Introduction
Hepatocellular carcinoma (HCC) is one of the most
common types of malignancies, and ranks as the fifth most prevalent
cancer worldwide (1,2). Tumorigenesis is a complicated process,
physical, chemical, and biological factors could lead to mutations
in genes and the formation of tumors (3). HCC is common in China with an incidence
rate of 250.28/100,000 and accounts for >50% of cancer cases
worldwide (4,5), which causes a heavy economic and
physical burden to patients. Despite progress in conventional
therapies, including surgical resection, percutaneous ablation,
chemotherapy and embolization treatment, the 5-year survival rate
of patients with HCC remains poor (6). Tumors have the characteristic of
metastasis, which impedes of the ability of complete surgical
resection, and recurrence is the biggest obstacle for the treatment
of tumors (7). This suggests that
diagnosis of HCC at an early stage is crucial for effective therapy
and good prognosis, and highlights the urgent requirement for
identification of biomarkers of HCC.
Corticotropin releasing hormone binding protein
(CRHBP) is a 37-kDa oligopeptide, and is a component of the
hypothalamic-pituitary-adrenal axis, involved in regulation of
physiological reactions (8).
Expression of CRHBP has been detected in numerous tissue types
(9,10). CRHBP prevents inappropriate activation
of corticotropin releasing hormone (CRH) by binding the CRH complex
(11).
CRHBP has been investigated in the field of normal
physiologic, metabolic function and oncogenesis (12,13). It
was reported that Caucasians and African Americans with low CRHBP
expression were associated with an increased risk of breast cancer
(14). Low CRHBP mRNA and protein
expression has been reported in prostatic and bladder cell
carcinoma (15,16).
Because the study of CRHBP expression in HCC tissue
is unclear, the present study used immunohistochemistry, western
blotting and reverse transcription-quantitative polymerase chain
reaction (RT-qPCR) and bioinformatics analysis to evaluate the
expression level of CRHBP in HCC. The association between CRHBP
expression and clinicopathology and overall survival time was also
investigated.
Materials and methods
Patients and samples
A total of 169 tumor tissue samples and 151 adjacent
non-tumorous liver tissue samples were collected at surgery from
patients with HCC at Zhejiang Provincial People's Hospital
(Zhejiang, China) between January 2010 and December 2016. The
patient cohort included 142 males and 27 females, with an age range
of 25–90 years old and a mean age of 58 years old. All tissues were
fixed with 4% formalin for 24 h at room temperature,
paraffin-embedded, and diagnosis was confirmed by pathologists at
Zhejiang Provincial People's Hospital (Zhejiang, China).
Information regarding tumor size, number, location, Edmondson Grade
and tumor metastasis were collected from patient medical records.
Overall survival (OS) was defined as the time from the date of
surgery to the date of mortality or the last day of follow up. The
research was approved by the Zhejiang Provincial People's Hospital
Ethics Committee (Zhejiang, China), and informed consent was
obtained from each participant.
Immunohistochemical staining
Using the paraffin-embedded specimens, two tissue
microarrays were designed and constructed by BioChip Company
(Shanghai, China, www.shbiochip.com). Briefly, two tissue microarrays in
(5-µm thick) were incubated at 70°C for 2 h, then deparaffinized in
xylene 10 min for three times, prior to rehydration in a graded
ethanol series (100, 95, 85 and 75% for 5 min). For antigen
retrieval, the microarrays were boiled at 120°C with TE buffer
(1.21 g/l Tris, 0.37 g/l EDTA, 0.5 ml/l Tween-20) at high pressure
for 3 min. The sections were then treated with 3% hydrogen peroxide
for 15 min at room temperature to eliminate endogenous peroxidase
activity, and blocked with 10% goat serum (reagent A;
Histostain-plus Bulk kit; Thermo Fisher Scientific, Inc., Waltham,
MA, USA) at room temperature for 20 min. The sections were then
incubated with a CRHBP primary antibody (dilution, 1:200, AF2796;
R&D Systems, Inc., Minneapolis, MN, USA) overnight at 4°C.
Following 3 washes with PBS, the sections were incubated with a
biotinylated second antibody (reagent B; Histostain-plus Bulk kit)
for 15 min, prior to the addition of streptavidin-peroxidase
(reagent C, Histostain-plus Bulk kit) for 15 min at room
temperature, according to the manufacturer's protocol. Finally, a
chromogenic reaction was performed using a DAB color-substrate
solution (OriGene Technologies, Inc., Rockville, MD, USA),
according to the manufacturer's protocol. Color development was
stopped when the brown was observed obviously in the tissue
microarrays. Hematoxylin (cat. no. C0107; Beyotime Institute of
Biotechnology, Haimen, China) staining was performed for 3–5 min.
