Introduction
Liver cancer is one of the most prevalent human
tumors worldwide (1) and is ranked as
the third leading cause of cancer-associated mortality in China
(2). In the majority of cases, liver
cancer develops from chronic inflammation and cirrhosis caused by
infections from the hepatitis B and C viruses, ethanol or
aflatoxins (3). Despite recent
advances in the comprehension of the molecular basis of liver
cancer and the use of novel chemotherapeutic approaches, liver
cancer remains associated with a poor prognosis (4). This is primarily due to only a limited
number of patients being able to undertake potentially curative
treatments, including surgical resection followed by orthotopic
liver transplantation (5).
Furthermore, the mortality rate has declined only modestly, owing
to the chemoresistance of liver cancer (4). Therefore, there is an urgent requirement
for development of effective therapeutic strategies for patients
with liver cancer in the advanced stage of the disease.
The initiation of liver cancer has long been
established to be the result of different genetic alterations that
ultimately lead to malignant transformations (6). MicroRNAs (miRNAs/miRs) are endogenous,
small, non-coding regulatory RNAs that are ~22 nucleotides in
length (7). miRNAs are able to act as
post-transcriptional regulators to negatively regulate the
expression of genes by binding directly to the 3′-untranslated
regions (3′-UTRs) of target mRNAs in a sequence-specific manner,
leading to mRNA degradation (7).
Previous studies have demonstrated that miRNA-mediated regulation
of gene expression exhibits a role in the development,
differentiation, proliferation and apoptosis of cells. The
dysregulation of miRNAs is associated with a variety of types of
cancer, and miRNAs may also serve a role in tumorigenesis and
progression (8–11). miRNA targets include tumor suppressor
and oncogenes (8,9), for example, miRNA-449 has been
demonstrated to repress the DNA synthesis, mitotic entry and
proliferation of liver cancer cells (5). Mechanistically, in hepatoma cells,
miR-449 controls lipogenesis and cholesterogenesis by the
inhibition of SIRT1 and SREBP-1c expression, and the downregulation
of their targeted genes (5).
Sirtuin 1 (SIRT1) can function as either a tumor
suppressor or an oncogene during cancer development, and
upregulation of SIRT1 is able to suppress colon cancer growth
(12). SIRT1 is positively associated
with malignancy in other types of cancer (13) and was previously identified to be
abnormally upregulated in liver cancer, where it promoted tumor
growth (14). Consequently,
inhibiting SIRT1 activity alone or in combination with other
therapies has been suggested as a novel therapeutic strategy for
the treatment of liver cancer (15).
The miR-29 family is composed of members with
conserved miRNA sequences including miR-29a, miR-29b, miR-29c and
miR-29d (16). miR-29c was
demonstrated to inhibit cell growth, cell migration and invasion in
pancreatic cancer by targeting integrin subunit β1 (17). In bladder cancer, miR-29c
overexpression inhibited cell growth, suppressed cell migration and
resulted in an accumulation of cells in the G1 phase during the
cell cycle through the target gene cyclin dependent kinase 6
(18). miR-29c may function as a
tumor suppressor serving a crucial role in the development of liver
carcinoma by targeting protein phosphatase, Mg2+/Mn2+ dependent 1D
(19). miR-29c is downregulated in
gastric cancer tissues and cell lines, and the overexpression of
miR-29c inhibits cell proliferation, promotes apoptosis and arrests
the cell cycle at the G1/G0 phase by targeting nuclear
autoantigenic sperm protein (20).
The present study aimed to uncover the function of
miR-29c in chemoresistance and the mechanisms by which miR-29c
regulates the cisplatin (cis-diamminedichloroplatinum, CDDP)
resistance of liver cancer.
Materials and methods
Cell culture
The HepG2 cell line was obtained from the American
Type Culture Collection (Manassas, VA, USA) and passaged for a
period of <6 months. The HepG2 cell line was originally assumed
to be a hepatocellular carcinoma cell line, but was subsequently
identified to originate from a hepatoblastoma, hence the emphasis
of the present study on ‘liver cancer’ (21). The cells were cultured in Dulbecco's
modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) containing 10% fetal bovine serum (Gibco;
Thermo Fisher Scientific, Inc.) and 1% penicillin and streptomycin,
at 37°C in a humidified 5% CO2 incubator.
Selection of chemoresistant cell
line
CDDP resistant HepG2 cell line (CDDP-R) was derived
from original parental cell line (CDDP-S) by continuous exposure to
cisplatin (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) following
initial dose-response studies of cisplatin (0, 1.25, 2.5, 5, 10 µM)
over 72 h at 37°C from which half maximal inhibitory concentration
(IC50) values were obtained. Initially, HepG2 was
treated with cisplatin (4.7 µM) (IC50) for 72 h at 37°C.
The media (DMEM + 10% FBS, Gibco; Thermo Fisher Scientific, Inc.)
was removed and cells were allowed to recover for a further 72 h.
This development period was performed for ~6 months.
IC50 concentrations were reassessed. Cells were then
maintained continuously in the presence of cisplatin at the new
IC50 concentration (20.5 µM) at 37°C for a further 6
months.
Plasmid construction and
extraction
pVax-based SIRT1 overexpression plasmid was
purchased from Fulengen Bio Co., Ltd. (Guangzhou, China). The pVax
empty plasmid was used as transfection control. All plasmids were
transformed into DH5α cells (Genewiz, Inc., Suzhou, China) for
amplification and DNA was extracted by the EndoFree Plasmid kit
(Qiagen GmbH, Hilden, Germany), according to the manufacturer's
protocol. The concentration was determined by measuring the
A260/A280 ratio using a Thermo ND 2,000 spectrophotometer (Thermo
Fisher Scientific, Inc.). The plasmid was stored at −20°C until
further use. The plasmid transfection was conducted using 2 µg
plasmid per well using Lipofectamine 3000 (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's
protocol.
Luciferase assay
The SIRT1-3′UTR containing the miR-29c binding site
and the miR-29c mutant binding site were purchased from Genewiz,
Inc., and extracted using the EndoFree Plasmid kit (Qiagen GmbH.).
The plasmids (2 µg/well) were co-transfected with miR-29c into
HepG2 cells using Lipofectamine 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the protocol previously described by
Luo et al (22). A luciferase
reporter assay (Promega Corporation, Madison, WI, USA) was
purchased and used to measure luciferase activity at 4 h
post-transfection, according to the manufacturer's protocol. The
relative luciferase activity was normalized to the miR-NC
group.
