Introduction
Gliomas arise from astrocytes, oligodendrocytes or
their progenitor cells. The malignant transformation of all these
cells are collectively called gliomas. Glioblastoma multiforme
(GBM) is the most common and invasive primary central nervous
system tumor in adults, with a 2-year survival rate of 3–5%.
Currently, the standard treatment for GBM is multimodal
comprehensive treatment including surgery combined with
radiotherapy and chemotherapy. Surgically eliminating GBM is
impossible, and GBMs are resistant to a variety of chemotherapy
drugs including Temozolomide. Thus, tumor relapses are unavoidable,
making the medial survival of GBM patients only 12 to 15 months
(1).
The chloride transporter family is widely expressed
in tissues and organs across the entire body (2). One member of the chloride voltage-gated
channel (CLCN) family, CLCN3, is predominantly expressed in acidic
intracellular compartments, especially lysosomes (3). CLCN3 participates in vesicular
acidification, chloride accumulation, and drug resistance (4–7). Recent
studies have shown that CLCN3 is highly expressed in GBM and plays
significant roles in cellular survival, proliferation and
malignancy (8,9).
Cisplatin (cis-diamminedichloroplatinum) is a
DNA damaging agent that is widely used to treat a variety of
malignant tumors, including gliomas (10,11).
Fluorescence tagging assays indicate that cisplatin is taken up by
cells, and then combines with membrane proteins, accumulating in
endosomes, lysosomes and the Golgi through endocytic recycling
compartments (12,13). Recent studies also indicated that cell
internalization, membrane recycling, organelle acidification, and
cell externalization are involved in the metabolism of cisplatin
(13–15). However, the mechanism of resistance to
cisplatin is complex and still not fully understood.
In this study, we utilized the human glioma cell
line U251, which is relatively insensitive to cisplatin, to
determine whether suppressing CLCN3 enhances the sensitivity of
glioma cells to cisplatin. We investigated the effects of combined
cisplatin and CLCN3 antisense oligonucleotide treatment on U251
cells and compared the results with cells treated with cisplatin
alone. Cytotoxicity, apoptosis induction, cell invasion and the
expression of relative genes were assayed. We further elucidated
the possible mechanisms involved in the different susceptibilities
of U251 cells to cisplatin-induced apoptosis. The results indicated
that inhibiting CLCN3 increased the sensitivity of U251 cells to
cisplatin through lysosomal dysfunction. These data provide new
evidence that intracellular CLCN proteins could be a target to
modulate chemosensitivity.
Materials and methods
Cell line and culture
All experiments were performed in U251 cells, which
were maintained at 37°C with 95% air and 5% CO2, and in
DMEM (HyClone; GE Healthcare Life Sciences, Logan, UT, USA)
supplemented with 10% fetal bovine serum (FBS; HyClone; GE
Healthcare Life Sciences), 50 U/ml penicillin and 50 µg/ml
streptomycin (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany).
Antisense and nonsense oligonucleotide
transfection
Phosphorothioate-modified antisense oligonucleotide
primers were purchased from Takara Biotechnology Co., Ltd. (Dalian,
China). The CLCN3 antisense oligonucleotide primer sequence used
was: 5′-TCCATTTGTCATTGT-3′. The CLCN3 antisense was shown to
eliminate both the short and long forms of CLCN3 (16). A nonsense primer sequence was
constructed from 15 randomized bases (5′-CCGTATGACCGCGCC-3′) and
served as an experimental control (17). Oligonucleotide primers were
transfected into U251 cells using Lipofectamine® 2000
transfection reagent (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) according to manufacturer's instructions. The
final concentration of oligonucleotide primers was 0.5–2 µg/ml. For
reverse transcription-quantitative polymerase chain reaction
(RT-qPCR) and western blotting, 24 h after transfection, cells were
treated with cisplatin (Sigma-Aldrich; Merck KGaA) for 24 h, and
then were harvested.
RT-qPCR
Total RNA was extracted from U251 cells using TRIzol
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
according to the manufacturer's protocol. Avian myeloblastosis
virus reverse transcriptase (Takara Biotechnology Co., Ltd.) was
used to reverse transcribe 2 µg RNA into cDNA. The PCR primers for
matrix metalloprotease 2 (MMP2) (Genebank NM
004530.2) and MMP9 (Genebank NM 004994.2) were created with
Primer 5 software (Premier Biosoft International, Palo Alto, CA,
USA); CLCN3, CCND1 and ACTB primers have been
published previously (18). The
primers were synthesized by Takara. Primers sequences and related
parameters are presented in Table I.
