.D1698D |
|
|
|
|
|
|
|
| ARID1A | p.D1850fs*4 | ARID1A | p.D1850fs*4 |
| CG 243 | ARID1A | p.D1850fs*4 | ARID1A | p.D1850fs*4 |
|
|
|
| FLT3 | p.L561L | FLT3 | p.L561L |
|
|
|
| SMARCB1 | p.T372T | SMARCB1 | p.T372T |
|
|
|
|
|
| NOTCH1 | p.D1698D | NOTCH1 | p.D1698D |
|
|
|
| STK11 | p.L282fs*3 | STK11 | p.L282fs*3 |
| CG 244 | MSH2 | Unknown |
|
|
|
|
|
| PDGFRA | p.V824V |
|
|
|
|
|
| STK11 | p.L282fs*3 |
|
|
|
|
|
|
|
| ARID1A | p.K1072fs*21 | ARID1A | p.K1072fs*21 |
|
|
|
| NOTCH1 | p.D1698D | NOTCH1 | p.D1698D |
|
|
|
| FLT3 | p.L561L | FLT3 | p.L561L |
| CG 245 | MSH2 | Unknown |
|
|
|
|
|
| STK11 | p.L282fs*3 |
|
|
|
|
|
|
|
| MYCN | p.P358L |
|
|
|
|
|
| APC | p.R554* |
|
|
|
|
|
| NOTCH1 | p.D1698D |
|
|
|
|
|
| PTEN | p.L57fs*6 |
|
|
Discussion
Patients with early-stage hormone receptor-positive
breast cancer may have several effective treatment options;
however, multiple patients also develop recurrence and metastasis.
Therefore, the early diagnosis of cancer, prognostication and
monitoring of the genomic characteristics of tumor cells are
essential (3). However, biopsies of
tumor tissues are not always easy to repeat. CTCs may be able to
overcome this limitation of tumor tissue biopsies as CTCs have
similar characteristics to those of primary tumors, and may cause
metastasis (16). Due to the presence
of CTCs at low concentrations (1 in 1×109) (17), it is important to enrich or isolate
them from the blood effectively. CellSearch (Menarini Silicon
Biosystems, Bologna, Italy), a well-known commercial device,
isolates CTCs through EpCAM+ selection (18). It is not possible to achieve this
EpCAM+ selection technique when tumor cells downregulate
EpCAM expression (19). In addition,
a previous study reported that a significant portion of CTCs are
EpCAM− (20).
Our group has developed a novel CTC enrichment
technique based on cell size difference and double negative
selection, which removes nontargeted cells with an antibody complex
against WBCs (13). Using this
technique, CTCs were effectively isolated at a purity that was
sufficient for genomic analysis (Fig.
2). In addition, COSMIC mutations were detected even in
patients lacking EpCAM+ cells (Table I), demonstrating that this technology
was able to effectively isolate EpCAM− CTCs. Among the
CTC-specific COSMIC gene mutations that were identified, EZH2,
NOTCH1 and PTEN have been reported to affect breast cancer status
(21–24). EZH2 mutations cause abnormal DNA
methylation and promote mammary stem cell expansion and metastasis
(21). NOTCH1 has been reported to
regulate the epithelial-mesenchymal transition and to promote the
migration and invasion of breast cancer cells (22). Furthermore, NOTCH1 expression in
breast tumor tissues is higher than in normal tissues (23). Mutated PTEN is not able to inhibit the
phosphoinositide 3-kinase/protein kinase B/mechanistic target of
rapamycin pathway, thereby losing its tumor suppressor activity
(24).
There was a previous report on single-gene mutation
analysis of CTCs and WBCs (25), and
another study performed cancer panel analysis of CTCs without WBCs
as controls (26). However, to the
best of our knowledge, the present study is the first attempt at a
CCP using CTCs in conjunction with WBCs. CTC-specific COSMIC
mutations were identified, and genomic information that may be
useful for precision medicine was provided.
In conclusion, the CTC isolation technique used by
the present study was effective, providing sufficient purity for
genomic analysis, and demonstrated that CCP analysis is a potential
application for precision medicine.