The process was completed with dehydration in 75, 85, 95, and 100%
ethyl alcohol for 5 min respectively, transparency three times in
xylene for 10 min and mounting with gelatin resin. All procedures
were performed at room temperature unless otherwise specified.
Evaluation of the
immunohistochemistry
The results of immunohistochemical CRHBP staining
were interpreted by 2 pathologists of the Pathology Department of
Zhejiang Provincial People's Hospital, considering the staining
intensity and the proportion of stained cells. The staining
intensity was scored as follows: 0, negative staining; 1, weak
staining; 2, medium staining, and 3, strong staining. The
proportion of stained cells were scored as follows: 0, no cells
stained; 1, 1–25% cells stained; 2, 26–50% cells stained; 3, 51–75%
cells stained, and 4, >75% cells stained. We multiplied the
staining intensity and proportion scores to confirm the level of
CRHBP expression: 0 was considered to indicate negative expression,
and ≥1 was considered to indicate positive expression.
Cell culture
Human liver cancer cell lines HCCLM3, MHCC97H, Huh7,
Hep3B, HepG2 were obtained from cell bank of Shanghai Academy of
Sciences (Shanghai, China), and cultured in DMEM (SH30243.01;
HyClone; GE Healthcare Life Sciences, Logan, UT, USA) containing
10% fetal bovine serum (16000–044; Gibco; Thermo Fisher Scientific,
Inc., Waltham, MA, USA), 1% penicillin-streptomycin solution
(SV30010; Hyclone). All cells were maintained in a humidified
incubator with 5% CO2 at 37°C.
Western blot analysis
CRHBP expression was analyzed in three paired HCC
and adjacent non-cancerous tissues and five liver cancer cell lines
(HCCLM3, MHCC97H, Huh7, Hep3B, HepG2). Proteins were extracted with
RIPA buffer (cat. no. P0013B; Beyotime Institute of Biotechnology)
and concentration determined by BCA Kit (cat. no. P0009; Beyotime
Institute of Biotechnology). Then, proteins were heat-inactivated
at 100°C 10 min, 12% SDS-PAGE (20 µg protein/lane) and transferred
to polyvinylidene fluoride membranes by electrophoresis. The
membranes were blocked in 5% skim milk for 1.5 h at room
temperature. A CRHBP primary antibody (dilution, 1:1,000, AF2796;
R&D) was incubated with the membranes overnight at 4°C. The
membranes were washed in Tris-buffered saline with Tween three
times prior to addition of a horseradish peroxidase conjugated-goat
anti-Mouse IgG secondary antibody (dilution, 1:5,000, HA1006; HuaAn
Biotechnology Co., Ltd., Hangzhou, China) for 1.5 h at room
temperature. Finally, protein expression levels were analyzed via
grey level was using Quantity One software (version 4.6.2; Bio-Rad
Laboratories, Inc., Hercules, CA, USA).
RT-qPCR analysis
Total RNA was isolated from five liver cancer cell
lines (HCCLM3, MHCC97H, Huh7, Hep3B, HepG2) using TRIzol Reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. cDNAs were synthesized from 1 µg of
DNase-treated total RNA using the PrimeScriptTM RT reagent kit with
gDNA Eraser (Takara Bio, Inc., Otsu, Japan), under 65°C, 5 min,
42°C, 60 min and 70°C, 10 min of reverse transcription. The primers
of CRHBP were 5′-CACACCAGCATCGAAACTGC-3′ (forward) and
5′-TGAAGACCATTTACGTGTCCCA-3′ (reverse). The Primers for GAPDH were
5′-TGAAGGTCGGAGTCAACGG-3′ (forward) and 5′-CTGGAAGATGGTGATGGGATT-3′
(reverse). qPCR was carried out using a Bio-Rad CFX Connect
Real-Time system (Bio-Rad Laboratories, Inc) using the thermal
cycling condition: 98°C for 2 min followed by 39 amplification
cycles at 98°C for 15 sec and 60°C for 15 sec. At the end of the
PCR cycles, melting curve analysis were performed. The expression
of CRHBP was normalized to GAPDH using 2−ΔΔCq method
(17).