PCR results were visualized using a Tanon-1600 figure gel image
processing system and analyzed with GIS 1D gel image system
software (Tanon, Shanghai, China).
 | Table I.Primer sequences and related
parameters. |
Table I.
Primer sequences and related
parameters.
| Gene | Sequence | Annealing
temperature | Product |
|---|
| CLCN3 | Sense:
5′-CCTCTTTCCAAAGTATAGCAC-3′ | 55°C | 552 bp |
|
| Antisense:
5′-TTACTGGCATTCATGTCATTTC-3′ |
|
|
| MMP-2 | Sense:
5′-GTGCTGAAGGACACACTAAAGAAGA-3′ | 55°C | 605 bp |
|
| Antisense:
5′-GGATGTTGAAACTCTTCCTACCGTT-3′ |
|
|
| MMP-9 | Sense:
5′-CACTGTCCACCCCTCAGAGC-3′ | 57°C | 243 bp |
|
| Antisense:
5′-GGAATAGCGGCTGTTCACCG-3′ |
|
|
| β-actin | Sense:
5′-GTGGGGCGCCCCAGGCACCA-3′ | 55°C | 538 bp |
|
| Antisense:
5′-CTCCTTAATGTCACGCACGATTTC-3′ |
|
|
Western blot analysis
Cells were harvested and lysed in RIPA lysis buffer
(Beyotime, Shanghai, China) that included a protease inhibitor
cocktail (19) (Sigma-Aldrich; Merck
KGaA). Protein quantification was performed using the Bio-Rad
Protein assay dye reagent (Bio-Rad Laboratories, Inc., Hercules,
CA, USA). Proteins were electrophoresed and separated by SDS-PAGE,
and then transferred from gels onto PVDF membranes (EMD Millipore,
Billerica, MA, USA). After incubation with primary (4°C, overnight)
and secondary antibodies (room temperature, 2 h), the blots were
developed with DAB (3,3′-diaminobenzidine). Western blots results
were analyzed with GIS 1D gel image system software. The following
primary antibodies were used: anti-CLCN3 (1:100, sc-17572),
anti-Bcl-2 (1:200, sc-783), anti-Bax (1:200, sc-7480),
anti-pro-caspase 3 (1:200, sc-7148), anti-cleaved-caspase 3 (1:200,
sc-22171), anti-cathepsin D (1:200, sc-136282), anti-β-actin
(1:400, sc-47778; Santa Cruz Biotechnology, Inc., Dallas, TX, USA).
Secondary HRP-conjugated antibodies were purchased from Thermo
Fisher Scientific, Inc. Densitometry was used to calculate the
ratio of CLCN3 to β-actin, cleaved caspase 3 to pro-caspase 3, and
BCL2 to BAX signal in each lane.
MTT Assay
Cell viability was detected by the MTT assay. MTT
(3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) was
purchased from Sigma-Aldrich; Merck KGaA. Cells were grown in
96-well plates (1×104 cells/ml and 100 µl/well)
maintained at 37°C with 95% air and 5% CO2 for 22–24 h.
After oligonucleotide transfection and cisplatin treatment, 20 µl
of MTT (5 mg/ml) was added to each well for 4 h. After removing the
medium, 100 µl of DMSO was added to each well, and the absorbance
at 570 nm was measured with a microplate reader (BioTek
Instruments, Inc., Winooski, VT, USA).
Morphological determination of
apoptosis by TUNEL
Apoptotic morphological changes in cells were
detected using a TUNEL assay according to the protocol of the In
Situ Cell Death Detection kit (Roche Applied Science, Penzberg,
Germany). Cells were fixed in 4% formaldehyde (4°C, 20 min), and
incubated by TUNEL reagent (keep in dark, 37°C, 60 min). Glycerol
was used as mounting medium. The results were observed using an
Olympus fluorescence microscope (BX60; Olympus Inc., Tokyo, Japan)
with 450–500 nm excitation wavelength and 515–565 nm (20) emission wavelength. Six fields of view
observed under microscope.
