Introduction
Gastric cancer is the most common cancer, with the
fourth highest incidence rate of all gastric adenocarcinomas and
>723,000 mortalities every year worldwide (1). Despite the decreased incidence and
mortality rates due to the major improved diagnosis and treatment,
the ≤5-year survival rate is <20% (1). This may be due to lack of understanding
of the mechanisms underlying the development and prognosis of
gastric cancer. Therefore, it is urgent to develop effective
therapeutic approaches for gastric cancer.
Nuclear receptor subfamily 1 group D member 1
(REV-ERBα) belongs to the nuclear hormone receptor family (2), which is abundantly expressed in liver,
adipose, muscle and brain tissue. REV-ERBα serves an important role
in regulating lipid metabolism, inflammatory responses and
circadian rhythm (2–4). Previous studies have demonstrated that
REV-ERBα may modulate the proliferation and apoptosis of
HER2+ breast cancer cells, which is associated with poor
clinical outcomes and survival (5,6). However,
to the best of our knowledge, there are no studies regarding the
regulation of REV-ERBα in gastric cancer. It is also unknown
whether REV-ERBα alterations are associated with the
clinicopathological factors and prognosis of human gastric cancer.
We hypothesize that the levels of REV-ERBα in gastric cancer are
altered compared with normal tissues, and is associated with the
clinicopathological features and prognosis of this disease. To
examine this hypothesis, samples from patients who were diagnosed
with gastric cancer were utilized, and the REV-ERBα gene and
protein levels in normal and cancer tissues in those patients were
compared. The associations between REV-ERBα expression with the
Tumor-Node-Metastasis (TMN) stages and survival times were also
analyzed in patients with gastric cancer. Finally, human gastric
cancer cells were treated with REV-ERBα activator GSK4112 to
determine its effects on apoptosis. Conclusively, the reduction of
REV-ERBα was associated with clinical features and prognosis in
gastric cancer, and REV-ERBα agonist resulted in apoptosis in
gastric cancer cells.
Materials and methods
Patients and tissues collection
All samples were obtained from 74 patients with
diagnosed gastric cancer who underwent surgery (surgical resection)
at The First Affiliated Hospital of Anhui Medical University in
2014, as previously described (7).
The median age of the study population was 63.4 years (range, 33–84
years) and the sex distribution was 58 males and 16 females. The
clinical characteristics are summarized in Table I. None of the patients had received
any other therapies, including radiotherapy or chemotherapy prior
to surgery. The hepatic, renal and bone marrow functions of all
patients were normal, and the Eastern Cooperative Oncology Group
Performance Status scores were between 0–2 (8). Patients with abnormal function rest
results, and those who were pregnant or breast-feeding were
excluded.
 | Table I.Association between REV-ERBα
expression and clinicopathological variables. |
Table I.
Association between REV-ERBα
expression and clinicopathological variables.
|
|
| REV-ERBα expression
level, n |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
variables | Numbers of patients,
n (n=74) | Low (n=43) | High (n=31) | P-value |
|---|
| Sex |
|
|
|
|
| Male | 58 | 35 | 23 | 0.322 |
|
Female | 16 | 8 | 8 |
|
| Age, years |
|
|
|
|
|
<60 | 23 | 16 | 7 | 0.138 |
| ≥60 | 51 | 27 | 24 |
|
| Primary tumor
site |
|
|
|
|
| Gastric
cardia or fundus | 36 | 22 | 14 | 0.392 |
| Gastric
antrum or body | 38 | 21 | 17 |
|
| Diameter of tumor,
cm |
|
|
|
|
|
<5 | 31 | 18 | 13 | 0.995 |
| ≥5 | 43 | 25 | 18 |
|
| Level of
differentiation |
|
|
|
|
|
Moderate | 22 | 8 | 14 | 0.009 |
| Poor | 52 | 37 | 15 |
|
| T stage |
|
|
|
|
|
T1-T2 | 21 | 5 | 16 | 0.001 |
|
T3-T4 | 53 | 39 | 14 |
|
| TNM stage |
|
|
|
|
| I–II | 18 | 4 | 14 | 0.001 |
|
III–IV | 56 | 40 | 16 |
|
| Lymph node
metastasis |
|
|
|
|
|
Present | 58 | 41 | 17 | 0.007 |
|
Absent | 16 | 5 | 11 |
|
All patients provided written informed consent. The
study protocol was approved by the Clinical Research Ethics
Committee of Anhui Medical University (Hefei, China). All methods
were performed in accordance with the human ethics guidelines in
the clinical research project (9).
Cell culture
The human gastric cancer SGC-7,901 [American Type
Culture Collection (ATCC), Manassas, VA, USA] and BGC-823 (ATCC)
cell lines and human normal gastric epithelial GES-1 (ATCC) cell
line were employed to detect REV-ERBα expression. Cells were
maintained in Dulbecco's modified Eagle's medium (DMEM; Gibco;
Thermo Fisher Scientific, Inc., Waltham, MA, USA) containing high
glucose, 1 mmol/l L-glutamine, pyridoxine hydrochloride, 110 mg/l
sodium pyruvate and bicarbonate. Additionally, 10% heat inactivated
foetal calf serum (Clark Bioscience, Richmond, VA, USA), 100 U/ml
penicillin and 100 µg/ml streptomycin were added to the DMEM. Cells
were cultured at 37°C in a humidified atmosphere of 5%
CO2.