Bioinformatics analysis using online
databases and search tool for the retrieval of interacting
genes/proteins
CRHBP mRNA expression in HCC and normal tissues was
compared using the online Oncomine database (https://www.oncomine.org) with the following filtering
conditions: gene, CRHBP; analysis type, cancer vs. normal; cancer
type, hepatocellular carcinoma; data type, mRNA. The 10-year
survival analysis was performed using OncoLnc (http://www.oncolnc.org/). Gene enrichment analysis of
CRH was performed using STRING (https://string-db.org/) as there was not enough CRHBP
data in the online database, and the top 20 enriched genes analyzed
to investigate its biological function. Target signaling pathway
was searched on KEGG database (www.genome.jp).
Statistical analysis
Statistical analysis was performed using SPSS
software (version 16.0; SPSS, Inc., Chicago, IL, USA). χ2
test was used to analyze the association between CRHBP
expression and clinicopathological parameters. The overall survival
curve was generated using the Kaplan-Meier method and further
analyzed using the log-rank test. P<0.05 was considered to
indicate a statistically significant difference.
Results
Clinical characteristics of
patients
The majority of the patients who participated in the
present study had a history of hepatitis B virus infection (79.3%).
Upon the last day of follow-up, 84 patients were alive and 41
mortalities had occurred. A total of 44 patients were lost to
follow-up (Table I).
 | Table I.Associations between expression of
CRHBP protein and clinicopathological characteristics in 169 cases
of hepatocellular carcinoma. |
Table I.
Associations between expression of
CRHBP protein and clinicopathological characteristics in 169 cases
of hepatocellular carcinoma.
|
|
| CRHBP expression |
|
|---|
|
|
|
|
|
|---|
| Clinical
parameters | Number | negative | Positive | P-value |
|---|
| Age (years) |
|
|
| 0.460 |
| ≥55 | 107 | 53 | 54 |
|
|
<55 | 62 | 32 | 30 |
|
| Sex |
|
|
| 0.513 |
| Male | 142 | 71 | 71 |
|
|
Female | 27 | 14 | 13 |
|
| Tumor diameter |
|
|
| 0.013 |
| (cm) |
|
<5 | 99 | 42 | 57 |
|
| ≥5 | 67 | 41 | 26 |
|
| Tumor number |
|
|
| 0.485 |
|
Single | 138 | 70 | 68 |
|
|
Multiple | 31 | 15 | 16 |
|
| Location |
|
|
| 0.614 |
|
Left | 28 | 16 | 12 |
|
|
Right | 103 | 51 | 52 |
|
| Left +
right | 10 | 4 | 6 |
|
| Edmondson
grade |
|
|
| 0.002 |
|
I+II | 97 | 39 | 58 |
|
|
III | 72 | 46 | 26 |
|
| Metastasis |
|
|
| 0.215 |
| M0 | 150 | 74 | 76 |
|
| M1 | 14 | 9 | 5 |
|
| Microvascular
invasion |
|
|
| 0.246 |
|
Absent | 70 | 35 | 35 |
|
|
Present | 59 | 34 | 25 |
|
| HBV antigen |
|
|
| 0.020 |
|
Negative | 34 | 23 | 11 |
|
|
Positive | 130 | 60 | 70 |
|
| Cirrhosis |
|
|
| 0.217 |
|
Negative | 52 | 29 | 23 |
|
|
Positive | 117 | 56 | 61 |
|
| AFP (ng/ml) |
|
|
| 0.014 |
|
<50 | 91 | 36 | 55 |
|
|
≥50 | 78 | 45 | 33 |
|
| Survival status at
end of follow-up |
|
|
| 0.009 |
| Not
alive | 41 | 27 | 14 |
|
|
Alive | 84 | 35 | 49 |
|
CRHBP expression in HCC
Positive immunohistochemical staining of CRHBP was
detected in 84/169 (49.7%) HCC tissues and 142/151 (94.0%) adjacent
non-tumorous tissues. The expression of CRHBP in tumor tissues was
significantly lower than that in adjacent non-tumorous tissues
(P<0.001; Table II; Fig. 1A and B). A bar chart illustrating the
score values of the immunostaining of CRHBP expression in HCC
tissues (Fig. 1C).
 | Table II.Expression of CRHBP protein in HCC
and normal liver tissues. |
Table II.
