Introduction
Oral squamous cell carcinoma (OSCC) is the
predominant histological types of oral cancer (1). Alcohol drinking, tobacco smoking, bad
oral hygiene, and human papilloma virus infections are the main
risk factors of OSCC (2,3). In China, there were 34.319 new cases
diagnosed as oral cavity cancer in 2010, including 23,096 males and
11,223 females (4).
Early detection oral of cancer is currently
challenging because although conventional oral examination can
detect tumors located in the oral cavity from the early stages, it
fails to identify all oral premalignant lesions (5).
Surgery is the primary therapy for oral cancer,
especially for patients in advanced stages. Chemotherapy and
radiation therapy are often combined with surgery in treating
patients with advanced oral cancer. Nevertheless, the 5-year
survival rate of patients with OSCC is only ~50% (6). Therefore, the mechanism underlying of
OSCC development should be elucidated, to optimize treatment and
improve patient survival.
c-Myc is a known oncogene, as well as target gene,
in various types of cancers, including lung (7–10) and
breast cancer (10–12), and hepatocellular carcinoma (13,14).
Previous studies showed that the c-Myc protein was overexpressed in
80% of the OSCC (15), Genome-wide
profiling of OSCC has also shown that, among other genes, gain of
c-Myc gene occurred at a high frequency (16).
In the present study, we investigated the function
of miR-1294 in OSCC, and tried to provide a potential therapeutic
target of OSCC.
Materials and methods
Tissues samples
For the present study, 24 OSCC tissues samples and
their matched adjacent normal tissues were acquired from the
Department of Stomatology, Stomatological Hospital of Jingmen City,
Hubei Province, China. The pathological diagnosis of all OSCC
patients was confirmed by the senior pathologists of the
Stomatological Hospital of Jingmen City. Tissues were frozen
immediately, and preserved in a −80°C freezer for further analysis,
including the detection of miR-1294 and c-Myc mRNA. Written
informed consent was obtained from all patients, and the present
study was approved by the Ethics Committees of the Stomatological
Hospital of Jingmen City.
HE staining
The 24 OSCC tissues was processed in standard
protocol for hematoxylin and eosin (HE) staining. Briefly, 4-µm
thick sections were cut. After deparaffinization (dunk for 45 sec
in xylene) and hydration (dunk for 45 sec in propanol), the slides
were staining in hematoxylin for 3–5 min, and washed in running
water for 5 min. After differentiated in 1% acid alcohol (30 sec),
the slides were staining in 1% eosin Y for 10 min. Then, these
slides were dehydrated in increasing concentration of alcohol (each
for 5 min) and clear in xylene for observation. These slides were
observed optical microscope (CX23; Olympus, Shinjuku, Tokyo,
Japan).
Cell culture
Overall, 4 OSCC cell lines (HSC2, HSC4, SAS and KON)
were acquired from the Cell Bank of the Chinese Academy of Medical
Sciences. The 4 OSCC cell lines were cultured in Dulbecco's
modified Eagles medium F-12 HAM (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) with 10% fetal bovine serum (FBS; Gibco; Thermo
Fisher Scientific, Inc., Waltham, MA, USA). Primary gingival
keratinocytes (PGK; ATCC® PCS-200-014™) were purchased
from American Type Culture Collection (ATCC; Manassas, VA, USA) and
cultured in Dermal Cell Basal Medium (ATCC®
PCS-200-030™) plus one Keratinocyte Growth Kit (ATCC®
PCS-200-040™).