In vitro proliferation and colony
formation assay
For proliferation assays, cells were seeded at
2×103 cells per well in 96-well plates, as previously
described (23). Cell Counting Kit-8
(Dojindo Molecular Technologies, Inc., Shanghai, China) was used
and absorbance was measured at 450 nm for each well at different
time points (0, 24, 48 and 72 h) using a microplate reader (Thermo
Fisher Scientific, Inc.). For colony formation assays, cells were
plated at 500–1,000 cells per well into 6-well plates and cultured
for ~14 days, followed by crystal violet (0.5% w/v) staining for 30
min at room temperature, and counted using a light Stereomicroscope
(×4).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
miRNAs were obtained using the mirVana miRNA
Isolation kit according to the manufacturer's protocol (Ambion;
Thermo Fisher Scientific, Inc.). RT-qPCR was performed for miR-29c
using miRNA primers obtained from Exiqon A/S (Vedbæk, Denmark).
β-actin was used as a loading control. The primers of miR-29c and
β-actin were not supplied according to the rules of the company
(Exiqon A/S, Vedbæk, Denmark). First, cDNA was synthesized from all
miRNA samples according to the manufacturer's protocol (Exiqon
A/S). Synthesized cDNAs were used as templates for gene-expression
analysis through RT-qPCR with SYBR Green (Applied Biosystems;
Thermo Fisher Scientific, Inc.). The qPCR conditions were as
follows: Denaturation at 94°C for 2 min, amplification for 30
cycles at 94°C for 0.5 min, annealing at 60°C for 0.5 min and
extension at 72°C for 1 min, followed by a terminal elongation step
at 72°C for 10 min. Data were analyzed with the 2−ΔΔCq
method (24).
Apoptosis analysis
Cells (2×105) transfected with miRNA and
treated by CDDP were harvested at 48 h post-transfection and
stained with Annexin V-FITC/PI Apoptosis Detection kit I (BD
Pharmingen; BD Biosciences, San Jose, CA, USA). Apoptotic cells
were assessed in triplicate and the experiment was repeated three
times independently by flow cytometry (FACS Calibur; BD
Biosciences) with FACSComp software (version 5.1; BD
Biosciences).
Xenograft tumors in nude mice
CDDP-R cells stably expressing miR-NC or miR-29c
(5×106 cells in 100 µl DMEM) were injected
subcutaneously into the flanks of Balb/c nude mice (4 mice/group)
(5 weeks old, 18–20 g, male; Vital River Laboratories, Beijing,
China). Mice were kept in a specific pathogen-free environment, on
a 12 h light/12 h dark cycle at a room temperature of 22±2°C with
ad libitum access to food and water. CDDP (20 nmol in 100 µl
saline) was injected every 3 days (5 times). An equal volume of
normal saline was injected as a negative control. Tumor volumes
were measured every 5 days. Tumor weights were measured immediately
after sacrificing the mice, and tumor samples were harvested for
whole protein lysates and embedded in paraffin for sectioning, as
previously described by Dai et al (25). Procedures involving animals conformed
to the guidelines of the Institutional Animal Care and Use
Committee of Sichuan Academy of Medical Sciences and Sichuan
Provincial People's Hospital (Sichuan, Chengdu, China) and study
approval was obtained.
TUNEL assay
Tumor tissue sections were examined for the presence
of apoptotic cells using TUNEL assay, in which fragmented DNA from
apoptotic cells is end-labeled with the fluorophore. The biopsy
samples were fixed in 10% phosphate-buffered formalin (Thermo
Fisher Scientific, Inc.) for 24 h at room temperature, processed
and then embedded in paraffin. Serial 4-µm thick tissue sections
were analyzed using the DeadEnd™ Fluorometric TUNEL system (Promega
Corporation) according to the manufacturer's protocol. Following
deparaffinization and rehydration, sections were fixed,
permeabilized with proteinase K for 8–10 min, and repeatedly fixed.
The sections were then covered with 50 ml terminal deoxynucleotidyl
transferase mix for 1 h at 37°C in a humidified chamber. The
coverslips were removed and the sections were immersed in 2X SSC
buffer for 15 min, washed with PBS and mounted with medium that
included DAPI (1.5 µg/ml) (Vectashield®; Vector
Laboratories Inc., Burlingame, CA, USA) at room temperature
(22–25°C) for 15 min. Fluorescence images of three different fields
of view were captured using a fluorescence microscope (Olympus
Corporation, Tokyo, Japan).
Western blot analysis
The proteins were extracted with ice-cold lysis
buffer containing 1 mM EDTA, 20 mM Tris-HCl (pH7.5), 1 mM
dithiothreitol, 0.1 mM phenylmethylsulfonyl fluoride, 5 mM
MgCl2 and a protease inhibitor cocktail (1:100) (Pierce,
Thermo Fisher Scientific Inc.), and then centrifuged at 12,000 × g
for 20 min at 4°C. Protein concentration of the supernatant from
the extract was measured with the Bicinchoninic Acid assay kit
(Beyotime Institute of Biotechnology, Beijing, China). Equivalent
amounts (30 µg) of proteins were loaded on 12% SDS-PAGE and
transferred to polyvinylidene difluoride membranes (Merck KGaA).
The membranes were blocked in Tris-buffered saline-Tween 20 and
probed with anti-SIRT1 at 4°C overnight (dilution, 1:1,500; catalog
no. ab32441; Abcam, Cambridge, UK). Following washing, the
membranes were incubated with horseradish peroxidase-conjugated
secondary antibodies (dilution, 1:10,000; cat no. ZB-5301; OriGene
Technologies, Inc., Beijing, China) for 60 min at room temperature.
All blots were probed with antibodies against β-actin at 4°C
overnight (dilution 1:3,000; cat no. MABT825; Merck KGaA) as a
loading control. Immobilon® ECL Ultra Western HRP
Substrate (cat no. WBULS0500; EMD Millipore, Billerica, MA, USA)
was used for detection with X-ray film. The densitometry was
measured by Image J software (Version 1.48; National Institutes of
Health, Bethesda, MD, USA).
Bioinformatics analysis
The potential target genes of miR-29c were predicted
using three different online programs with databases of different
algorithms, including TargetScan (http://www.targetscan.org/), MicroRNA.org (http://www.microrna.org/) and miRDB (http://mirdb.org/) using h-miR-29c as a keyword on the
8th November 2016. The predicted targets were listed.