Immunohistochemistry
REV-ERBα expression in human gastric cancer and
normal tissues (5 cm from the tumor site) was measured by
immunohistochemistry as described previously (7). Immunohistochemistry was performed on
4-µm-thick sections from 10% formalin-fixed paraffin-embedded
tissue specimens. Sections were deparaffinized with 100% xylene at
25°C for 10 min and removed xylene through a graded series (100, 85
and 70% ethanol) and were subjected to microwaving with 10 mm
citrate buffer (pH=6.0) at 100°C for 5 min. Briefly, the sections
were blocked with 3% hydrogen peroxide and 10% normal goat serum
(Clark Bioscience) each for 10 min at room temperature, and then
incubated with REV-ERBα rabbit antibody (cat no. AB174309; 1:100
dilution; Abcam, Cambridge, MA, USA) overnight at 4°C. Following
incubation with a biotin-conjugated secondary antibody (cat. no.
PV6000; 1:100 dilution; ZSGB-BIO; OriGene Technologies, Inc.,
Beijing, China), the tissue slides were incubated with a
streptavidin-biotin horseradish peroxidase complex for 30 min at
room temperature, followed by incubation with 3,3′-diaminobenzidine
(ZSGB-BIO; OriGene Technologies, Inc.) for 5 min at room
temperature. The counterstaining with 20% hematoxylin was then
performed for 60 sec at room temperature, and images of stained
samples were captured in a single-blinded manner under a
fluorescent microscope at a magnification of ×200. The relative
protein expression of all images was calculated using mean optical
density units, and the sequences were analyzed using IPWIN
Application software version 6.0.0260 (Media Cybernetics, Inc.,
Rockville, MD, USA). The staining intensity was scored according to
the number of cells: 0, no staining; 1 (≤25%), weakly stained; 2
(25–50%), moderately stained; or 3 (≥50%), markedly stained. A low
REV-ERBα expression was defined as score ‘0’, ‘1’ or ‘2’, and a
high REV-ERBα expression was defined as score ‘3’. The patients
were divided into two groups: The low expression group (n=43) and
high expression group (n=31), as summarized in Table I.
Western blot analysis
Gastric tissues and cells were lysed in lysis buffer
(25 mM HEPES, 2 mM MgCl2, 2 mM DTT, 1 mM EDTA, 1 mM PSMF, 5 µg/ml
leupeptin, pH 7.4). Subsequent to freeze-thawing the suspension
liquid containing the extracted protein 3 times, the lysates were
centrifuged at 32,869.2 × g at 4°C for 30 min. The concentration of
all the extracted protein was determined by the BCA assay. The
protein extracts (10 µl) were separated by 12% SDS-PAGE and
transferred to polyvinylidene fluoride (PVDF) membranes. Following
non-specific blocking with 5% skimmed milk at room temperature for
2 h, PVDF membranes were washed 2 times with TBST (TBS contained
0.1% Tween-20) for 10 min each time. The membranes were then
incubated with primary antibodies against: REV-ERBα (rabbit; cat
no. AB174309; 1:500 dilution; Abcam); cleaved caspase-3 (mouse; cat
no. SC70497; 1:500 dilution; Santa Cruz Biotechnology, Inc.,
Dallas, TX, USA); B-cell lymphoma 2 (Bcl-2; mouse; cat no. SC23960;
1:500 dilution; Santa Cruz Biotechnology, Inc.); Bcl-2-associated X
protein (Bax; mouse; cat no. SC23959; 1:500 dilution; Santa Cruz
Biotechnology, Inc.); and β-actin (mouse; cat no. AB8226; 1:1,000
dilution; Abcam) overnight at 4°C, and then washed 3 times with
TBST for 10 min each time. The membranes were incubated with the
corresponding Goat anti-mouse horseradish peroxidase-conjugated
secondary antibody (mouse; cat no. AP124P; 1:1,000 dilution; EMD
Millipore, Billerica, MA, USA) or rabbit anti-Goat horseradish
peroxidase-conjugated secondary antibody (rabbit; cat no. AP106P;
1:1,000 dilution; EMD Millipore) for 2 h at 20°C, and then washed 3
times with TBST again for 10 min each time. Finally, the detection
of the molecules of interest was performed using enhanced
chemiluminescence (Beyotime Institute of Biotechnology, Haimen,
China). The bands were quantified to calculate relative protein
expression levels using Quantity one software version 4.99.5.2.0
(Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Reverse transcription quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from human tissues and cells
using TRIzol® (Life Technologies; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. Total
RNA was reverse transcribed by cDNA synthesis using a PrimeScript
RT Reagent kit with gDNA Eraser (Perfect Real Time; Takara Bio,
Inc., Otsu, Japan) at 37°C for 30 min and 85°C for 5 sec. qPCR was
performed using a 7,900 Thermal Cycler (ABI, Applied Biosystems;
Thermo Fisher Scientific, Inc.) with GoTaq® Green Master
Mix (Promega Corporation, Madison, WI USA) at an initial
denaturation at 95°C for 30 sec, followed by 40 cycles of
denaturation for 5 sec at 95°C, annealing for 30 sec at 60°C and
extension for 15 sec at 72°C. The qPCR primers for REV-ERBα were
5′-ACAGAATCGAACTCTGCACTTCT-3′ (forward) and
5′-GGGGAGGGAGGCAGGTATT-3′ (reverse) (10). The primers for β-actin were
5′-CATGTACGTTGCTATCCAGGC-3′ (forward) and
5′-CTCCTTAATGTCACGCACGAT-3 (reverse). The cycle threshold (Cq)
values were obtained in each sample. Relative levels of mRNA were
determined using the 2−ΔΔCq method (10). β-actin was used as an internal gene
for normalization.