Detection of miR-1294 in OSCC tissue
samples and SAS cells
The levels of miR-1294 in the 24 OSCC tissues cells
and HSC2, HSC4, SAS and KON cells were detected by qRT-PCR. In
detail, the total RNA was extracted from the 24 specimens using the
TRIzol reagent, according to the manufacturers' s protocol
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA). The
level of miR-1294 was then detected by TaqMan miRNA RT Real-Time
PCR (17). Single-stranded cDNA was
synthesized by the TaqMan MicroRNA Reverse Transcription Kit
(Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA,
USA), and then amplified by TaqMan Universal PCR Master Mix
(Invitrogen; Thermo Fisher Scientific, Inc.) and miRNA-specific
TaqMan MGB probes (Applied Biosystems). The U6 snRNA was used for
normalization (18). The c-Myc mRNA
levels in the OSCC tissues were assayed by real-time qPCR (delta Cq
method of quantification) using the SYBR-Green reagent (Shanghai
Shengong Biotechnology Co., Ltd., Shanghai, China) (19,20). The
primers used for c-Myc (human) are listed as follows: F,
5′-AATGAAAAGGCCCCCAAGGTAGTTATCC-3 and R,
5′-GTCGTTTCCGCAACAAGTCCTCTTC-3; GAP DH F,
5′-CTGACTTCAACAGCGACACC-3′ and R, 5′-TAGCCAAATTCGTTGTCATACC-3′. The
following thermocycling conditions were maintained: 94°C for 5 min;
30 cycles of 94°C for 45 sec, 55°C for 45 sec and 72°C for 1 min;
and 72°C for 10 min (21).
Overexpression and downregulation of
miR-1294 in SAS cells
miR-1294 was overexpressed by miR-1294 mimics and
decreased by miR-1294 antisense oligonucleotides (ASO), as
described in a previous study (18).
miR-1294 mimics, miR-1294 ASO and control miRNA were purchased from
Shanghai Shengong Biotechnology Co., Ltd. miRNAs were transfected
into cells using the Lipofectamine® 2000 reagent (Thermo
Fisher Scientific, Inc.).
Cell proliferation assay
Cellular growth was analyzed by the MTT assay, as
previously described (22–25). Briefly, SAS cells were cultured in
96-flat well plates overnight, at a cells density of
5×105/well. The MTT reagent (0.1 mg/ml) was added into
the medium for 5 min, followed by the addition of 100 µl of DMSO.
The optical density (OD) value was measured on a microplate reader,
with a 570 nm filter.
Cell migration assay
Transwell systems were used to assess cell
migration. In detail, the Transwell chambers (8.0 µm pore;
Sigma-Aldrich; Merck KGaA) were placed in 24-well plates. The
miR-1294 mimics or ASO-transfected SAS cells were FBS deprived for
12 h, and subsequently added to the upper chambers. Medium
containing 10% FBS was placed in the lower chambers and, after 24
h, the migrating SAS cells were counted.
Prediction of the putative targets of
miR-1294
The putative targets of miR-1294 were predicted by
the online software Targetscan (http://www.targetscan.org/vert_71/). TargetScan
predicts biological targets of miRNAs by searching for the presence
of 8mer, 7mer, and 6mer sites that match the seed region of each
miRNA (26).
Dual luciferase reporter assays
SAS cells were seeded at 1×105
cells/well and were serum-starved for 6 h
pre-transfection. Mutants of c-Myc 3′ untranslated region (3′UTR)
were generated using the Site-Directed Mutagenesis Kit (Huijun
Company, China). The 3′UTR of c-Myc and mutated controls were
cloned and inserted into the reporter plasmid (500 ng) (Promega
Corporation, Madison, WI, USA). miR-1294 mimics were then
transfected into the SAS cells containing either the wild-type or
mutant 3′UTR plasmids, using Lipofectamine 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.). Twenty-four hours later, cells were
harvested and the luciferase activity was measured using the
Dual-Luciferase Reporter Assay System (Huijun Company).
Plasmid transfection
c-Myc overexpression plasmid (pcDNA3.1-c-Myc) was
constructed by Sangon Biotech Co., Ltd. (Shanghai, China. Then,
miR-1294 mimics and pcDNA3.1-c-Myc plasmid were co-transfected into
SAS cells, using Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). In details, cells should be 70–90% confluent at
the time of transfection. Diluted DNA and diluted Lipofectamine
2000 were mixed gently and incubated for 20 min at room
temperature. Then, the 100 µl of complexes were added into each
well containing cells and medium. Then, incubated SAS cells at at
37°C in a CO2 incubator for 18–48 h prior to testing for
transgene expression.