Expression of CRHBP protein in HCC
and normal liver tissues.
|
|
| CRHBP
expression |
|
|---|
|
|
|
|
|
|---|
| Tissue type | Number | Negative | Positive | P-value |
|---|
| HCC | 169 | 85 | 84 | P<0.001 |
| Adjacent
non-tumorous | 151 | 9 | 142 |
|
The association between CRHBP
expression and clinicopathological characteristics
Tumor diameters ≥5 cm were significantly associated
with low expression of CRHBP (P=0.013). Low CRHPB expression was
also significantly associated with Edmondson Grade (P=0.002), high
α-fetoprotein (AFP) levels (P=0.014) and hepatitis B virus (HBV)
infection (P=0.020). No significant association was identified
between CRHBP expression and age, sex, tumor location, metastasis
status, microvascular invasion or cirrhosis (P>0.05; Table I).
Association between CRHBP expression
and prognosis
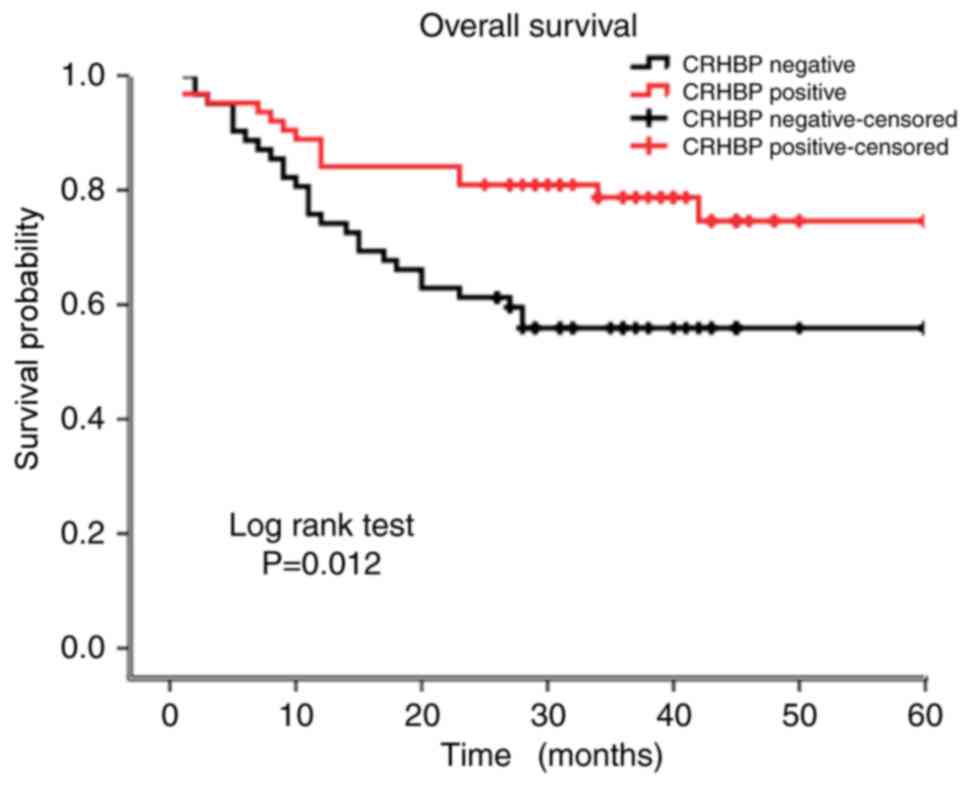
To evaluate the prognostic value of CRHBP
expression, Kaplan-Meier curves were constructed. The 5-year
overall survival rate was significantly different between patients
exhibiting low CRHBP expression (39.20 months) and patients
exhibiting high CRHBP expression (49.18 months) (P=0.012; log-rank
test; Fig. 2). Thus, it was inferred
that patients exhibiting low CRHBP expression in the liver tissue
had a reduced survival time compared with those exhibiting high
CRHBP expression.