Morphological measurement of
apoptosis
The morphological changes associated with apoptosis
were assayed using fluorescence microscopy following Hoechst33258
staining. The number of cells was counted in five random fields
under a fluorescence microscope at a magnification of ×200.
Briefly, SGC-7901 and BGC-823 cells (3,500 cells/well) were fixed
for 30 min at room temperature in 70% ethanol, followed by
Hoechst33258 (10 µg/ml) staining for 2 min at 37°C and then
visualization using the UV fluorescence microscope. Apoptotic cells
were considered as cells exhibiting nuclear and cytoplasmic
shrinkage, chromatin condensation and apoptotic bodies. A minimum
of 400 cells were counted, and the percentage of apoptotic cells,
or the apoptotic index, was calculated as described previously
(11).
Statistical analysis
Data are expressed as the mean ± standard deviation.
Data analysis was performed using SPSS 17.0 software (SPSS, Inc.,
Chicago, IL, USA). Comparison between different groups was
performed using analysis of variance. The Student-Newman-Keuls test
was the post-hoc test used following analysis of variance. The
χ2 test was utilized to analyze the associations between
REV-ERBα expression levels and clinicopathological variables and
survival time. A Kaplan-Meier test was utilized to describe
survival curves and the log-rank test was used to analyze survival
curves. P<0.05 was considered to indicate a statistically
significant difference.
Results
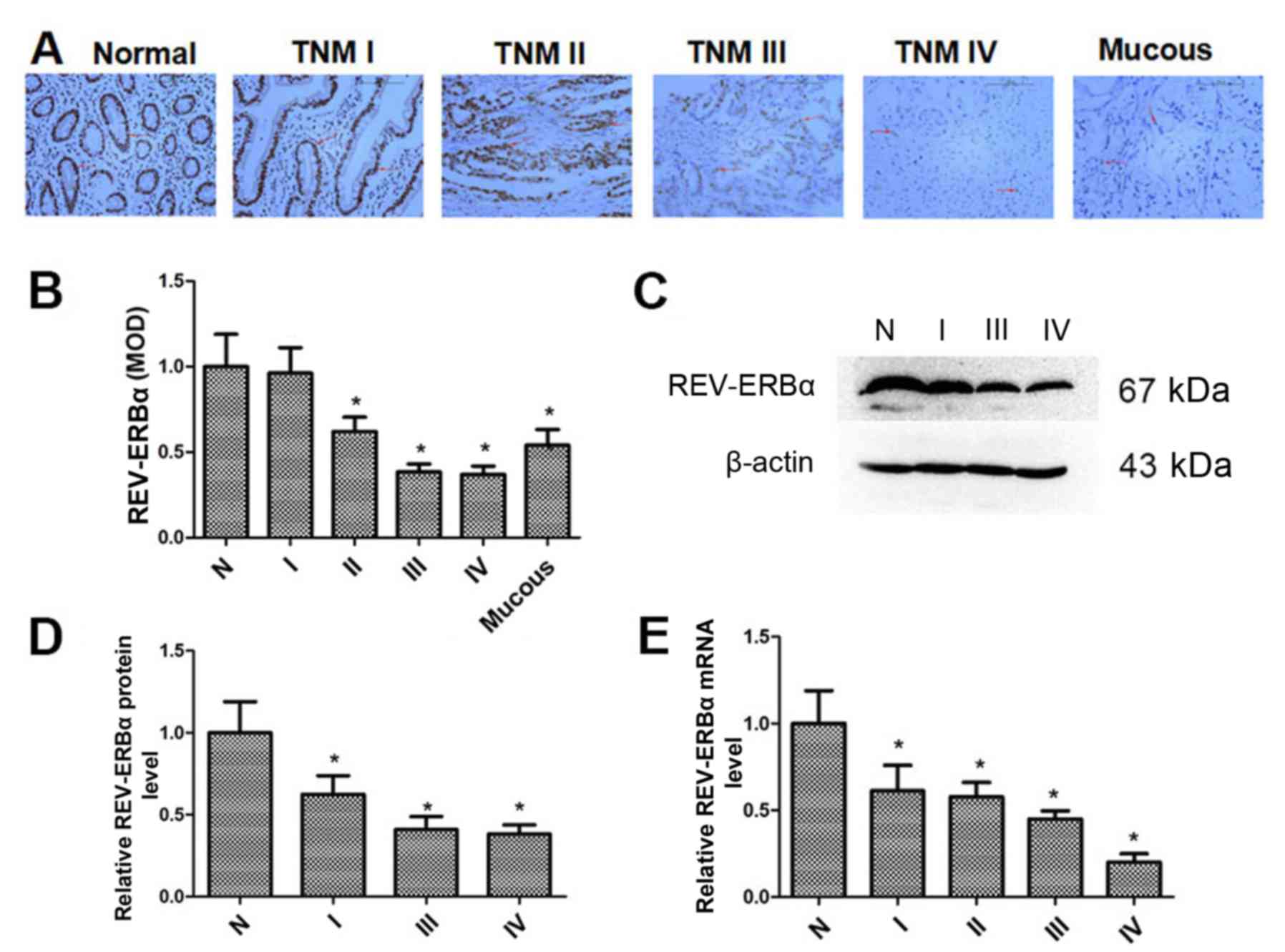
REV-ERBα expression is decreased in
human gastric cancer tissues
The REV-ERBα expression levels in normal gastric and
mucous gastric cancer tissues, and gastric cancer tissues with
different TNM stages, were determined by immunohistochemistry. As
demonstrated in Fig. 1A and B,
REV-ERBα protein levels were decreased in gastric cancer tissues,
which was associated with increased TNM stage. Additionally,
REV-ERBα expression in mucous gastric cancer tissues was also
decreased compared with normal gastric tissues. Furthermore, the
REV-ERBα expression in normal gastric and gastric cancer tissues
was confirmed by western blot analysis (Fig. 1C and D). The mRNA levels of REV-ERBα
were also measured by RT-qPCR (Fig.