Western blot analysis
SAS Cells were thawed and lysed in lysis buffer (150
mM NaCl, 50 mM Tris-HCI, 1% Triton X-100 and 0.1% SDS) with the
Protease Inhibitor Cocktail (Sigma-Aldrich; Merck KGaA) and the
Phosphatase Inhibitor Cocktail (Sigma-Aldrich; Merck KGaA). Protein
was separated by 10% SDS-PAGE gel and was transferred into PVDF
membrane. Then, the membrane was blocked in 5% bovine serum albumin
(BSA; A1933; Sigma-Aldrich; Merck KGaA) for 1 h. For c-Myc
analysis, an anti-c-Myc antibody (ab11917; Abcam, Cambridge, UK)
was prepared in 5% BSA (1:500). The membrane was incubated with
c-Myc antibody in 4°C for overnight. After washed by 3 times, the
membrane was incubated with a peroxidase-linked antibody to rabbit
antibody IgG (ab218695; 1:2,000 dilution; Abcam) in room
temperature for 2 h. Proteins were detected with the ECL Western
Blotting Detection Reagents (GE Healthcare, Chicago, IL, USA).
Images were analyzed using ImageJ [National Institutes of Health
(NIH) Bethesda, MD, USA, USA]. β-actin was used as an internal
control. For β-actin detection, an anti-β-actin antibody (ab8226;
Abcam) was prepared in 5% BSA (1:2,000). The following steps were
same with the steps of detecting c-Myc.
Statistical analysis
All experiments were repeated 3 times. Data are
shown as mean ± SD. Two-tailed Student's t-test was used to analyze
the mean value between two groups; one-way ANOVA was used to test
the mean value among 3 groups or more with post hoc contrasts by
Student-Newman-Keuls test. The correlation between c-Myc and
miR-1294 levels were tested by Pearson correlation coefficient
analysis. The indicator of statistical significance was P<0.05.
All calculations were performed using the SPSS software (version
16.0) (SPSS, Inc., Chicago, IL, USA).
Results
miR-1294 levels in OSCC tissues
Initially, the 6 preventatives image of OSCC tissues
are shown (Fig. 1A). The expression
levels of miR-1294 in 24 OSCC tissues and their matched adjacent
normal tissues were analyzed by qRT-PCR. Results indicated lower
levels of miR-1294 in OSCC tissues (Fig.
1B). This was reflected by the mean value of miR-1294 in the 24
OSCC tissues, which was lower than the mean value in the 24 normal
adjacent tissues (Fig. 1C). Then, we