Quantification of apoptosis by flow
cytometry
Treated cells were collected and incubated with
propidium iodide (PI) and Annexin V (both from Keygene, Nanjing,
China), and then assayed using a fluorescence activated cell
sorting system (FACS). Early apoptotic cells were only labelled
with Annexin V (21). Samples were
stained using the manufacturer's protocol and analyzed using a BD
FACSCalibur flow cytometer (BD Biosciences, Franklin Lakes, NJ,
USA). The software used for flow cytometry analysis is BD
CellQuest™ Pro (The Premier Acquisition and Analysis Software;
Apple Computer, Inc., Cupertino, CA, USA).
Measuring Cathepsin D activity
Treated cells were collected and Cathepsin D
activity was detected using the protocol supplied with the
Cathepsin D activity assay kit (BioVision, Inc., Milpitas, CA,
USA). Briefly, 1×106 cells were harvested, washed with
ice cold PBS, resuspended in 100 µl of PBS, and centrifuged for 2–5
min at 4°C at full speed using a cold microcentrifuge to remove
insoluble material. The cells were then resuspended in 200 µl of
Cell Lysis Buffer, incubated on ice for 10 min and centrifuged for
2–5 min at 4°C at full speed using a cold microcentrifuge to remove
insoluble material. The cleared cell lysate was then transferred
into a new tube. We then added 52 µl of the Reaction Mix to each
well and incubated at 37°C for 1–2 h in the dark. Fluorescence was
detected using a Packard Bioscience Fusion™ instrument (Packard
BioScience Co., Arvada, CO, USA) with a 328 nm excitation
wavelength and a 460 nm emission wavelength.
Measuring intracellular acridine
orange (AO) emission spectra
Our previous study found that AO accumulates in
acidic compartments such as lysosomes, where it fluoresces red
(22,23). After transfection and treatment, cells
were incubated with AO (final concentration 5 µM). The granular red
(lysosomal) fluorescence was measured using laser confocal
micro-spectrofluorometry (FluoView™ FV300; Olympus Inc.).
Transwell invasion and mobility
assay
Chambers with 8 µm pore filters (Becton Dickinson)
were placed into a 24-well plate. Treated cells were harvested,
resuspended, and then added to the upper compartment of the chamber
(5×104 cells/200 µl). Matrigel-coated filters were used
to detect cell invasion. Cells that invaded through the Matrigel
were counted on the underside of the filter according to the
manufacturer's instructions. The filter membranes with cells were
stained by 0.1% crystal violet at room temperature for 10 min, and
observed by optical microscope. Three independent experiments were
performed.
Statistical analysis
All results are presented as mean ± standard error.
The number of independent experiments are shown as n values.
One-way ANOVA was used for all statistical analyses followed by
Tukey's test. All data are carried out using the Student's t-test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Effects of nonsense oligonucleotide
and CLCN3 antisense oligonucleotide transfection on CLCN3 mRNA and
protein expression in U251 cells
Initially, we tested whether CLCN3 antisense
oligonucleotides could effectively suppress CLCN3 mRNA and protein
expression. RT-qPCR and western blot assays were performed to
detect the effects of transfecting nonsense and CLCN3 antisense
oligonucleotide on CLCN3 mRNA and protein expression in U251 cells.
The results showed that CLCN3 antisense oligonucleotides
significantly decreased CLCN3 mRNA and protein expression. This
indicated that CLCN3 antisense oligonucleotides could effectively
suppress CLCN3 expression (Fig.
1).
Silencing CLCN3 further reduces the
viability of cisplatin-treated U251 cells
Changes in the viability of cisplatin-treated U251
cells were used to evaluate the cytotoxic effects of silencing
CLCN3. Cells were transfected with nonsense or CLCN3 antisense
oligonucleotide for 24 h followed by treatment with cisplatin for
24 h. We observed a significant enhancement of the
cisplatin-induced cytotoxicity following CLCN3 antisense
transfection in U251 cells. Suppressing CLCN3 expression reduced
cell viability at all concentrations of cisplatin tested (Fig. 2A). We also investigated whether CLCN3
antisense transfection enhanced cisplatin-induced apoptosis in U251
cells. Apoptosis quantification by flow cytometry was used to
determine the apoptotic rate of cells treated with cisplatin and
CLCN3 antisense. Early apoptotic cells were only labelled with
Annexin V and shown in Q4 quadrant. The results showed that
transfecting CLCN3 antisense before cisplatin treatment (15
µmol/l), increased the population of early apoptotic cells.