Statistical analysis
Continuous normally distributed variables are
represented graphically as the mean ± standard deviation. For
statistical comparison of quantitative data between groups,
analysis of variance (ANOVA) with Dunnett's multiple comparisons or
Student's t-test was performed. All statistical analyses were
performed using SPSS 22.0 statistical software (IBM Corp., Armonk,
NY, USA). P<0.05 was considered to indicate a statistically
significant difference.
Results
miR-29c is downregulated in
CDDP-resistant liver cancer cells
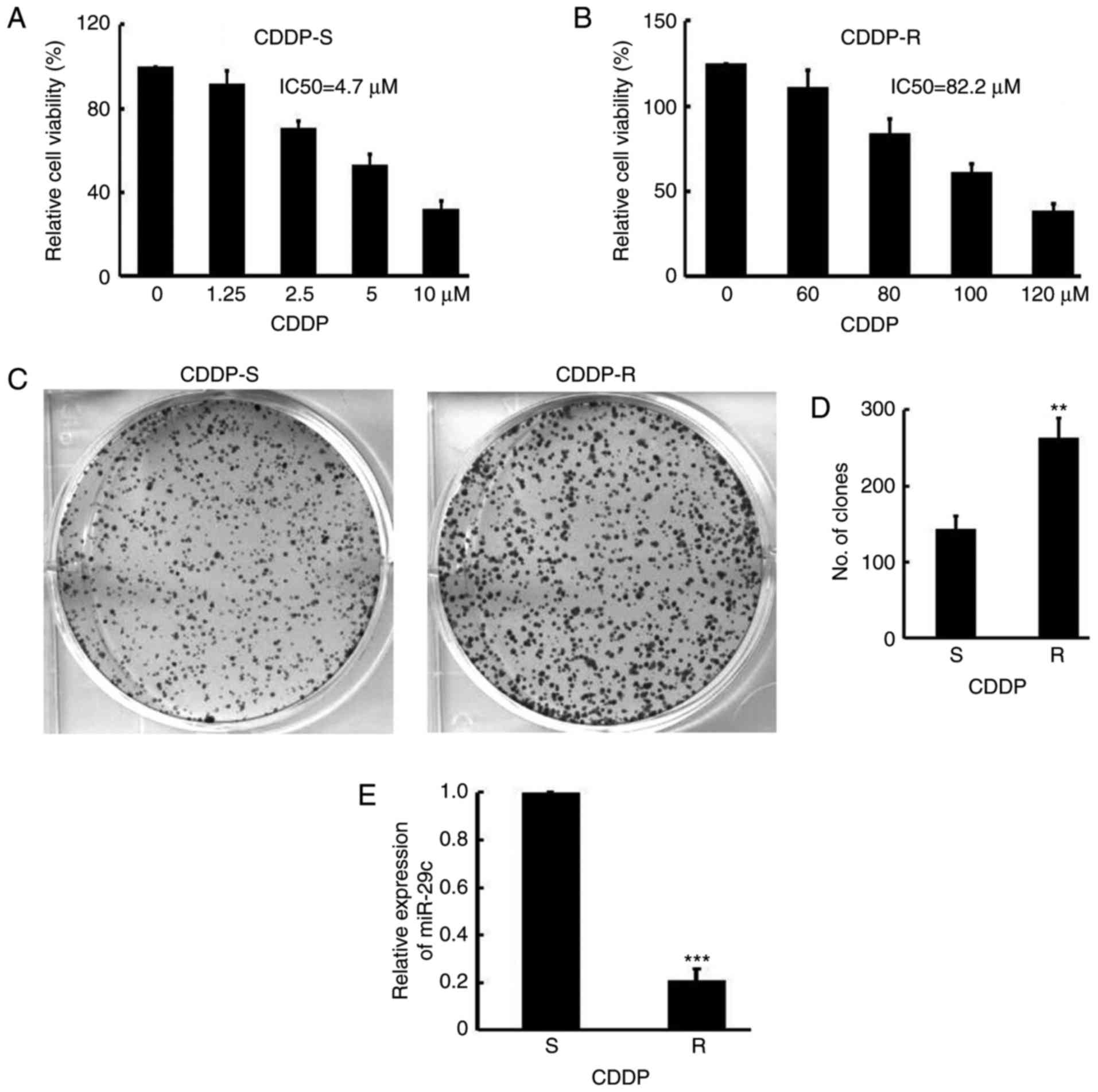
To assess the expression of miR-29c in CDDP-R liver
cancer, CDDP-R HepG2 cell lines were selected by gradually
increasing CDDP concentration in the culture medium. The
IC50 value for CDDP in the parent HepG2 CDDP-sensitive
(CDDP-S) line and the derived CDDP-resistant HepG2 (CDDP-R) cells
was calculated. It was demonstrated that the IC50 value
in the CDDP-S cells (IC50, 4.7±0.4 µM) (Fig. 1A) was significantly lower than that in
the CDDP-R cells (IC50, 82.2±3.1 µM) (P<0.05)
(Fig. 1B). The colony formation
abilities of the two cell lines was analyzed, and the CDDP-R cells
were demonstrated to exhibit increased colony formation abilities
(CDDP-S, 143±18 vs. CDDP-R, 264±25; P<0.01) (Fig. 1C-D). Notably, miR-29c expression
levels, as detected by qPCR, were significantly lower in CDDP-R
cells than in CDDP-S cells (CDDP-R, 0.21±0.05; P<0.001)
(Fig. 1E). These results suggested
that miR-29c is downregulated in CDDP-R cancer cells.
Overexpression of miR-29c restores
CDDP sensitivity in liver cancer cells in vitro
As miR-29c expression was reduced following
acquisition of resistance to CDDP, the present study investigated
the role of miR-29c in chemoresistance. A plasmid expressing
miR-29c was introduced and it was validated that the plasmid
effectively enhanced miR-29c levels in the CDDP-R cells (miR-29c
vs. miR-negative control (NC), 14.2±3.2 vs. 1.0±0.02) (Fig. 2A). Overexpression of miR-29c along
with CDDP treatment (20 µM) in CDDP-R cells resulted in
significantly reduced viability compared with that in the untreated
group and the miR-NC plus CDDP-treated group (decreased 45.0%;
miR-29c vs. miR-NC, 0.82±0.13 vs. 1.49±0.21) (Fig. 2B). The colony formation assay results
corroborated these results; treatment with miR-29c and CDDP (20 µM)
resulted in reduced colony numbers in CDDP-R cells (decreased
72.9%; miR-29c vs. miR-NC, 67±15 vs. 247±27 mm3)
(Fig. 2C and D). Furthermore, an
improved rate of apoptosis in CDDP-R cells was observed following
treatment with miR-29c and CDDP (increased 8.0 times; miR-29c vs.
miR-NC, 21.83±4.25 vs. 2.73±0.72%) (Fig.