Acknowledgements
Not applicable.
Funding
The present study was supported by a grant from the
National R&D Program, Ministry of Trade, Industry and Energy,
Republic of Korea (grant no., 10045947).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
SHC, MSK JL and BHJ directed and designed the study
with contributions from CHL and SJL. DHL, DYH and PSP analyzed the
circulating tumor cells from the patients. MSC and HKL maintained
the cell lines and performed the spike tests. SHA, BHS, JWL and JHY
provided patients' blood samples and clinical information. NJK, WCL
and KSY performed the CCP (comprehensive cancer panel) analyses.
CHL and MSK wrote the manuscript with contributions from JL and
BHJ. All authors have read and approved the manuscript.
Ethics approval and consent to
participate
The present study was approved by the Institutional
Review Board of Asan Medical Center (Institutional Review Board no.
2013-1048). All blood samples and medical data used in the present
study were irreversibly anonymized.
Consent for publication
Not applicable.
Competing interests
CHL, SJL, SHC, DHL, DYH, MSC, PSP, HKL, MSK, JL and
BHJ are employees of Cytogen, Inc. (Seoul, Korea), and the CTC
isolation kit was supplied courtesy of Cytogen, Inc. The authors
report no other competing interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jung KW, Won YJ, Kong HJ, Oh CM, Cho H,
Lee DH and Lee KH: Cancer statistics in Korea: Incidence,
mortality, survival, and prevalence in 2012. Cancer Res Treat.
47:127–141. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lorusso G and Rüegg C: New insights into
the mechanisms of organ-specific breast cancer metastasis. Semin
Cancer Boil. 22:226–233. 2012. View Article : Google Scholar
|
|
4
|
Suzuki M and Tarin D: Gene expression
profiling of human lymph node metastases and matched primary breast
carcinomas: Clinical implications. Mol Oncol. 1:172–180. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Von Minckwitz G, du Bois A, Schmidt M,
Maass N, Cufer T, de Jongh FE, Maartense E, Zielinski C, Kaufmann
M, Bauer W, et al: Trastuzumab beyond progression in human
epidermal growth factor receptor 2-positive advanced breast cancer:
A german breast group 26/breast international group 03–05 study. J
Clin Oncol. 27:1999–2006. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Peake BF and Nahta R: Resistance to
HER2-targeted therapies: A potential role for FOXM1. Breast Cancer
Manag. 3:423–431. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Blackwell KL, Burstein HJ, Storniolo AM,
Rugo H, Sledge G, Koehler M, Ellis C, Casey M, Vukelja S, Bischoff
J, et al: Randomized study of Lapatinib alone or in combination
with trastuzumab in women with ErbB2-positive,
trastuzumab-refractory metastatic breast cancer. J Clin Oncol.
28:1124–1130. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Modi S, Stopeck A, Linden H, Solit D,
Chandarlapaty S, Rosen N, D'Andrea G, Dickler M, Moynahan ME,
Sugarman S, et al: HSP90 inhibition is effective in breast cancer:
A phase II trial of tanespimycin (17-AAG) plus trastuzumab in
patients with HER2-positive metastatic breast cancer progressing on
trastuzumab. Clin Cancer Res. 17:5132–5139. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Franken B, de Groot MR, Mastboom WJ,
Vermes I, van der Palen J, Tibbe AG and Terstappen LW: Circulating
tumor cells, disease recurrence and survival in newly diagnosed
breast cancer. Breast Cancer Res. 14:R1332012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cohen SJ, Punt CJ, Iannotti N, Saidman BH,
Sabbath KD, Gabrail NY, Picus J, Morse M, Mitchell E, Miller MC, et
al: Relationship of circulating tumor cells to tumor response,
progression-free survival, and overall survival in patients with
metastatic colorectal cancer. J Clin Oncol. 26:3213–3221. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Van de Stolpe A, Pantel K, Sleijfer S,
Terstappen LW and den Toonder JM: Circulating tumor cell isolation
and diagnostics: Toward routine clinical use. Cancer Res.