Morphological determination of apoptosis by TUNEL staining also
showed an increased induction of apoptosis following combined
treatment (Fig. 2B and C).
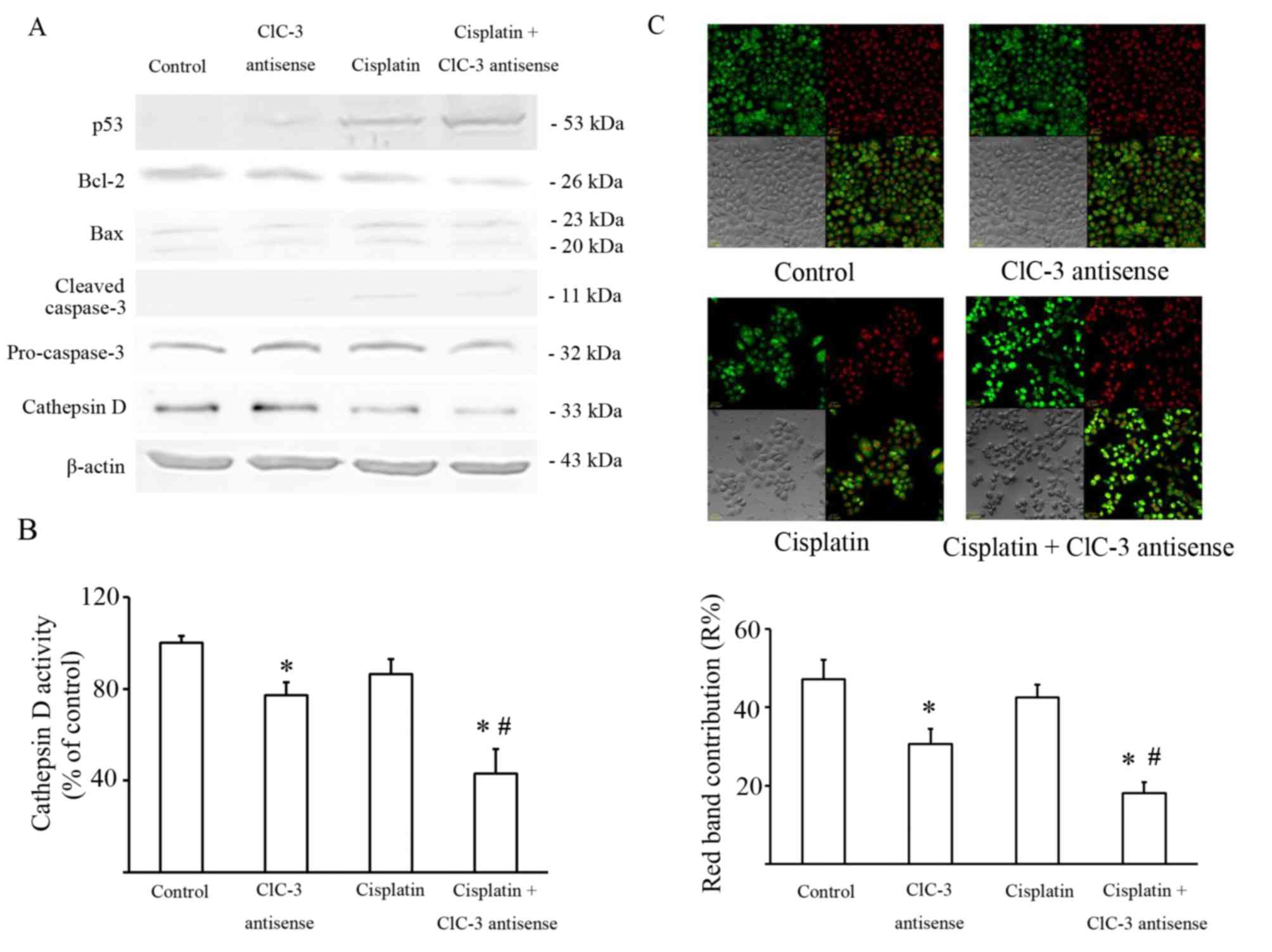
Effect of CLCN3 antisense transfection
on cisplatin cytotoxicity and lysosome dysfunction
It is well known that cisplatin damages DNA by
forming DNA adducts that induce the tumor suppressor TP53, which
may lead to the induction of a number of processes, including
apoptosis. The level of TP53 expression was detected by western
blot, and the results showed that suppressing CLCN-3 induced a
greater accumulation of TP53 in U251 cells treated with cisplatin.