2E and F). These results indicate that overexpression of
miR-29c could restore the CDDP sensitivity of CDDP-R cells in
vitro.
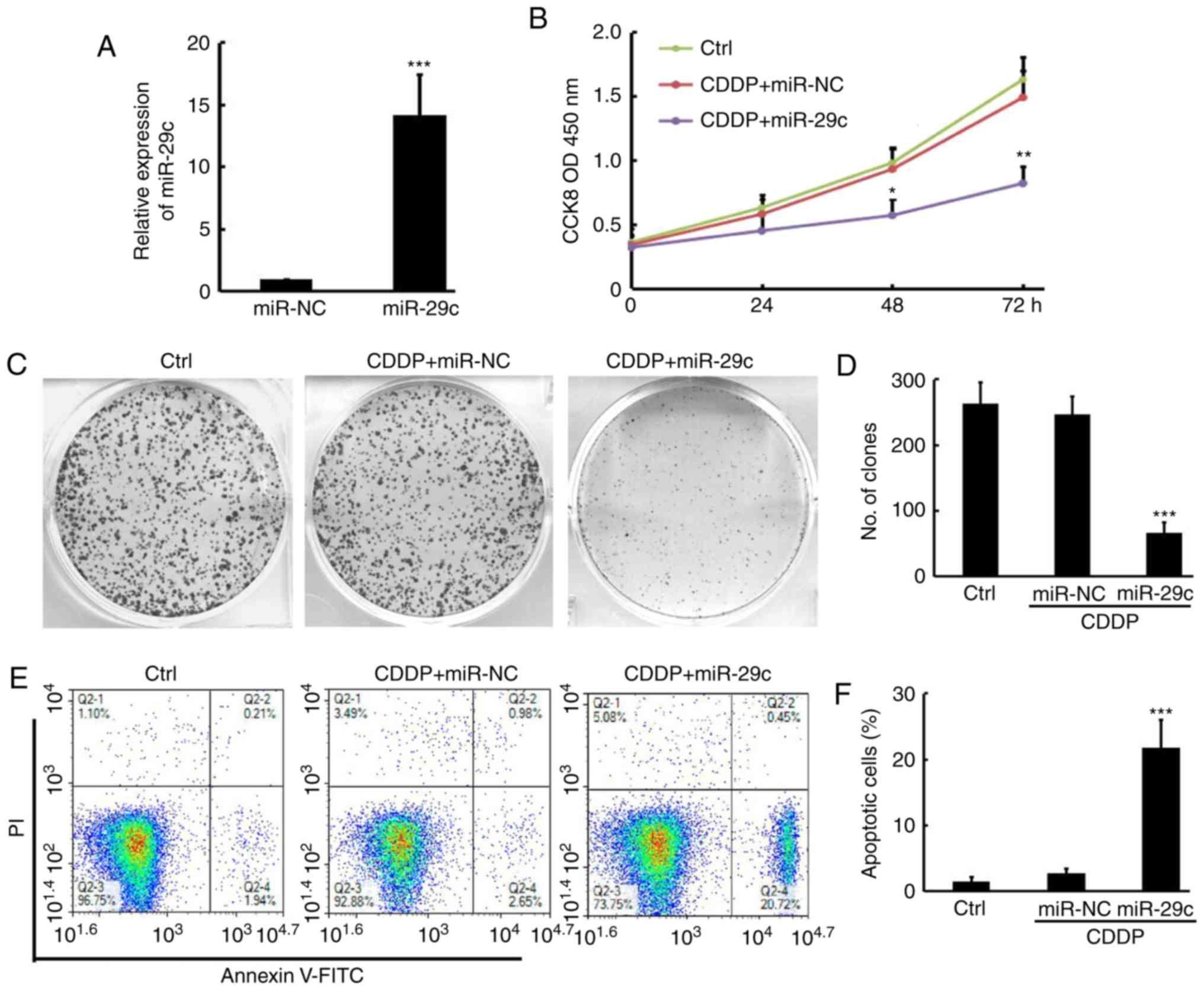 | Figure 2.Overexpression of miR-29c restores
CDDP sensitivity in liver cancer cells in vitro. (A)
Overexpression of miR-29c in CDDP-R cells by quantitative
polymerase chain reaction following transfection of miR-29c plasmid
compared with miR-NC (n=3). (B) Overexpression of miR-29c and CDDP
(20 µM) treatment inhibited cell proliferate compared with that of
CDDP (20 µM) + miR-NC and non-treated control (Ctrl) in CDDP-R
cells (n=3). *P<0.05 and **P<0.01 vs sthe CDDP+miR-NC group.
(C) Crystal violet staining of colony formation for CDDP (20 µM)
plus miR-NC or miR-29c combination treatments or non-treated
control in CDDP-R cells after 10 days, with (D) statistical
analysis of colony numbers for triplicate experiments (D) (n=3).