CRHBP is downregulated in liver cancer
tissues and cell lines
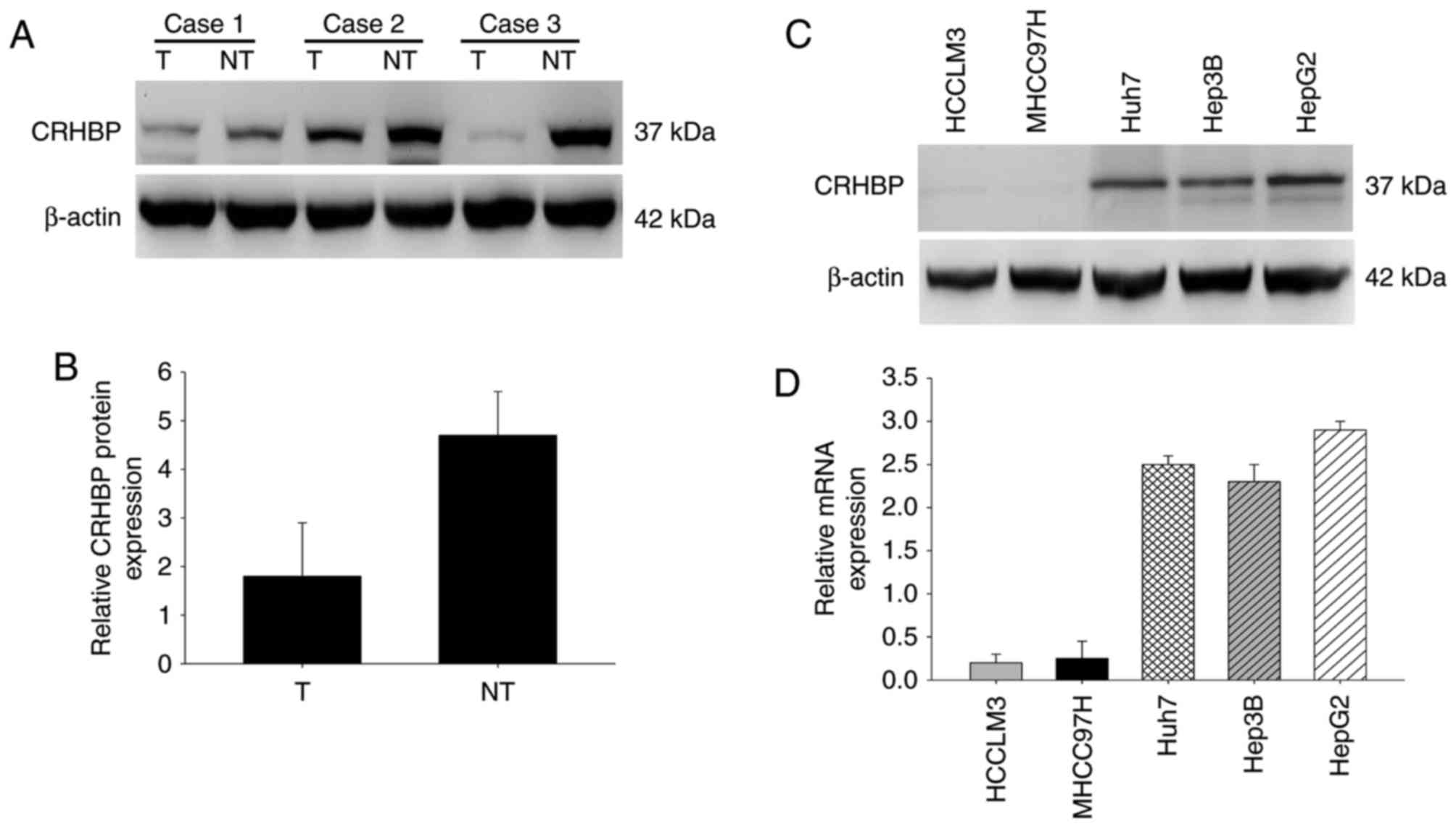
To further confirm the expression pattern of CRHBP
in HCC, the protein expression level of CRHBP was determined in
paired HCC and adjacent non-cancerous liver tissues by western
blotting. Compared with the adjacent non-tumor tissues, significant
downregulation of CRHBP protein expression was observed in HCC
tissues (Fig. 3A and B). Furthermore,
CRHBP levels were lower in highly metastatic liver cancer cell
lines, including HCCLM3 and MHCC97H cells, than in those with low
metastatic potential, including Huh7 and Hep3B cells, and was
highest in HepG2 hepatic cancer cells as determined by western
blotting and RT-qPCR (Fig. 3C and D).
These results suggest that reduced CRHBP expression is common in
patients with HCC, and that this may be associated with
tumorigenesis.