The ratio of BCL2 and BAX expression was also significantly
decreased in U251 cells exposed to cisplatin with or without CLCN3
antisense transfection compared with the control group and the
CLCN3 antisense group. Caspase 3 activity was significantly
increased in U251 cells exposed to cisplatin with or without CLCN3
antisense compared with the control group (Fig. 3A; Table
II).
 | Table II.Status of different apoptosis
relative genes in U251 cells. |
Table II.
Status of different apoptosis
relative genes in U251 cells.
| Groups | Control | CLCN3
antisense | Cisplatin | Cisplatin+CLCN3
antisense |
|---|
| p53/β-actin | 1.0±2.4 |
18.5±3.7a |
102.3±15.4a |
121.7±10.9a,b |
| Bcl-2/Bax | 1.0±0.12 | 1.02±0.24 |
0.36±0.14a |
0.24±0.08a,b |
| Pro-caspase
3/cleaved-caspase 3 | 1.0±0.47 | 1.10±0.32 |
4.49±0.91a |
6.68±0.84a,b |
The intracellular chloride channel CLCN3 is
expressed mainly in acidic intracellular compartments, especially
lysosomes, and participates in their acidification (8). Lysosomal cathepsins, such as Cathepsin
D, play key roles in the degradative function of lysosomes. So, the
expression and the activity of lysosomal cathepsins are significant
markers of lysosome function. Thus, Cathepsin D expression was also
evaluated by western blot. These data showed that Cathepsin D
expression decreased in U251 cells transfected by CLCN3 antisense
with or without cisplatin (Fig. 3A).
Cathepsin D activity was consistent with its expression (Fig. 3B).
AO preferentially accumulates in lysosomes and
fluoresces red. When AO relocates from the lysosomes to the cytosol
it fluoresces green. Staining with AO revealed that red granular
fluorescence was decreased by CLCN3 antisense, but was not
significantly altered by cisplatin. Furthermore, the combination of
CLCN3 antisense transfection with cisplatin reduced red granular
fluorescence more than cisplatin treatment alone (Fig. 3C).
Effects of cisplatin and CLCN3
antisense treatment on MMP expression, invasion and mobility in
U251 cells
The cell invasion and mobility-related genes,
MMP2 and MMP9, were downregulated by cisplatin
treatment with or without CLCN3 antisense transfection (Fig. 4A). Additionally, MMP2 and
MMP9 expression were decreased by CLCN3 antisense
transfection. Cell invasion and mobility were reduced by cisplatin
with or without CLCN3 antisense transfection and by CLCN3 antisense
alone (Fig. 4B; Table III).
 | Table III.Correlation analysis of MMP-2 and
MMP-2 mRNA expression, cell invasion and cell mobility of U251
cells. |
Table III.
Correlation analysis of MMP-2 and
MMP-2 mRNA expression, cell invasion and cell mobility of U251
cells.
|
| Relative
values |
|
|
|---|
|
|
|
|
|
|---|
| Treatment | MMP-2/β-actin | MMP-9/β-actin | Invasion cell
number | Mobility optical
density |
|---|
| Control | 1.00±0.14 | 1.00±0.11 | 81.0±4.6 | 0.81±0.02 |
| ClC-3
antisense |
0.61±0.17a |
0.64±0.25a |
42.0±4.4a |
0.58±0.13a |
| Cisplatin |
0.59±0.05a |
0.63±0.14a |
48.3±2.5a |
0.42±0.07a |
| Cisplatin+ClC-3
antisense |
0.59±0.27a |
0.29±0.09a,b |
12.7±5.1a,b |
0.31±0.11a,b |
Discussion
Cisplatin was the first clinically-applied
platinum-based drug and has a potent DNA-damaging anticancer
effect. It is well known that the primary cytotoxic action of
cisplatin is due to its ability to form adducts with DNA and induce
apoptosis (24). However, cancer
cells with high intrinsic or acquired resistance to cisplatin are a
major hurdle to its clinical success (25).