(E) Flow cytometry analysis of apoptotic cells for CDDP (20 µM)
plus miR-NC or miR-29c combination treatments or non-treated
control in CDDP-R cells after 48 h, with (F) statistical analysis
of apoptosis cells for triplicate experiments (n=3). **P<0.01
and ***P<0.001 vs. the miR-NC group. NC, negative control; CCK8,
Cell Counting Kit-8; OD, optical density; Ctrl, non-treated
control; CDDP, cisplatin; PR, propidium iodide; FITC, fluorescein
isothiocyanate; miR, microRNA. |
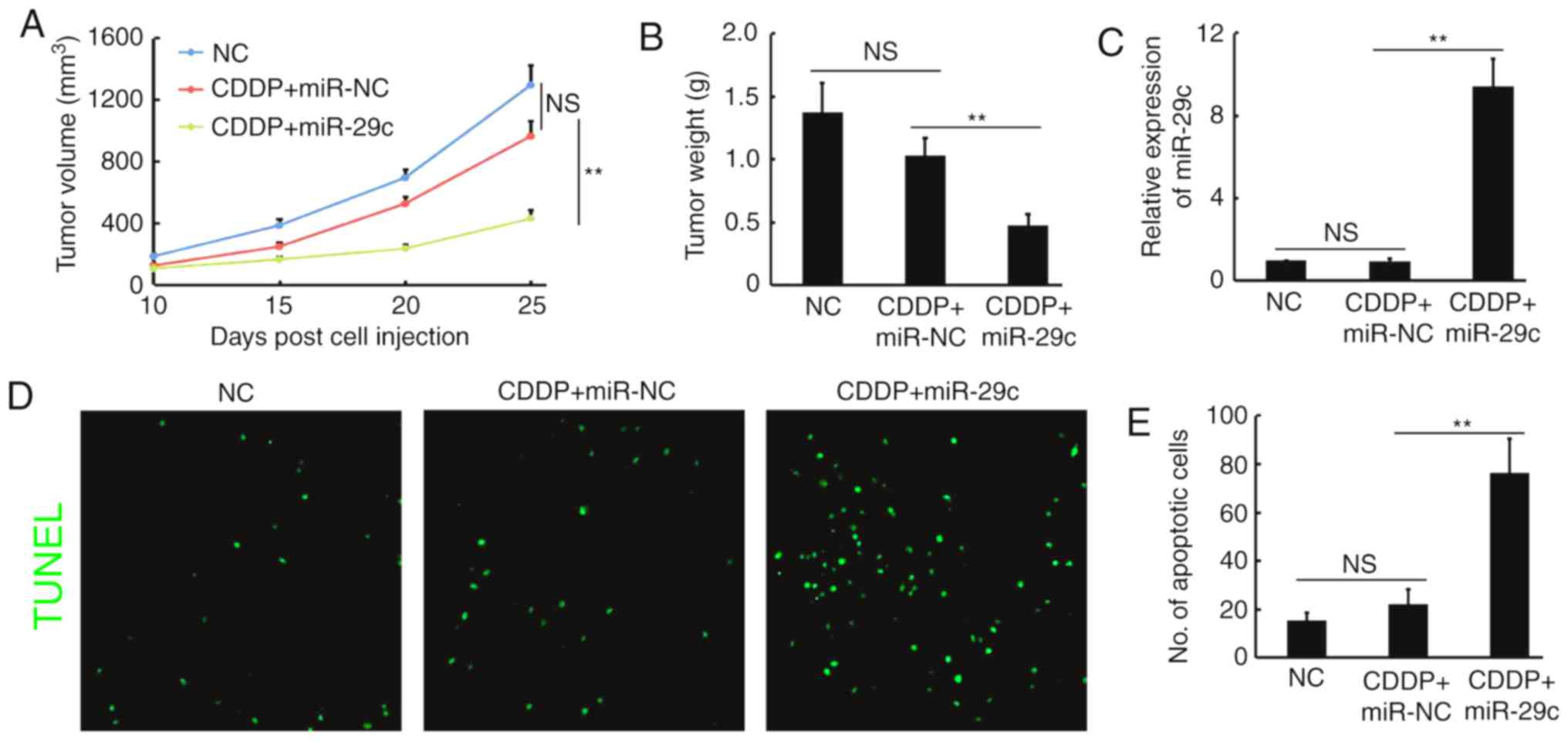
Overexpression of miR-29c restores
CDDP sensitivity in liver cancer in vivo
To validate the observed phenomenon in vivo,
a xenograft tumor model in nude mice was established, using the
selected CDDP-resistant HepG2 cell line with CDDP treatment and
stable expression of either miR-NC or miR-29c. The present study
demonstrated that restoring miR-29c in CDDP-R cells markedly
reduced xenograft tumor growth, including tumor volume (decreased
55.5%; miR-29c vs. miR-NC, 428.4±59.2 vs. 963.2±102.3
mm3) and tumor weight (decreased 53.4%; miR-29c vs.
miR-NC, 0.48±0.09 vs. 1.03±0.14 g), whereas there was no
significant difference between the CDDP + miR-NC and normal saline
control groups (Fig. 3A and B).
RT-qPCR analysis demonstrated that the expression of miR-29c in the
CDDP + miR-29c-treated tumors was 9.42 times higher than that in
the control groups (Fig. 3C). TUNEL
staining revealed a markedly increased number of apoptotic cells
upon CDDP + miR-29c treatment (increased 3.45 times; miR-29c vs.
miR-NC: 76.3±14.3 vs. 22.1±6.2) (Fig. 3D
and E). These results indicated that overexpression of miR-29c
may sensitize CDDP-resistant liver cancer to CDDP in
vivo.
miR-29c directly targets SIRT1 to
enhance CDDP sensitivity of liver cancer
To investigate the possible mechanisms by which
miR-29c restores liver cancer sensitivity to CDDP,
bioinformatics-based prediction was performed using TargetScan and
miRDB (http://mirdb.org), and it was demonstrated that
miR-29c may potentially target the SIRT1 3′-UTR. To investigate if
this was a possible mechanism for miR-29c-mediated restoration of
liver cancer sensitivity to CDDP, the protein level of SIRT1 was
determined and substantially increased SIRT1 protein levels were
observed upon acquisition of CDDP resistance (Fig. 4A). A luciferase assay indicated that
luciferase expression in SIRT1-3′UTR constructs was significantly
affected by miR-29c, whereas no significant reduction was observed
in SIRT1-3′UTR mutant constructs (Fig.
4B). A SIRT1 expression vector was ectopically expressed and
verified that SIRT1 levels were restored (Fig. 4C). The present study assayed for cell
viability and colony formation ability and identified that
overexpression of SIRT1 relieves the effect of miR-29c on cell
proliferation (CDDP + miR-NC, 1.74±0,18; CDDP + miR-29c, 0.89±0.11;
CDDP + miR-29c + SIRT1, 1.82±0.13) and colony numbers (CDDP +
miR-NC, 213±25; CDDP + miR-29c, 42±8; CDDP + miR-29c + SIRT1,
243±34) (Fig. 4D-F). Additionally,
overexpression of SIRT1 restores the effect of miR-29c, which
promotes apoptosis in CDDP-R cell lines (CDDP + miR-NC, 3.38±0.62%;
CDDP + miR-29c, 23.52±4.21%; CDDP+miR-29c+SIRT1, 3.04±0.76%)
(Fig. 4G-H). Collectively, these
findings revealed that miR-29c can target and suppress SIRT1, and
restore sensitivity to CDDP in CDDP-R liver cancer cell lines.