1E). It was identified that the levels of REV-ERBα mRNA were
also decreased in gastric cancer tissues, which was associated with
incremental TNM stage. These results suggest that REV-ERBα levels
are decreased significantly in gastric cancer tissues with higher
TNM stages.
Association between REV-ERBα
expression and clinicopathological factors in gastric cancer
The association between the REV-ERBα expression and
clinicopathological factors was analyzed using immunohistochemistry
data (Table I). A low expression of
REV-ERBα was significantly associated with poor differentiation
(P=0.009), T stage (P=0.001), TMN stage (P=0.001) and lymph node
metastasis (P=0.007). The results indicated that REV-ERBα level is
associated with the progression of gastric cancer.
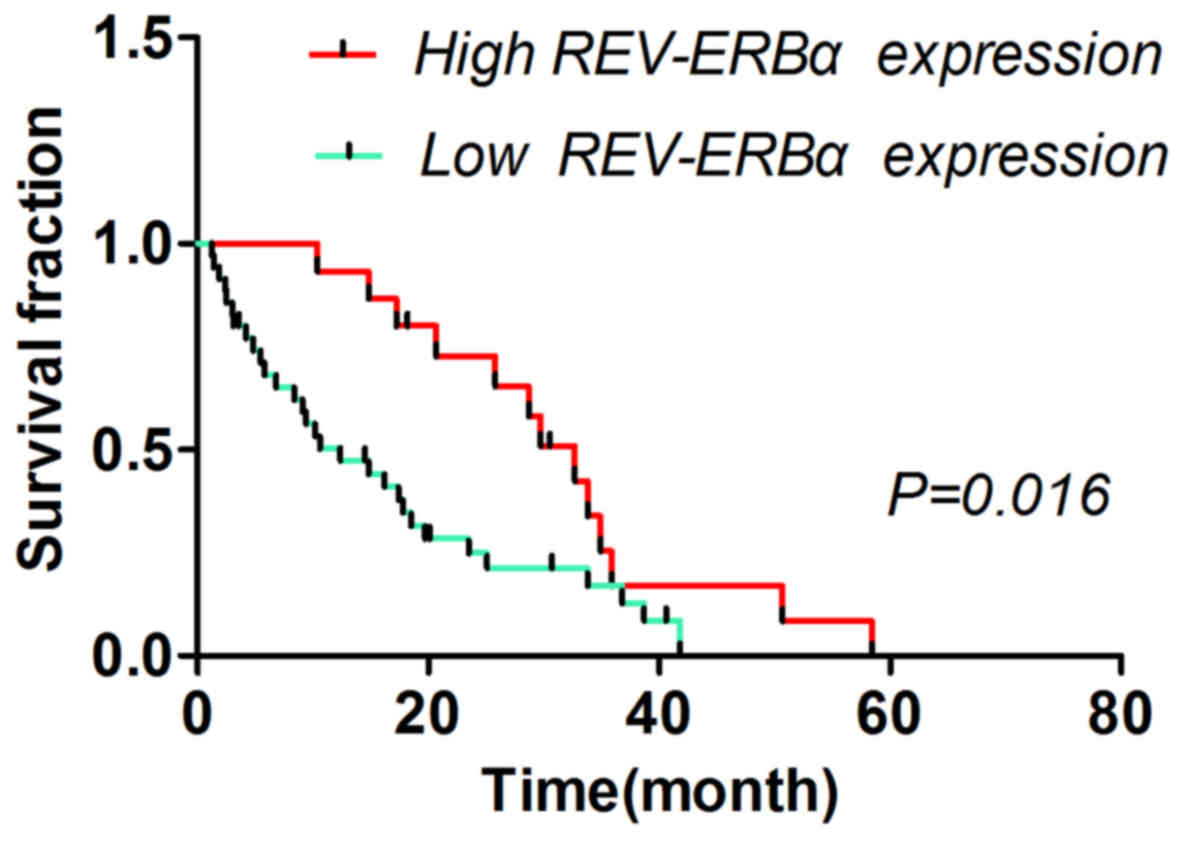
Association between REV-ERBα
expression and survival time of patients with gastric cancer
The different survival times of patients with low
and high REV-ERBα expression levels are summarized in Table II. The 3- and 5-year survival times
of patients were significantly associated with REV-ERBα expression
(P=0.009 and P=0.002, respectively). The patients with low REV-ERBα
expression exhibited poor prognosis (P<0.05) compared with
patients with high REV-ERBα expression (Fig. 2).
 | Table II.Association between REV-ERBα
expression and survival time of patients with gastric cancer. |
Table II.