CLCN3 is primarily expressed on acidic intracellular
compartments, especially lysosomes (3,8). It has
been shown that CLCN3 protein is expressed at high levels in some
tumors, including gliomas (26–29). These
studies indicate that the function of CLCN3 is to facilitate
lysosomal acidification and that it plays roles in proliferation,
invasion, migration, and drug-resistance (30). The direct or indirect link between
lysosomes and cisplatin resistance has been implied by several
studies (31–33). Chloride channel blockers, such as
4-acetamido-4′-isothiocyanostilbene-2,2′-disulfonic acid (SITS)
(34), and
4–4′-diisothiocyanostilbene-2,2′-disulfonic acid (DIDS) (35), are known to weaken cisplatin
resistance; therefore, we investigated the relationship between
cisplatin-induced apoptosis and chloride channel activity in human
glioma cells.
In this study, we hypothesized that CLCN3 plays a
role in cisplatin resistance in human glioma U251 cells via
lysosomal dysfunction. To test our hypothesis, we utilized CLCN3
antisense oligonucleotide transfection to suppress CLCN3 mRNA and
protein expression in U251 cells. There are two classes of
apoptosis induced by DNA-damaging agents such as cisplatin: 1) the
extrinsic receptor-dependent pathway (36) and 2) the intrinsic mitochondrial
pathway (37). Cisplatin damages DNA
by forming DNA adducts, which may then induce apoptotic TP53
responses (38). After DNA damage, if
the damage is irreversible, apoptosis is triggered. Lysosomal
dysfunction and membrane permeabilization cause the release of
lysosomal enzymes into the cytosol in response to the action of
cathepsins following p53 activation, thereby mediating cell death
(39,40).
We investigated the effects of combined CLCN3
antisense oligonucleotide transfection and cisplatin treatment on
U251 cells. Our results showed that cisplatin not only decreased
cell viability but also induced apoptosis. We detected membrane
phosphatidylserine translocation by Annexin V-fluorescein
isothiocyanate (FITC) staining, DNA degradation by TUNEL staining,
and p53 upregulation and caspase 3 activation by western blot. Our
results illustrated that cisplatin induced cellular DNA
fragmentation and triggered p53 accumulation. The altered BCL2/BAX
expression ratio indicated activation of mitochondrial membrane
permeabilization. Red granular fluorescence was decreased by CLCN3
antisense transfection, and was not significantly altered by
cisplatin as shown by AO staining. CLCN3 antisense induced the
lysosomal pH increasing in cisplatin-treated U251 cells. Moreover,
Cathepsin D activity was decreased by CLCN3 suppression in U251
cells. These data suggested that CLCN3 suppression leaded to
lysosomal dysfunction involved in mechanism of cisplatin-resistance
in U251 cells. But the link between the lysosomal function and
chemotherapeutic sensitivity still need further investigation.
In our previous study, we found that CLCN3 plays
dual roles in the mechanisms of cisplatin resistance in U251 cells.
On one hand, CLCN3 promotes the Akt/mTOR pathway through generating
ROS via Nox, while on the other hand, as CLCN3 has indispensable
roles in the acidification of acidic intracellular compartments
such as late-endosomes, lysosomes and mature autophagosomes, CLCN3
deficiency probably induces autophagy (23). As a follow-up from the previously
published article (23), in this
study, we furthered our investigation into the possible roles of
CLCN3 and discovered that suppressing CLCN3 induced lysosomal
dysfunction, through increasing pH and decreasing Cathepsin D
activity. Although our data indicated that CLC3 may provide
possible targets for anticancer drugs, but one of limitations of
our work must be pay attention to is that we only employed U251
cells in our study, so more research depending on various types
cancer cells should be done to define the molecular mechanism of
its activation.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81672948 and
81202552), Jilin Provincial Research Foundation for International
Science and Technology Cooperation Projects, China, (grant no.
20160414005GH), Jilin University Bethune Plan B Projects (grant no.
2015222).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JS and LZ conceived the idea and paper, reviewed the
literature and examined the data. YHZ performed the vector
construction experiments and wrote the manuscript. JJZ and LCZ
participated in the qPCR and western blot experiments. XYY
participated in the experiments of apoptosis evaluation. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sontheimer H: Malignant gliomas:
Perverting glutamate and ion homeostasis for selective advantage.