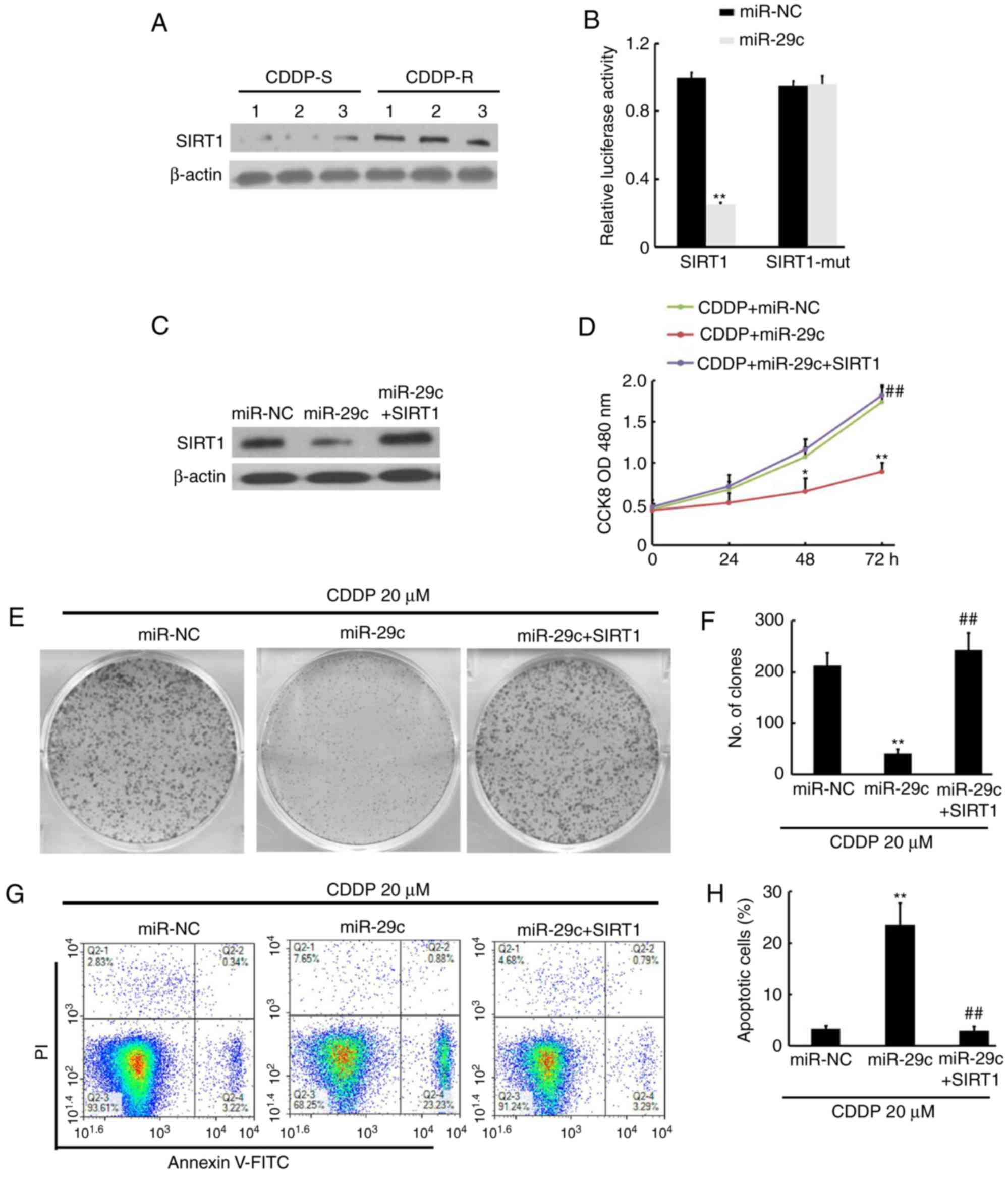 | Figure 4.miR-29c directly targets SIRT1 to
enhance CDDP sensitivity. (A) SIRT1 expression was upregulated in
CDDP-R cells compared with that in CDDP-S cells for triplicate
samples. β-actin was used as a loading control. The relative
expression of SIRT1 was quantified. (B) Relative repression of
luciferase expression was standardized to a transfection control.
**P<0.01 vs. miR-NC. (C) SIRT1 was downregulated by miR-29c and
restored by co-transfection of SIRT1 in CDDP-R cells. The relative
expression of SIRT1 was quantified. (D) miR-29c + CDDP (20 µM)
treatment inhibited cell proliferation compared with CDDP (20 µM) +
miR-NC treatment, and overexpression of SIRT1 restored the cell
proliferation compared with CDDP + miR-29c in CDDP-R cells (n=3).
(E) Crystal violet staining of colony formation for CDDP (20 µM)
plus miR-NC or miR-29c combination treatments or SIRT1 in CDDP-R
cells after 10 days as indicated, with (F) statistical analysis of
colony numbers for triplicate experiments (n=3). (G) Flow
cytometric analysis of apoptosis cells for CDDP (20 µM) plus miR-NC
or miR-29c combination treatments or SIRT1 in CDDP-R cells after 48
h as indicated, with (H) statistical analysis of apoptotic cells
for triplicate experiments (n=3). **P<0.01 (vs. CDDP + miR-NC),
##P<0.01 (vs. CDDP + miR29c). CDDP, cisplatin; S,
sensitive; R, resistant; SIRT1, silent mating type information
regulation 2 homolog 1; NC, negative control run on the same
membranes as the other proteins; FITC, fluorescein isothiocyanate;
miR, microRNA; OD, optical density. |
Discussion
Previous studies have indicated that miRNAs serve a
crucial role in human cancer development (7,26), with
expression profiling of miRNAs being utilized for the
classification of tumor stages and prognoses (27,28). In
the present study, miRNA expression patterns of liver cancer were
screened and miR-29c was identified to be associated with
chemoresistance. Further analysis demonstrated that miR-29c
expression was downregulated in CDDP-R liver cancer cell lines and
tissues compared with that in their CDDP-S counterparts.