71:5955–5960. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Giuliano M, Giordano A, Jackson S, De
Giorgi U, Mego M, Cohen EN, Gao H, Anfossi S, Handy BC, Ueno NT, et
al: Circulating tumor cells as early predictors of metastatic
spread in breast cancer patients with limited metastatic
dissemination. Breast Cancer Res. 16:4402014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kim EH, Lee JK, Kim BC, Rhim SH, Kim JW,
Kim KH, Jung SM, Park PS, Park HC, Lee J, et al: Enrichment of
cancer cells from whole blood using a microfabricated porous
filter. Anal Biochem. 440:114–116. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
American Joint Committee on Cancer: AJCC
Cancer Staging Manual. 7th edition. Springer; 2010
|
|
15
|
Forbes SA, Bindal N, Bamford S, Cole C,
Kok CY, Beare D, Jia M, Shepherd R, Leung K, Menzies A, et al:
COSMIC: Mining complete cancer genomes in the catalogue of somatic
mutations in cancer. Nucleic Acids Res. 39:D945–D950. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lang JM, Casavant BP and Beebe DJ:
Circulating tumor cells: Getting more from less. Sci Transl Med.
4:141ps132012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mostert B, Sleijfer S, Foekens JA and
Gratama JW: Circulating tumor cells (CTCs): Detection methods and
their clinical relevance in breast cancer. Cancer Treat Rev.
35:463–474. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Riethdorf S, Fritsche H, Müller V, Rau T,
Schindlbeck C, Rack B, Janni W, Coith C, Beck K, Jänicke F, et al:
Detection of circulating tumor cells in peripheral blood of
patients with metastatic breast cancer: A validation study of the
cellsearch system. Clin Cancer Res. 13:920–928. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Casavant BP, Mosher R, Warrick JW, Maccoux
LJ, Berry SM, Becker JT, Chen V, Lang JM, McNeel DG and Beebe DJ: A
negative selection methodology using a microfluidic platform for
the isolation and enumeration of circulating tumor cells. Methods.
64:137–143. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Giordano A, Gao H, Anfossi S, Cohen E,
Mego M, Lee BN, Tin S, De Laurentiis M, Parker CA, Alvarez RH, et
al: Epithelial-mesenchymal transition and stem cell markers in
patients with HER2-positive metastatic breast cancer. Mol Cancer
Ther. 11:2526–2534. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wu J and Crowe DL: The histone
methyltransferase EZH2 promotes mammary stem and luminal progenitor
cell expansion, metastasis and inhibits estrogen receptor-positive
cellular differentiation in a model of basal breast cancer. Oncol
Rep. 34:455–460. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shao S and Zhao X, Zhang X, Luo M, Zuo X,
Huang S, Wang Y, Gu S and Zhao X: Notch1 signaling regulates the
epithelial-mesenchymal transition and invasion of breast cancer in
a Slug-dependent manner. Mol Cancer. 14:282015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yuan X, Zhang M, Wu H, Xu H, Han N, Chu Q,
Yu S, Chen Y and Wu K: Expression of Notch1 correlates with breast
cancer progression and prognosis. PLoS One. 10:e01316892015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen Z, Trotman LC, Shaffer D, Lin HK,
Dotan ZA, Niki M, Koutcher JA, Scher HI, Ludwig T, Gerald W, et al:
Crucial role of p53-dependent cellular senescence in suppression of
PTEN-deficient tumorigenesis. Nature. 436:725–730. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fernandez SV, Bingham C, Fittipaldi P,
Austin L, Palazzo J, Palmer G, Alpaugh K and Cristofanilli M: TP53
mutations detected in circulating tumor cells present in the blood
of metastatic triple negative breast cancer patients. Breast Cancer
Res. 16:4452014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu S, Wang H, Zhang L, Tang C, Jones L,
Ye H, Ban L, Wang A, Liu Z, Lou F, et al: Rapid detection of
genetic mutations in individual breast cancer patients by
next-generation DNA sequencing. Hum Genomics. 9:22015. View Article : Google Scholar : PubMed/NCBI
|