Trends Neurosci. 26:543–549. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jentsch TJ: Discovery of CLC transport
proteins: Cloning, structure, function and pathophysiology. J
Physiol. 593:4091–4109. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Leanza L, Biasutto L, Managò A, Gulbins E,
Zoratti M and Szabò I: Intracellular ion channels and cancer. Front
Physiol. 4:2272013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li X, Wang T, Zhao Z and Weinman SA: The
ClC-3 chloride channel promotes acidification of lysosomes in
CHO-K1 and Huh-7 cells. Am J Physiol Cell Physiol. 282:C1483–C1491.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hara-Chikuma M, Yang B, Sonawane ND,
Sasaki S, Uchida S and Verkman AS: ClC-3 chloride channels
facilitate endosomal acidification and chloride accumulation. J
Biol Chem. 280:1241–1247. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Weylandt KH, Nebrig M, Jansen-Rosseck N,
Amey JS, Carmena D, Wiedenmann B, Higgins CF and Sardini A: ClC-3
expression enhances etoposide resistance by increasing
acidification of the late endocytic compartment. Mol Cancer Ther.
6:979–986. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zifarelli G and Pusch M: CLC chloride
channels and transporters: A biophysical and physiological
perspective. Rev Physiol Biochem Pharmacol. 158:23–76.
2007.PubMed/NCBI
|
|
8
|
Hong S, Bi M, Wang L, Kang Z, Ling L and
Zhao C: CLC-3 channels in cancer (review). Oncol Rep. 33:507–514.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lui VC, Lung SS, Pu JK, Hung KN and Leung
GK: Invasion of human glioma cells is regulated by multiple
chloride channels including ClC-3. Anticancer Res. 30:4515–4524.
2010.PubMed/NCBI
|
|
10
|
Rosenberg B: Fundamental studies with
cisplatin. Cancer. 55:2303–23l6. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Madajewicz S, Chowhan N, Tfayli A, Roque
C, Meek A, Davis R, Wolf W, Cabahug C, Roche P, Manzione J, et al:
Therapy for patients with high grade astrocytoma using
intraarterial chemotherapy and radiation therapy. Cancer.
88:2350–2356. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Safaei R, Katano K, Larson BJ, Samimi G,
Holzer AK, Naerdemann W, Tomioka M, Goodman M and Howell SB:
Intracellular localization and trafficking of fluorescein-labeled
cisplatin in human ovarian carcinoma cells. Clin Cancer Res.
11:756–767. 2005.PubMed/NCBI
|
|
13
|
Liang XJ, Mukherjee S, Shen DW, Maxfield
FR and Gottesman MM: Endocytic recycling compartments altered in
cisplatin-resistant cancer cells. Cancer Res. 66:2346–2353. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Safaei R and Howell SB: Copper
transporters regulate the cellular pharmacology and sensitivity to
Pt drugs. Crit Rev Oncol Hematol. 53:13–23. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liao C, Hu B, Arno MJ and Panaretou B:
Genomic screening in vivo reveals the role played by vacuolar H+
ATPase and cytosolic acidification in sensitivity to DNA-damaging
agents such as cisplatin. Mol Pharmacol. 71:416–425. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shimada K, Li X, Xu G, Nowak DE, Showalter
LA and Weinman SA: Expression and canalicular localization of two
isoforms of the ClC-3 chloride channel from rat hepatocytes. Am J
Physiol Gastrointest Liver Physiol. 279:G268–G276. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Olsen ML, Schade S, Lyons SA, Amaral MD
and Sontheimer H: Expression of voltage-gated chloride channels in
human glioma cells. J Neurosci. 23:5572–5582. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lu SJ, Quan C, Li F, Vida L and Honig GR:
Hematopoietic progenitor cells derived from embryonic stem cells:
Analysis of gene expression. Stem Cells. 20:428–437. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mizel SB: Interleukin 1 and T cell
activation. Immunol Rev. 63:51–72. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Niinuma A, Higuchi M, Takahashi M, Oie M,
Tanaka Y, Gejyo F, Tanaka N, Sugamura K, Xie L, Green PL and Fujii
M: Aberrant activation of the interleukin-2 autocrine loop through
the nuclear factor of activated T cells by nonleukemogenic human
T-cell leukemia virus type 2 but not by leukemogenic type 1 virus.
J Virol. 79:11925–11934. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Duan L, Aoyagi M, Tamaki M, Nakagawa K,
Nagashima G, Nagasaka Y, Ohno K, Yamamoto K and Hirakawa K:
Sensitization of human malignant glioma cell lines to tumor
necrosis factor-induced apoptosis by cisplatin. J Neurooncol.