Association between REV-ERBα
expression and survival time of patients with gastric cancer.
|
| 3-year survival
time | 5-year survival
time |
|---|
|
|
|
|
|---|
| REV-ERBα
expression | Survival, n | Mortality, n | P-value | Survival, n | Mortality, n | P-value |
|---|
| Low | 12 | 31 | 0.009 | 8 | 35 | 0.002 |
| High | 18 | 13 | – | 16 | 15 | – |
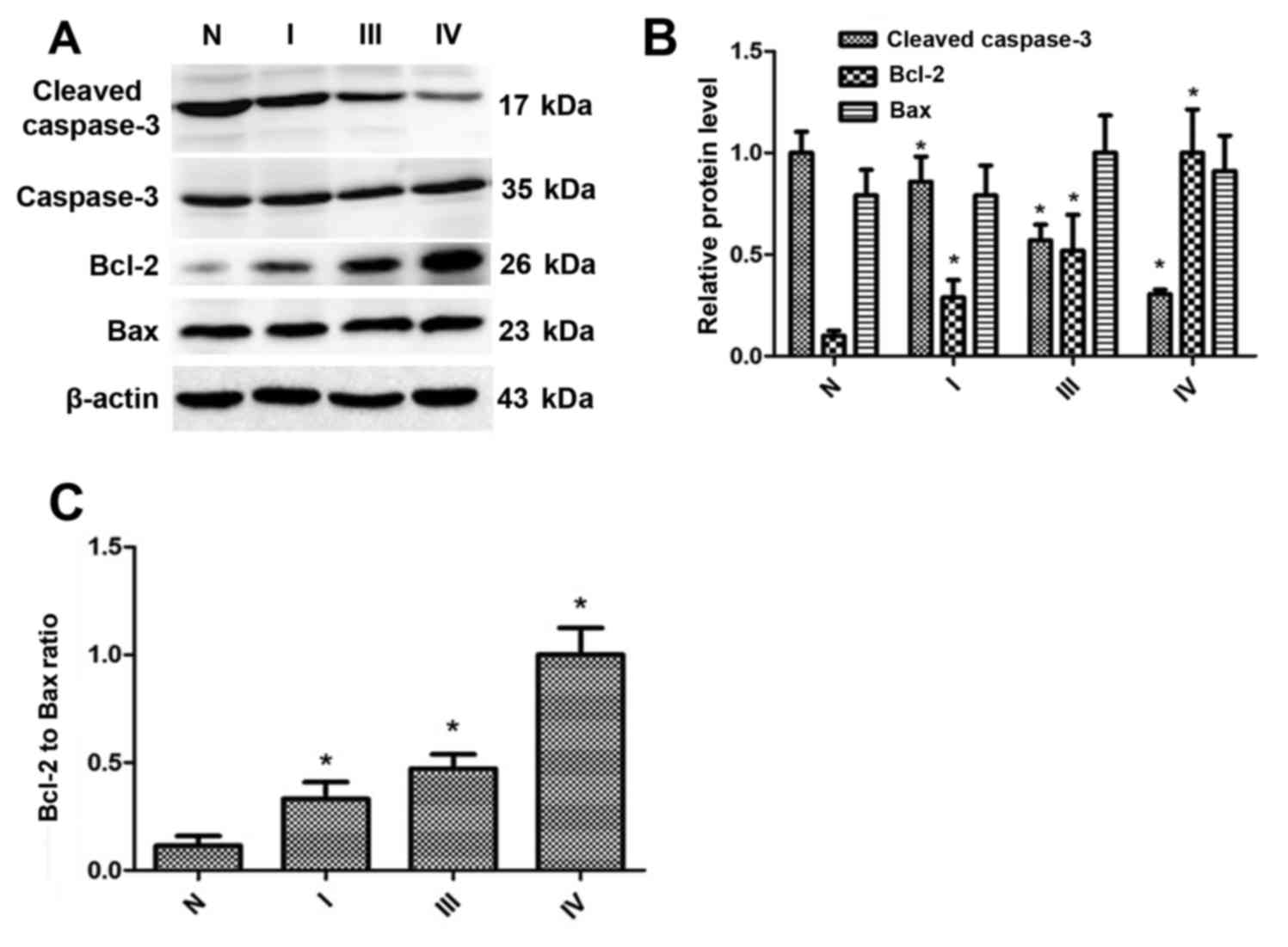
Cleaved caspase-3 expression is
downregulated, whereas the Bcl-2 expression and Bcl-2/Bax are
upregulated, in gastric cancer tissues
To determine whether REV-ERBα was associated with
the expression of cleaved caspase-3, Bcl-2 and Bax, western blot
analysis was employed to detect the levels of these proteins. As
indicated in Fig. 3A and B, the
expression levels of cleaved caspase-3 were downregulated in
gastric cancer tissues, which was associated with enhanced TNM
stage. The Bcl-2 expression levels were upregulated in gastric
cancer tissues, which were also associated with enhanced TNM stage.
Additionally, the ratio of Bcl-2 to Bax, according to the
densitometry of the western blot analysis bands (Fig. 3C), was increased in gastric cancer
tissues compared with normal gastric tissues. Therefore, these
results suggest that the level of REV-ERBα is decreased in human
gastric cancer, which is associated with decreased levels of
apoptosis.