The members of the miR-29 family function as tumor
suppressors and are downregulated in several human cancers,
including colon, lung, prostate, and breast cancer (29–32). The
family includes miR-29a, miR-29b and miR-29c, which differ in their
last few 3′-end nucleotides. Earlier studies have demonstrated that
miR-29c acts as a tumor suppressor in gallbladder cancer by
modulating levels of cell cycle regulator proteins (33).
The average miRNA has ~100 target sites and
regulates a large fraction of protein-coding genes (34). miR-29c, which inhibits cell
proliferation, promotes apoptosis and arrests cell cycle at G1/G0
phase by targeting the Nuclear autoantigenic sperm protein, is
downregulated in gastric cancer tissues and cell lines (20). miR-29c inhibits proliferation,
migration and invasion in lung cancer cell lines by targeting
vascular endothelial growth factor A in vitro (35). The present study provides evidence
that miR-29c downregulates SIRT1 by targeting the 3′-UTR of SIRT1
mRNA. Using a series of in vitro and in vivo assays
of liver cancer, cancer cell growth and colony formation were
demonstrated to be significantly decreased by overexpression of
miR-29c, whereas apoptosis was significantly increased, suggesting
that it serves roles in chemoresistant cell proliferation,
apoptosis and tumor growth. The antiproliferative effect of miR-29c
overexpression appears to be associated with a change in SIRT1
expression level in chemoresistant cells over time.
Previous studies demonstrated that SIRT1 expression
levels were positively correlated with tumor grade (36). Depletion of SIRT1 reduced the colony
formation ability of liver cancer cells on soft agar, and xenograft
growth in mice (14,37). Furthermore, patients with
SIRT1-positive liver cancer have a lower survival rate than those
with SIRT1-negative liver cancer (38). Overexpression of SIRT1 has been
demonstrated to contribute to chemoresistance in serous epithelial
ovarian cancer, where it may be a potential prognostic indicator
for patient survival outcome (39).
SIRT1 is one among other genes involved in DNA repair that are
upregulated in platinum-resistant epithelial ovarian cancer
(40).
Collectively, data from the present and previous
studies support a pro-tumorigenic and chemoresistant role for
SIRT1, which may be targeted by miR-29c in liver cancer. As an
miRNA may inhibit more than one target gene, a single gene could be
targeted by multiple miRNAs, the results of the present study
demonstrate only one point of the regulating network that could
impact liver tumor progression.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Author's contributions
PL was involved in the acquisition of the data and
the analysis and interpretation of the data. WZ was involved in the
conception and design of the present study.
Ethics approval and consent to
participate
Procedures involving animals conformed to the
guidelines of the Institutional Animal Care and Use Committee of
Sichuan Academy of Medical Sciences and Sichuan Provincial People's
Hospital (Sichuan, China).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Islami F, Miller KD, Siegel RL, Fedewa SA,
Ward EM and Jemal A: Disparities in liver cancer occurrence in the
United States by race/ethnicity and state. CA Cancer J Clin.
67:273–289. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Herold C, Reck T, Fischler P, Ott R,
Radespiel-Troeger M, Ganslmayer M, Hohenberger W, Hahn EG and
Schuppan D: Prognosis of a large cohort of patients with
hepatocellular carcinoma in a single European centre. Liver.
22:23–28. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Xiong H, Ni Z, He J, Jiang S, Li X, He J,
Gong W, Zheng L, Chen S, Li B, et al: LncRNA HULC triggers
autophagy via stabilizing Sirt1 and attenuates the chemosensitivity
of HCC cells. Oncogene. 36:3528–3540. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang H, Feng Z, Huang R, Xia Z, Xiang G
and Zhang J: MicroRNA-449 suppresses proliferation of hepatoma cell
lines through blockade lipid metabolic pathway related to SIRT1.
Int J Oncol. 45:2143–2152. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bhalla KN: Epigenetic and chromatin
modifiers as targeted therapy of hematologic malignancies. J Clin
Oncol. 23:3971–3993. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bertoli G, Cava C and Castiglioni I:
MicroRNAs: New biomarkers for diagnosis, prognosis, therapy
prediction and therapeutic tools for breast cancer. Theranostics.
5:1122–1143. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Brennecke J, Hipfner DR, Stark A, Russell
RB and Cohen SM: Bantam encodes a developmentally regulated
microRNA that controls cell proliferation and regulates the
proapoptotic gene hid in drosophila. Cell. 113:25–36. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hatfield SD, Shcherbata HR, Fischer KA,
Nakahara K, Carthew RW and Ruohola-Baker H: Stem cell division is
regulated by the microRNA pathway. Nature. 435:974–978. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Pekarsky Y, Santanam U, Cimmino A,
Palamarchuk A, Efanov A, Maximov V, Volinia S, Alder H, Liu CG,
Rassenti L, et al: Tcl1 expression in chronic lymphocytic leukemia
is regulated by miR-29 and miR-181. Cancer Res. 66:11590–11593.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fang F, Chang RM, Yu L, Lei X, Xiao S,
Yang H and Yang LY: MicroRNA-188-5p suppresses tumor cell
proliferation and metastasis by directly targeting FGF5 in
hepatocellular carcinoma. J Hepatol. 63:874–885. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Firestein R, Blander G, Michan S,
Oberdoerffer P, Ogino S, Campbell J, Bhimavarapu A, Luikenhuis S,
de Cabo R, Fuchs C, et al: The SIRT1 deacetylase suppresses
intestinal tumorigenesis and colon cancer growth. PLoS One.
3:e20202008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Guarente L: Calorie restriction and
sirtuins revisited. Genes Dev. 27:2072–2085. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen J, Zhang B, Wong N, Lo AW, To KF,
Chan AW, Ng MH, Ho CY, Cheng SH, Lai PB, et al: Sirtuin 1 is
upregulated in a subset of hepatocellular carcinomas where it is
essential for telomere maintenance and tumor cell growth. Cancer
Res. 71:4138–4149. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Portmann S, Fahrner R, Lechleiter A, Keogh
A, Overney S, Laemmle A, Mikami K, Montani M, Tschan MP, Candinas D
and Stroka D: Antitumor effect of SIRT1 inhibition in human HCC
tumor models in vitro and in vivo. Mol Cancer Ther. 12:499–508.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Dong CW, Wang YX, Du FT, Ding W and Hu SY:
Low miR-29c expression is a prognostic marker in hepatocellular
carcinoma. Genet Mol Res. 15:2016. View Article : Google Scholar
|
|
17
|
Lu Y, Hu J, Sun W, Li S, Deng S and Li M:
MiR-29c inhibits cell growth, invasion, and migration of pancreatic
cancer by targeting ITGB1. Onco Targets Ther. 9:99–109.