52:23–36. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Millot C, Millot JM, Morjani H, Desplaces
A and Manfait M: Characterization of acidic vesicles in
multidrug-resistant and sensitive cancer cells by acridine orange
staining and confocal microspectrofluorometry. J Histochem
Cytochem. 45:1255–1264. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Su J, Xu Y, Zhou L, Yu HM, Kang JS, Liu N,
Quan CS and Sun LK: Suppression of chloride channel 3 expression
facilitates sensitivity of human glioma U251 cells to cisplatin
through concomitant inhibition of Akt and autophagy. Anat Rec
(Hoboken). 296:595–603. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Strojan P, Vermorken JB, Beitler JJ, Saba
NF, Haigentz M Jr, Bossi P, Worden FP, Langendijk JA, Eisbruch A,
Mendenhall WM, et al: Cumulative cisplatin dose in concurrent
chemoradiotherapy for head and neck cancer: A systematic review.
Head Neck. 38 Suppl 1:E2151–E2158. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Borst P, Rottenberg S and Jonkers J: How
do real tumors become resistant to cisplatin? Cell Cycle.
7:1353–1359. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mao J, Chen L, Xu B and Wang L, Li H, Guo
J, Li W, Nie S, Jacob TJ and Wang L: Suppression of ClC-3 channel
expression reduces migration of nasopharyngeal carcinoma cells.
Biochem Pharmacol. 75:1706–1716. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sontheimer H: An unexpected role for ion
channels in brain tumor metastasis. Exp Biol Med (Maywood).
233:779–791. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Habela CW, Olsen ML and Sontheimer H: ClC3
is a critical regulator of the cell cycle in normal and malignant
glial cells. J Neurosci. 28:9205–9217. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Moreland JG, Davis AP, Bailey G, Nauseef
WM and Lamb FS: Anion channels, including ClC-3, are required for
normal neutrophil oxidative function, phagocytosis, and
transendothelial migration. J Biol Chem. 281:12277–12288. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Shimizu T, Lee EL, Ise T and Okada Y:
Volume-sensitive Cl(−) channel as a regulator of acquired cisplatin
resistance. Anticancer Res. 28:75–83. 2008.PubMed/NCBI
|
|
31
|
Safaei R, Larson BJ, Cheng TC, Gibson MA,
Otani S, Naerdemann W and Howell SB: Abnormal lysosomal trafficking
and enhanced exosomal export of cisplatin in drug-resistant human
ovarian carcinoma cells. Mol Cancer Ther. 4:1595–1604. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chauhan SS, Liang XJ, Su AW,
Pai-Panandiker A, Shen DW, Hanover JA and Gottesman MM: Reduced
endocytosis and altered lysosome function in cisplatin-resistant
cell lines. Br J Cancer. 88:1327–1334. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Garmann D, Warnecke A, Kalayda GV, Kratz F
and Jaehde U: Cellular accumulation and cytotoxicity of
macromolecular platinum complexes in cisplatin-resistant tumor
cells. J Control Release. 131:100–106. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yarbrough JW, Merryman JI, Barnhill MA and
Hahn KA: Inhibitors of intracellular chloride regulation induce
cisplatin resistance in canine osteosarcoma cells. In Vivo.
13:375–383. 1999.PubMed/NCBI
|
|
35
|
Lee EL, Shimizu T, Ise T, Numata T, Kohno
K and Okada Y: Impaired activity of volume-sensitive Cl-channel is
involved in cisplatin resistance of cancer cells. J Cell Physiol.
211:513–521. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ashkenazi A: Targeting death and decoy
receptors of the tumour-necrosis factor superfamily. Nat Rev
Cancer. 2:420–430. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
37
|
Jiang X and Wang X: Cytochrome C-mediated
apoptosis. Annu Rev Biochem. 73:87–106. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Burger H, Nooter K, Boersma AW, Kortland
CJ and Stoter G: Expression of p53, Bcl-2 and Bax in
cisplatin-induced apoptosis in testicular germ cell tumour cell
lines. Br J Cancer. 77:1562–1567. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Jäättelä M, Candé C and Kroemer G:
Lysosomes and mitochondria in the commitment to apoptosis: A
potential role for cathepsin D and AIF. Cell Death Differ.
11:135–136. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Boya P and Kroemer G: Lysosomal membrane
permeabilization in cell death. Oncogene. 27:6434–6451. 2008.
View Article : Google Scholar : PubMed/NCBI
|