assayed the miR-1294 levels in normal primary gingival
keratinocytes and 4 OSCC cell lines (HSC2, HSC4, SAS and KON), and
found that SAS cells showed the lowest levels of miR-1294 (Fig. 1D).
In vitro role of miR-1294 in SAS
cells
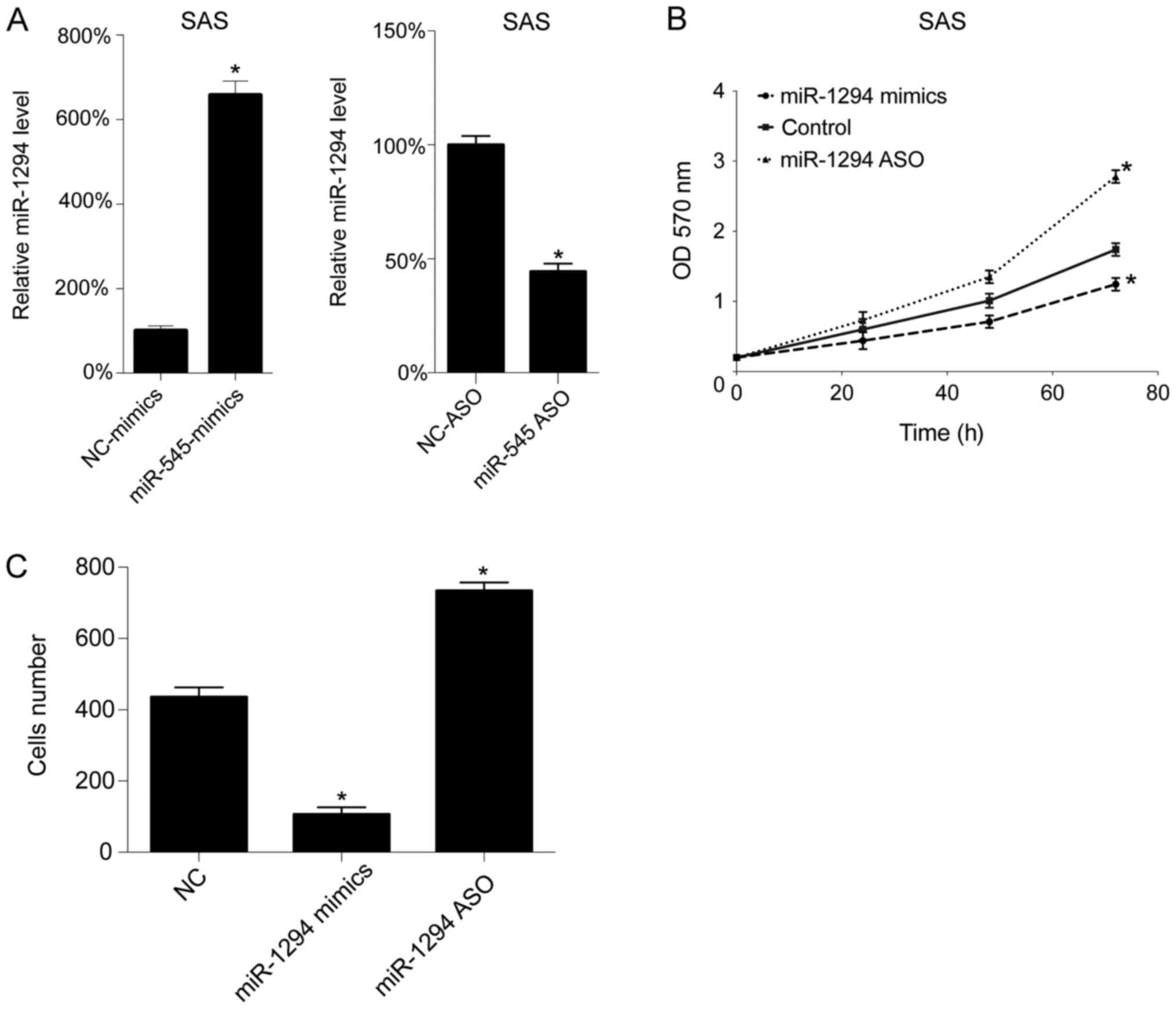
Next, we assessed the function of miR-1294 in
vitro. The levels of miR-1294 levels were upregulated and
downregulated by miR-1294 mimics and miR-1294 ASO transfection,
respectively. As expected, miR-1294 mimics transfection increased
the level of miR-1294 in SAS cells, whereas miR-1294 ASO
transfection decreased the level of miR-1294 in the SAS cells
(Fig. 2A). Next, cell growth
following transfection was assessed using by MTT assay. As seen in
Fig. 2B, upregulation of miR-1294
inhibited cell growth, whereas downregulation promoted cell growth
in SAS cells. Subsequently, we analyzed cell migration following
transfection, and found that miR-1294 mimics decreased the number
of migrating cells, whereas miR-1294 ASO increased the number of
migrating cells, in SAS cells (Fig.
2C).
c-Myc is a target gene of
miR-1294
Next we attempted to determine that whether c-Myc is
a target gene of miR-1294. c-Myc is a very important oncogene,
which shows a high amplification mutation across all various types
of tumors (Fig. 3A). The potential
binding sites of the 3′UTR of c-Myc were revealed by bioinformatics
methods (Fig. 3B). To establish
whether miR-1294 could target c-Myc, mutants of 3′UTR of c-Myc were
generated and, subsequently transfected into luciferase reporter
plasmids. miR-1294 mimics and c-Myc mutants were co-transfected
into SAS cells. At 24 h post-transfection, that miR-1294 mimics
reduced the luciferase activity of the 3′UTR of wild-type c-Myc,
but not that of mutated of 3′UTR of c-Myc (Fig. 3C). Forty-eight hours later, the c-Myc
protein levels were determined by western blot analysis. As shown
in Fig. 3D, miR-1294 mimics inhibited
the expression of c-Myc in SAS cells (Fig. 3D). Moreover, we assayed the c-Myc mRNA
expression levels in the 24 OSCC tissues, and correlated c-Myc mRNA
expression with miR-1294 expression. Obtained results revealed a
negative correlation between c-Myc mRNA expression and miR-1294
expression in the 24 OSCC tissue samples (Fig. 3E).