2015.PubMed/NCBI
|
|
18
|
Zhao X, Li J, Huang S, Wan X, Luo H and Wu
D: MiRNA-29c regulates cell growth and invasion by targeting CDK6
in bladder cancer. Am J Transl Res. 7:1382–1389. 2015.PubMed/NCBI
|
|
19
|
Wang B, Li D, Sidler C, Rodriguez-Juarez
R, Singh N, Heyns M, Ilnytskyy Y, Bronson RT and Kovalchuk O: A
suppressive role of ionizing radiation-responsive miR-29c in the
development of liver carcinoma via targeting WIP1. Oncotarget.
6:9937–9950. 2015.PubMed/NCBI
|
|
20
|
Yu B, Chen X, Li J, Gu Q, Zhu Z, Li C, Su
L and Liu B: microRNA-29c inhibits cell proliferation by targeting
NASP in human gastric cancer. BMC Cancer. 17:1092017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lopez-Terrada D, Cheung SW, Finegold MJ
and Knowles BB: Hep G2 is a hepatoblastoma-derived cell line. Hum
Pathol. 40:1512–1515. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Luo P, He T, Jiang R and Li G:
MicroRNA-423-5p targets O-GlcNAc transferase to induce apoptosis in
cardiomyocytes. Mol Med Rep. 12:1163–1168. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cheng L, Yang Q, Li C, Dai L, Yang Y, Wang
Q, Ding Y, Zhang J, Liu L, Zhang S, et al: DDA1, a novel oncogene,
promotes lung cancer progression through regulation of cell cycle.
J Cell Mol Med. 21:1532–1544. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Dai L, Cui X, Zhang X, Cheng L, Liu Y,
Yang Y, Fan P, Wang Q, Lin Y, Zhang J, et al: SARI inhibits
angiogenesis and tumour growth of human colon cancer through
directly targeting ceruloplasmin. Nat Commun. 7:119962016.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chang Y, Liu C, Yang J, Liu G, Feng F,
Tang J, Hu L, Li L, Jiang F, Chen C, et al: MiR-20a triggers
metastasis of gallbladder carcinoma. J Hepatol. 59:518–527. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yang X, Zhang XF, Lu X, Jia HL, Liang L,
Dong QZ, Ye QH and Qin LX: MicroRNA-26a suppresses angiogenesis in
human hepatocellular carcinoma by targeting hepatocyte growth
factor-cMet pathway. Hepatology. 59:1874–1885. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Altman DG, McShane LM, Sauerbrei W and
Taube SE: Reporting recommendations for tumor marker prognostic
studies (REMARK): Explanation and elaboration. PLoS Med.
9:e10012162012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cummins JM, He Y, Leary RJ, Pagliarini R,
Diaz LA Jr, Sjoblom T, Barad O, Bentwich Z, Szafranska AE,
Labourier E, et al: The colorectal microRNAome. Proc Natl Acad Sci
USA. 103:3687–3692. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Fabbri M, Garzon R, Cimmino A, Liu Z,
Zanesi N, Callegari E, Liu S, Alder H, Costinean S,
Fernandez-Cymering C, et al: MicroRNA-29 family reverts aberrant
methylation in lung cancer by targeting DNA methyltransferases 3A
and 3B. Proc Natl Acad Sci USA. 104:15805–15810. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Porkka KP, Pfeiffer MJ, Waltering KK,
Vessella RL, Tammela TL and Visakorpi T: MicroRNA expression
profiling in prostate cancer. Cancer Res. 67:6130–6135. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Iorio MV, Ferracin M, Liu CG, Veronese A,
Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M,
et al: MicroRNA gene expression deregulation in human breast
cancer. Cancer Res. 65:7065–7070. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Pang L, Gu W, Wang N, Hu J, Cui X, Zhang
J, Zhao J, Liu C, Zhang W, Zou H, et al: MicroRNA-29c-5p suppresses
gallbladder carcinoma progression by directly targeting CPEB4 and
inhibiting the MAPK pathway. Clin Exp Pharmacol Physiol.
24:445–457. 2017.(In Chinese).
|
|
34
|
Brennecke J, Stark A, Russell RB and Cohen
SM: Principles of microRNA-target recognition. PLoS Biol.
3:e852005. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liu L, Bi N, Wu L, Ding X, Men Y, Zhou W,
Li L, Zhang W, Shi S, Song Y and Wang L: MicroRNA-29c functions as
a tumor suppressor by targeting VEGFA in lung adenocarcinoma. Mol
Cancer. 16:502017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chen HC, Jeng YM, Yuan RH, Hsu HC and Chen
YL: SIRT1 promotes tumorigenesis and resistance to chemotherapy in
hepatocellular carcinoma and its expression predicts poor
prognosis. Ann Surg Oncol. 19:2011–2019. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wu Y, Meng X, Huang C and Li J: Emerging
role of silent information regulator 1 (SIRT1) in hepatocellular
carcinoma: A potential therapeutic target. Tumour Biol.
36:4063–4074. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Choi HN, Bae JS, Jamiyandorj U, Noh SJ,
Park HS, Jang KY, Chung MJ, Kang MJ, Lee DG and Moon WS: Expression
and role of SIRT1 in hepatocellular carcinoma. Oncol Rep.
26:503–510. 2011.PubMed/NCBI
|
|
39
|
Shuang T, Wang M, Zhou Y and Shi C:
Over-expression of Sirt1 contributes to chemoresistance and
indicates poor prognosis in serous epithelial ovarian cancer (EOC).
Med Oncol. 32:2602015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ziebarth AJ, Nowsheen S, Steg AD, Shah MM,
Katre AA, Dobbin ZC, Han HD, Lopez-Berestein G, Sood AK, Conner M,
et al: Endoglin (CD105) contributes to platinum resistance and is a
target for tumor-specific therapy in epithelial ovarian cancer.
Clin Cancer Res. 19:170–182. 2013. View Article : Google Scholar : PubMed/NCBI
|