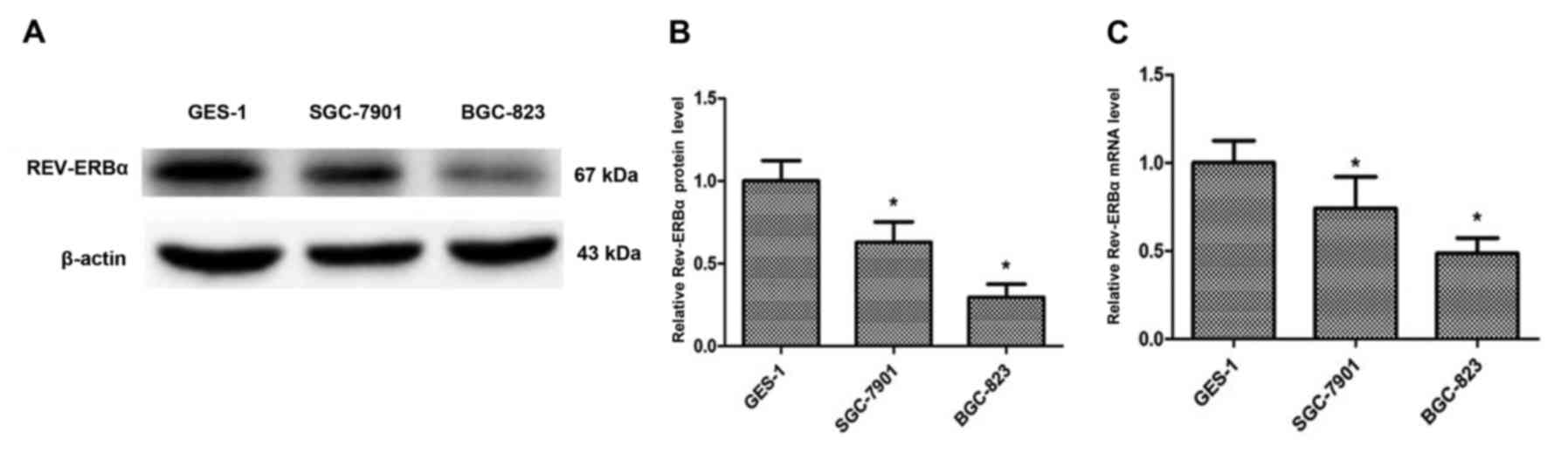
REV-ERBα expression is decreased in
human gastric cancer cells, and activation of REV-ERBα causes
apoptosis in gastric cancer cells
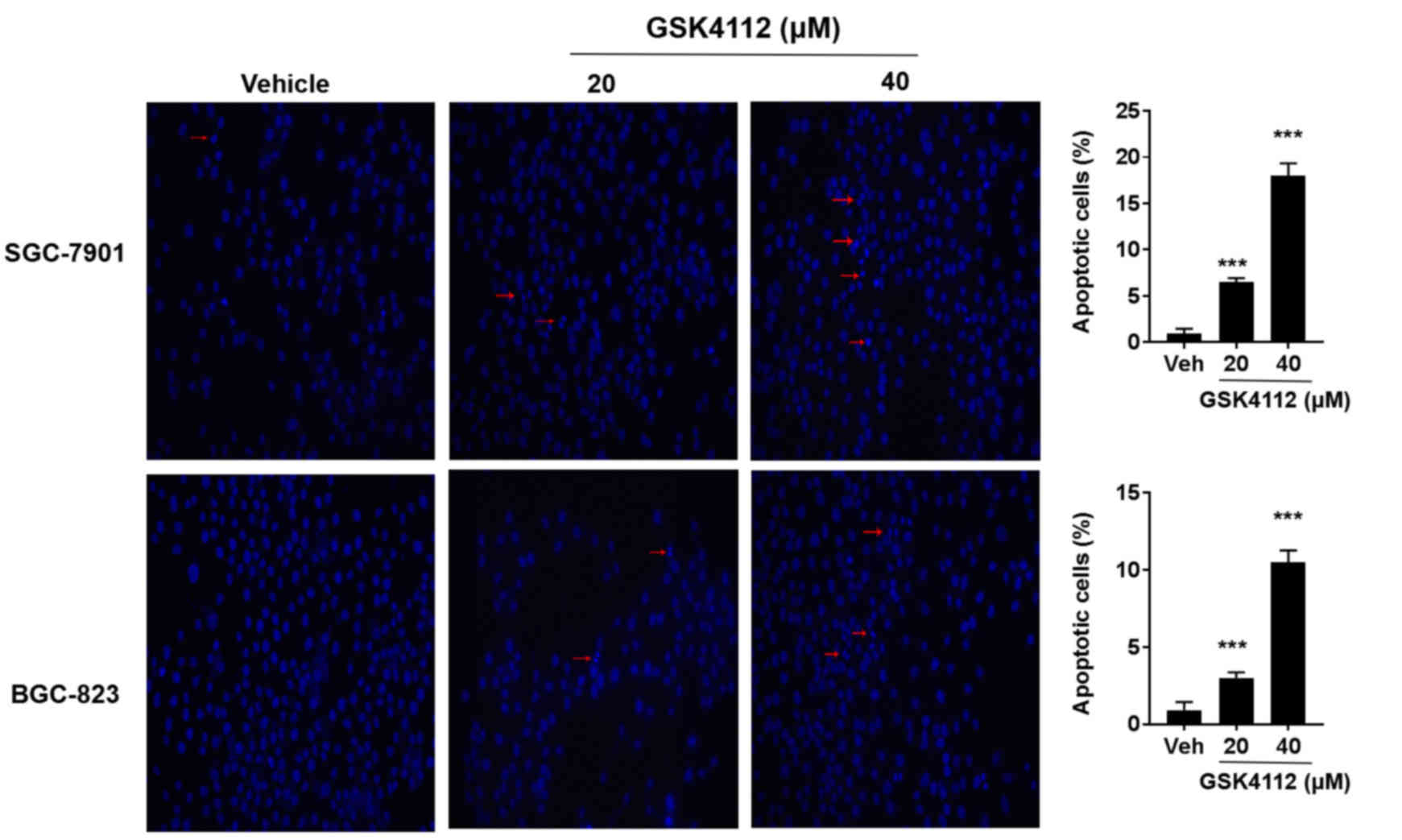
To additionally investigate the REV-ERBα expression
in gastric cancer, GES-1, SGC-7901 and BGC-823 cell lines were
employed to determine the expression level of REV-ERBα (Fig. 4A and B). The REV-ERBα expression
levels were significantly decreased in SGC-7901 (moderately
differentiated) and BGC-823 (undifferentiated) compared with the
GES-1 cell line. The mRNA levels of REV-ERBα were also decreased in
SGC-7901 and BGC-823 cells compared with the GES-1 cell line
(Fig. 4C). Furthermore, SGC-7901 and
BGC-823 cells were treated with the REV-ERBα activator GSK4112 (20
and 40 µM; MedChemExpress, Monmouth Junction, NJ, USA), and it was
identified that GSK4112 treatment for 48 h at 37°C caused an
increase in apoptosis in a dose-dependent manner (Fig. 5). Therefore, these results suggest
that REV-ERBα activation induces apoptosis in human gastric cancer
cells.