c-Myc reduces the inhibitory effect of
miR-1294
To confirm miR-1294 play its role via c-Myc, we
performed the rescue experiment including plasmid transfection and
cell migration assay. We overexpressed the c-Myc levels in SAS
cells by c-Myc overexpression plasmid transfection
(pcDNA3.1-c-Myc). Twenty-four hours later, the c-Myc mRNA levels in
SAS cells was analyzed by qRT-PCR, and we found that pcDNA3.1-c-Myc
increased the c-Myc levels in SAS cells (Fig. 4A). Next miR-1294 mimics and
pcDNA3.1-c-Myc plasmid were co-transfected into SAS cells, then the
cells growth were tested by MTT analyzed, and we found that
miR-1294 inhibited cells growth as expected, and c-Myc
overexpression plasmid restored the inhibitory effect of miR-1294
(Fig. 4B). Subsequently, we analyzed
cell migration following transfection, and found that miR-1294
mimics decreased the number of migrating cell, whereas c-Myc
overexpression plasmid restored the inhibitory effect of miR-1294,
in SAS cells (Fig. 4C).
TRL4, TLR6, TLR8 and TLR9 are target
genes of miR-1294
TLR2, TLR4 and TLR9 were expressed in primary
tumors, neck metastases as well as in recurrent tumors of OSCC. We
found that miR-1294 could target TRL4, TLR6, TLR8 and TLR9
(Fig. 5).
Discussion
In the present study, the function of miR-1294 in
OSCC was investigated. Our data showed that miR-1294 levels in OSCC
tissues were lower than the levels in matched adjacent normal
tissues. Overexpression of miR-1294 inhibited the proliferation and
migration of SAS cells, and vice versa. To the best of our
knowledge, this is the first report on the inhibitory function of
miR-1294 in OSCC. In esophageal squamous cell carcinoma, inhibitory
effect of miR-1294 on the translation of c-Myc has been already
described and proved by Liu et al (18). Our study showed the similar scenario
as this previous study described and stress the function of
miR-1294 in OSCC.
The role of c-Myc has been intensively studied in
head and neck cancers including OSCC. In OSCC, c-Myc level was
overexpressed (27), more
importantly, overexpression of c-Myc is correlated with poor
prognosis of OSCC (28). The
underlying mechanism is that c-Myc is elicited mainly via
activation of transcription of those c-Myc target genes that are
positive regulators of the cell cycle, such as cyclins D1, D2, E
and A, Cdk4, e2f1, e2f2, Cdc25A and B (29,30). Thus
it is possible that the low levels of miR-1294 may be also
correlated with poor prognosis of OSCC. However, whether c-Myc and
miR-1294 are correlated with the grade of pathology, whether c-Myc
play a role in cell migration are unclearly. In further study, we
will investigated these issues and analyze c-Myc protein in tissues
by western blotting and immunohistochemical. The correlation of the
expression of miR-1294 and the grade of pathology may be tested and
the morphology of migrated cells will be observed by microscope. We
will collect more OSCC specimen and analyzed this possibility in a
further study.
Bioinformatics methods predicted that TRL4, TLR6,
TLR8 and TLR9 are target genes of miR-1294 (Fig. 5). Toll-like receptors (TLRs) are cell
surface or intracellular transmembrane proteins, acting as
pattern-recognition receptor. TLRs are capable of recognizing
bacterial, viral, fungal and protozoal components (31–38). A
previous study showed that TLR2, TLR4 and TLR9 were expressed in
primary tumors, neck metastases as well as in recurrent tumors of
OSCC (39). As OSCC developed in an
immune cell-rich environment, immune cells interact with cancer
cells intensively, resulting in a network of regulation. For
example, TLR2 played a role in Treg expansion and their suppressive
capacity (40). Thus, we guessed that
TRL4, TLR6, TLR8 and TLR9 maybe be involved in the regulation of
immune microenvironments of OSCC, and miR-1294 may play its
antitumor role partly by targeting Toll-like receptors.
In conclusion, our data reveal the inhibitory role
of miR-1294 in OSCC.
Acknowledgements
The authors would like to thank Dr Zhumei Wu
(Department of Stomatology, Stomatological hospital of Jingmen
City, China) for the helpful discussion.
Funding
This work was supported by Funding of Hubei Province
(grant no. 020716040026).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Author's contributions
ZW and JY collected patient data and performed cell
experiments. JY and TZ performed PCR, western blotting and other
molecular experiment. HG contributed to study design and manuscript
writing.
Ethics approval and consent to
participate
The present study was approved by Ethic Committee of
Tongji Medical College, Huazhong University of Science and
Technology and each patient provided written informed consent.
Consent for publication
All patients gave informed consent for the use of
their tissues and publication of the data and images.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Rivera C: Essentials of oral cancer. Int J
Clin Exp Pathol. 8:11884–11894. 2015.PubMed/NCBI
|
|
2
|
Gupta PC: Mouth cancer in India: A new
epidemic? J Indian Med Assoc. 97:370–373. 1999.PubMed/NCBI
|
|
3
|
Mehrotra R, Singh M, Kumar D, Pandey A,
Gupta R and Sinha U: Age specific incidence rate and pathological
spectrum of oral cancer in Allahabad. Indian J Med Sci. 57:400–404.
2003.PubMed/NCBI
|
|
4
|
Zheng CM, Ge MH, Zhang SS, Tan Z, Wang P,
Zheng RS, Chen WQ and Xia QM: Oral cavity cancer incidence and
mortality in China, 2010. J Cancer Res Ther. 11 Suppl 2:C149–C154.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lingen MW, Kalmar JR, Karrison T and
Speight PM: Critical evaluation of diagnostic aids for the
detection of oral cancer. Oral Oncol. 44:10–22. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ries L, Eisner M, Kosary C, et al: Cancer
statistics review, 1975–2002. Bethesda, MD: National Cancer
Institute 21; 2005
|
|
7
|
Little CD, Nau MM, Carney DN, Gazdar AF
and Minna JD: Amplification and expression of the c-myc oncogene in
human lung cancer cell lines. Nature. 306:194–196. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yanaihara N, Caplen N, Bowman E, Seike M,
Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, et
al: Unique microRNA molecular profiles in lung cancer diagnosis and
prognosis. Cancer Cell. 9:189–198. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Engelman JA, Zejnullahu K, Mitsudomi T,
Song Y, Hyland C, Park JO, Lindeman N, Gale CM, Zhao X, Christensen
J, et al: MET amplification leads to gefitinib resistance in lung
cancer by activating ERBB3 signaling. Science. 316:1039–1043. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dubik D, Dembinski TC and Shiu RP:
Stimulation of c-myc oncogene expression associated with
estrogen-induced proliferation of human breast cancer cells. Cancer
Res. 47:6517–6521. 1987.PubMed/NCBI
|
|
11
|
Chen CR, Kang Y and Massagué J: Defective
repression of c-myc in breast cancer cells: A loss at the core of
the transforming growth factor beta growth arrest program. Proc
Natl Acad Sci USA. 98:992–999. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Escot C, Theillet C, Lidereau R, Spyratos
F, Champeme MH, Gest J and Callahan R: Genetic alteration of the
c-myc protooncogene (MYC) in human primary breast carcinomas. Proc
Natl Acad Sci USA. 83:4834–4838. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Peng SY, Lai PL and Hsu HC: Amplification
of the c-myc gene in human hepatocellular carcinoma: Biologic
significance. J Formos Med Assoc. 92:866–870. 1993.PubMed/NCBI
|
|
14
|
Hsu T, Möröy T, Etiemble J, Louise A,
Trépo C, Tiollais P and Buendia MA: Activation of c-myc by
woodchuck hepatitis virus insertion in hepatocellular carcinoma.
Cell. 55:627–635. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pai R, Pai S, Lalitha R, Kumaraswamy S,
Lalitha N, Johnston R and Bhargava M: Over-expression of c-Myc
oncoprotein in oral squamous cell carcinoma in the South Indian
population. Ecancermedicalscience. 3:1282009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen YJ, Lin SC, Kao T, Chang CS, Hong PS,
Shieh TM and Chang KW: Genome-wide profiling of oral squamous cell
carcinoma. J Pathol. 204:326–332. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Schmittgen TD, Lee EJ, Jiang J, Sarkar A,
Yang L, Elton TS and Chen C: Real-time PCR quantification of
precursor and mature microRNA. Methods. 44:31–38. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu K, Li L, Rusidanmu A, Wang Y and Lv X:
Down-regulation of miR-1294 is related to dismal prognosis of
patients with esophageal squamous cell carcinoma through elevating
c-Myc expression. Cell Physiol Biochem. 36:100–110. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bustin SA: Quantification of mRNA using
real-time reverse transcription PCR (RT-PCR): TRends and problems.