Discussion
REV-ERBs were originally regarded as orphan
receptors to regulate gene transcription in response to
multifarious environmental stimuli (12). The members of REV-ERB family,
including REV-ERBα and REV-ERBβ, exhibit abundant and overlapping
expression in adipose, muscle, brain and liver tissues (13). However, REV-ERBα is broadly expressed
at the similar level in a number of different tissues, whereas
REV-ERBβ is highly expressed in parts of the brain, including the
pineal gland and prefrontal cortex, thyroid, uterus and pituitary
(12).
REV-ERBα modulates the circadian rhythm by directly
activating the expression of key circadian clock genes, including
Clock circadian regulator, Brain and muscle ANRT-like 1,
Cryptochrome and Period (14–17). Epidemiological data indicate that
disruption of the circadian clock is associated with an increased
risk of development of breast cancer (18,19).
Dysregulation of the circadian clock genes may have complex effects
on energy homeostasis, potentially resulting in metabolic disorders
including cancer (20,21). The present study identified that
REV-ERBα expression was significantly decreased in human gastric
cancer tissues, which was associated with different
clinicopathological stages. This was in agreement with the results
that the levels of REV-ERBα were additionally decreased in
undifferentiated BGC-823 cells compared with moderately
differentiated SGC-7,901 cells. These data suggest the possibility
of REV-ERBα as a biomarker of gastric cancer.
It has also been demonstrated that the REV-ERBα
expression leading to apoptosis was affected by circadian rhythms
(22). REV-ERBα controls the
circadian clock genes and regulates Early growth response 1
expression, causing extensive expression of Tumor protein p73 (p73)
(23). p73 activates the
transcription of Bax and Bcl-2, causing the release of cytochrome c
from the mitochondria (24).
Apoptosome assembly, and the eventual cleavage and activation of
transducer caspase-9, in turn cleaves and activates executioner
caspase-3. Hence, the circadian clock has a regulating role in the
expression of apoptosis factors (Caspase-3, Bcl-2 and Bax) and the
activation of the apoptosis pathway (24). The present study identified that
REV-ERBα expression was decreased in human gastric cancer, and that
this decrease may protect against apoptosis by decreasing the
levels of cleaved caspase-3 and increasing Bcl-2 levels. However,
one limitation of the present study is that there were only 74
patients included. A larger human cohort is required to confirm
these results. It was also unknown how REV-ERBα regulates the
apoptosis pathway and expression of apoptosis-associated factors,
including cleaved caspase-3, Bcl-2 and Bax, in gastric cancer.
It has been demonstrated previously that REV-ERBα
regulates lipid metabolism during proliferation and apoptosis
(2,25,26). The
preliminary data from the present study suggested that REV-ERBα
decreased glycolysis levels in gastric cancer cells (data not
shown). Nevertheless, it is unclear whether a REV-ERBα-mediated
decrease of glycolysis is beneficial for therapy in gastric
cancer.
In summary, the present study employed
immunohistochemistry, western blot analysis and RT-qPCR methods,
and identified that REV-ERBα expression was decreased in human
gastric cancer tissues. The data indicated that REV-ERBα expression
was significantly associated with poor differentiation, T stage,
TMN stage and lymph node metastasis in human gastric cancer.
Additionally, the survival time of patients was significantly
associated with REV-ERBα expression, suggesting that REV-ERBα may
be an independent prognosis factor in gastric cancer. Furthermore,
REV-ERBα activation induced apoptosis in human gastric cancer
cells. Therefore, REV-ERBα is a potential biomarker for tumor
development and prognosis, and a potential therapeutic target for
gastric cancer.
Acknowledgements
Not applicable.
Funding
The present study was funded by the Natural Science
Foundation of Anhui Province (grant no. 1608085MH182).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
XSW and ZW designed the study. NW performed the
immunohistochemistry to determine the expression of REV-ERBα. XSW
and XW conducted the western blot analysis. XW also performed the
cell proliferation and morphological measurements of apoptosis. HY
performed RT-qPCR to detect the expression of REV-ERBα. XSW drafted
the manuscript.
Ethics approval and consent to
participate
The study protocol was approved by the Clinical
Research Ethics Committee of Anhui Medical University (Hefei,
China). Informed consent was obtained from all individual
participants included in the study.
Consent for publication
Written informed consent was obtained from all
patients for the publication of their data.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cho H, Zhao X, Hatori M, Yu RT, Barish GD,
Lam MT, Chong LW, DiTacchio L, Atkins AR, Glass CK, et al:
Regulation of circadian behaviour and metabolism by REV-ERB-α and
REV-ERB-β. Nature. 485:123–127. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mazzoccoli G, Cai Y, Liu S, Francavilla M,
Giuliani F, Piepoli A, Pazienza V, Vinciguerra M, Yamamoto T and
Takumi T: REV-ERBα and the clock gene machinery in mouse peripheral
tissues: A possible role as a synchronizing hinge. J Biol Regul
Homeost Agents. 26:265–276. 2012.PubMed/NCBI
|
|
4
|
Vieira E, Merino B and Quesada I: Role of
the clock gene Rev-erbα in metabolism and in the endocrine
pancreas. Diabetes Obes Metab. 17(Suppl 1): S106–S114. 2015.