J Mol Endocrinol. 29:23–39. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ren X, Zhang Z, Tian J, Wang H, Song G,
Guo Q, Tian J, Han Y, Liao Q, Liu G, et al: The downregulation of
c-Myc and its target gene hTERT is associated with the
antiproliferative effects of baicalin on HL-60 cells. Oncol Lett.
14:6833–6840. 2017.PubMed/NCBI
|
|
22
|
Gerlier D and Thomasset N: Use of MTT
colorimetric assay to measure cell activation. J Immunol Methods.
94:57–63. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fotakis G and Timbrell JA: In vitro
cytotoxicity assays: Comparison of LDH, neutral red, MTT and
protein assay in hepatoma cell lines following exposure to cadmium
chloride. Toxicol Lett. 160:171–177. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Twentyman PR and Luscombe M: A study of
some variables in a tetrazolium dye (MTT) based assay for cell
growth and chemosensitivity. Br J Cancer. 56:279–285. 1987.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ferrari M, Fornasiero MC and Isetta AM:
MTT colorimetric assay for testing macrophage cytotoxic activity in
vitro. J Immunol Methods. 131:165–172. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Field J: Oncogenes and tumour-suppressor
genes in squamous cell carcinoma of the head and neck. Eur J Cancer
B Oral Oncol. 28:67–76. 1992. View Article : Google Scholar
|
|
28
|
Field JK, Spandidos DA, Stell PM, Vaughan
ED, Evan GI and Moore JP: Elevated expression of the c-myc
oncoprotein correlates with poor prognosis in head and neck
squamous cell carcinoma. Oncogene. 4:1463–1468. 1989.PubMed/NCBI
|
|
29
|
de Nigris F, Sica V, Herrmann J,
Condorelli G, Chade AR, Tajana G, Lerman A, Lerman LO and Napoli C:
c-Myc oncoprotein: cell cycle-related events and new therapeutic
challenges in cancer and cardiovascular diseases. Cell Cycle.
2:325–328. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Dang CV: c-Myc target genes involved in
cell growth, apoptosis, and metabolism. Mol Cell Biol. 19:1–11.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Takeda K, Kaisho T and Akira S: Toll-like
receptors. Annu Rev Immunol. 21:335–376. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Moresco EM, LaVine D and Beutler B:
Toll-like receptors. Curr Biol. 21:R488–R493. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Akira S, Takeda K and Kaisho T: Toll-like
receptors: Critical proteins linking innate and acquired immunity.
Nat Immunol. 2:675–680. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
34
|
Fig TLR: Toll-like receptors. 2017.
|
|
35
|
Aderem A and Ulevitch RJ: Toll-like
receptors in the induction of the innate immune response. Nature.
406:782–787. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kawai T and Akira S: The role of
pattern-recognition receptors in innate immunity: Update on
Toll-like receptors. Nat Immunol. 11:373–384. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Takeda K and Akira S: Toll-like receptors
in innate immunity. Int Immunol. 17:1–14. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kawai T and Akira S: Toll-like receptors
and their crosstalk with other innate receptors in infection and
immunity. Immunity. 34:637–650. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Mäkinen LK, Ahmed A, Hagström J, Lehtonen
S, Mäkitie AA, Salo T, Haglund C and Atula T: Toll-like receptors
2, 4, and 9 in primary, metastasized, and recurrent oral tongue
squamous cell carcinomas. J Oral Pathol Med. 45:338–345. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sutmuller RP, den Brok MH, Kramer M,
Bennink EJ, Toonen LW, Kullberg BJ, Joosten LA, Akira S, Netea MG
and Adema GJ: Toll-like receptor 2 controls expansion and function
of regulatory T cells. J Clin Invest. 116:485–494. 2006. View Article : Google Scholar : PubMed/NCBI
|