View Article : Google Scholar
|
|
5
|
Kourtidis A, Jain R, Carkner RD, Eifert C,
Brosnan MJ and Conklin DS: An RNA interference screen identifies
metabolic regulators NR1D1 and PBP as novel survival factors for
breast cancer cells with the ERBB2 signature. Cancer Res.
70:1783–1792. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ka NL, Na TY, Na H, Lee MH, Park HS, Hwang
S, Kim IY, Seong JK and Lee MO: NR1D1 recruitment to sites of DNA
damage inhibits repair and is associated with chemosensitivity of
breast cancer. Cancer Res. 77:2453–2463. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang Z, Xiong F, Wang X, Qi Y, Yu H, Zhu Y
and Zhu H: Nuclear receptor retinoid-related orphan receptor alpha
promotes apoptosis but is reduced in human gastric cancer.
Oncotarget. 8:11105–11113. 2017.PubMed/NCBI
|
|
8
|
Yuan SQ, Nie RC, Chen YM, Qiu HB, Li XP,
Chen XJ, Xu LP, Yang LF, Sun XW, Li YF, et al: Glasgow Prognostic
Score is superior to ECOG PS as a prognostic factor in patients
with gastric cancer with peritoneal seeding. Oncol Lett.
15:4193–4200. 2018.PubMed/NCBI
|
|
9
|
Wang Z, Si X, Xu A, Meng X, Gao S, Qi Y,
Zhu L, Li T, Li W and Dong L: Activation of STAT3 in human gastric
cancer cells via interleukin (IL)-6-type cytokine signaling
correlates with clinical implications. PLoS One. 8:e757882013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) methods. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang Z, Dong L, Zhen Y, Wang Y, Qi D, Xu
A, Meng X and Li W: Astragalus extract inhibits proliferation but
enhances apoptosis in gastric cancer. Pak J Pharm Sci.
29:1473–1482. 2016.PubMed/NCBI
|
|
12
|
Sitaula S, Zhang J, Ruiz F and Burris TP:
Rev-erb regulation of cholesterologenesis. Biochem Pharmacol.
131:68–77. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wu Y, Qi Y, Liu H, Wang X, Zhu H and Wang
Z: AMPK activator AICAR promotes 5-FU-induced apoptosis in gastric
cancer cells. Mol Cell Biochem. 411:299–305. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Partch CL, Green CB and Takahashi JS:
Molecular architecture of the mammalian circadian clock. Trends
Cell Biol. 24:90–99. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Marciano DP, Chang MR, Corzo CA, Goswami
D, Lam VQ, Pascal BD and Griffin PR: The therapeutic potential of
nuclear receptor modulators for treatment of metabolic disorders:
PPARγ, RORs, and Rev-erbs. Cell Metab. 19:193–208. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lee J, Lee S, Chung S, Park N, Son GH, An
H, Jang J, Chang DJ, Suh YG and Kim K: Identification of a novel
circadian clock modulator controlling BMAL1 expression through a
ROR/REV-ERB-response element-dependent mechanism. Biochem Biophys
Res Commun. 469:580–586. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mazzoccoli G, de Cata A, Piepoli A and
Vinciguerra M: The circadian clock and the hypoxic response pathway
in kidney cancer. Tumour Biol. 35:1–7. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Reszka E and Przybek M: Circadian genes in
breast cancer. Adv Clin Chem. 75:53–70. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang Y, Kojetin D and Burris TP:
Anti-proliferative actions of a synthetic REV-ERBα/β agonist in
breast cancer cells. Biochem Pharmacol. 96:315–322. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fu L and Kettner NM: The circadian clock
in cancer development and therapy. Prog Mol Biol Transl Sci.
119:221–282. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gutierrez-Martinez P, Rossi DJ and Beerman
I: DNA damage and aging around the clock. Trends Mol Med.
22:635–637. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chomez P, Neveu I, Mansén A, Kiesler E,
Larsson L, Vennström B and Arenas E: Increased cell death and
delayed development in the cerebellum of mice lacking the
rev-erbA(alpha) orphan receptor. Development. 127:1489–1498.
2000.PubMed/NCBI
|
|
23
|
Tao W, Wu J, Zhang Q, Lai SS, Jiang S,
Jiang C, Xu Y, Xue B, Du J and Li CJ: EGR1 regulates hepatic clock
gene amplitude by activating Per1 transcription. Sci Rep.
5:152122015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lee JH and Sancar A: Circadian clock
disruption improves the efficacy of chemotherapy through
p73-mediated apoptosis. Proc Natl Acad Sci USA. 108:10668–10672.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bugge A, Feng D, Everett LJ, Briggs ER,
Mullican SE, Wang F, Jager J and Lazar MA: Rev-erbα and Rev-erbβ
coordinately protect the circadian clock and normal metabolic
function. Genes Dev. 26:657–667. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
He Y, Lin F, Chen Y, Tan Z, Bai D and Zhao
Q: Overexpression of the circadian clock gene rev-erbα affects
murine bone mesenchymal stem cell proliferation and osteogenesis.
Stem Cells Dev. 24:1194–1204. 2015. View Article : Google Scholar : PubMed/NCBI
|