Bioinformatics analysis of CRHBP in
patients with HCC
A total of 5 datasets: Wurmbach Liver Statistics
(18), Chen Liver Statistics
(19), Roessler Liver Statistics,
Roessler Liver 2 Statistics (20) and
Mas Liver Statistics (21), were
selected to analyze the expression of CRHBP in HCC vs. normal
tissues using Oncomine databases. It was demonstrated that the
expression of CRHBP mRNA was significantly lower in HCC tissues
than in normal tissues (Fig. 4;
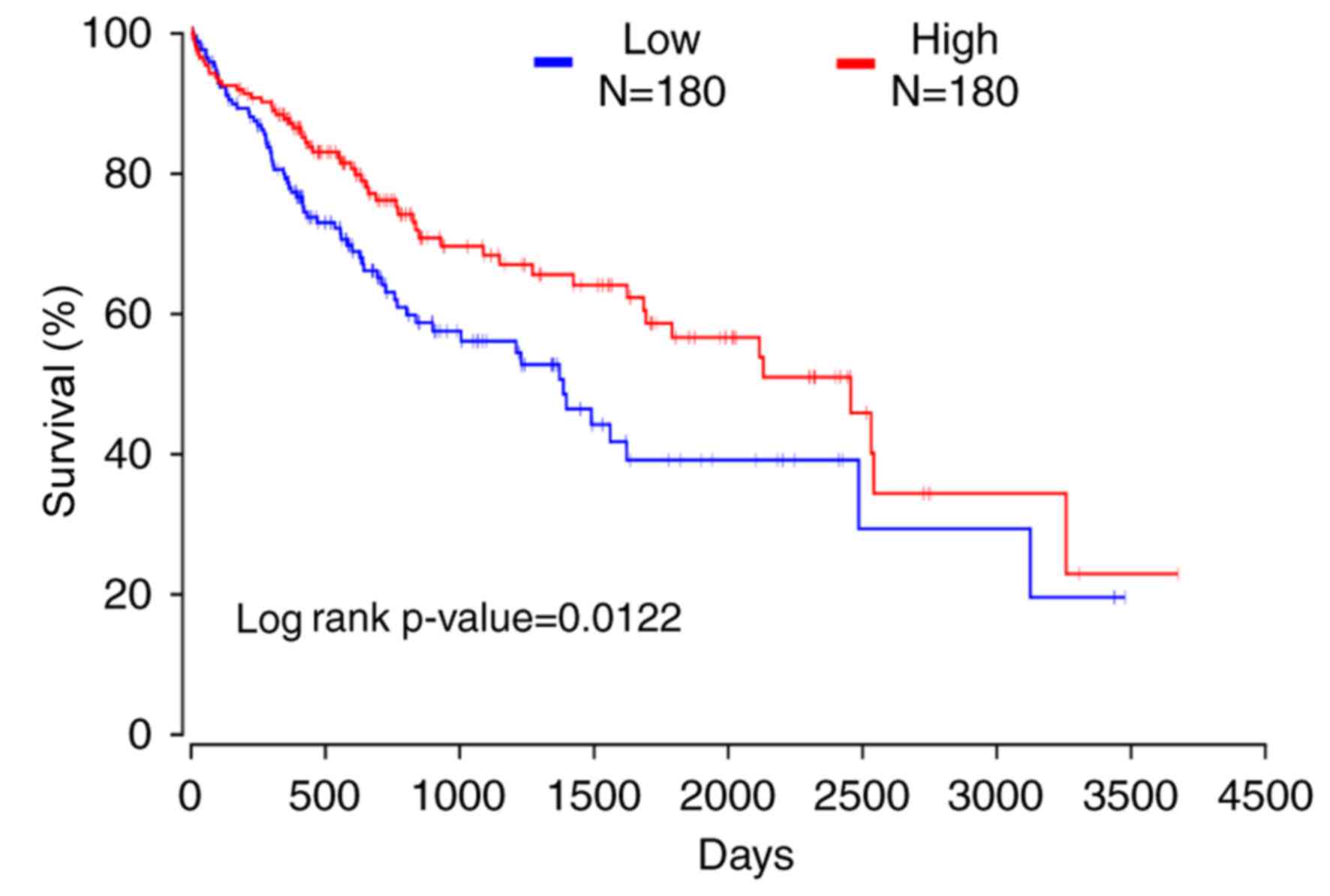
P<0.01). OncoLnc was used to reveal that patients exhibiting low
expression of CRHBP had a shorter 10-year survival time (Fig. 5; log-rank, P=0.0122). STRING was used
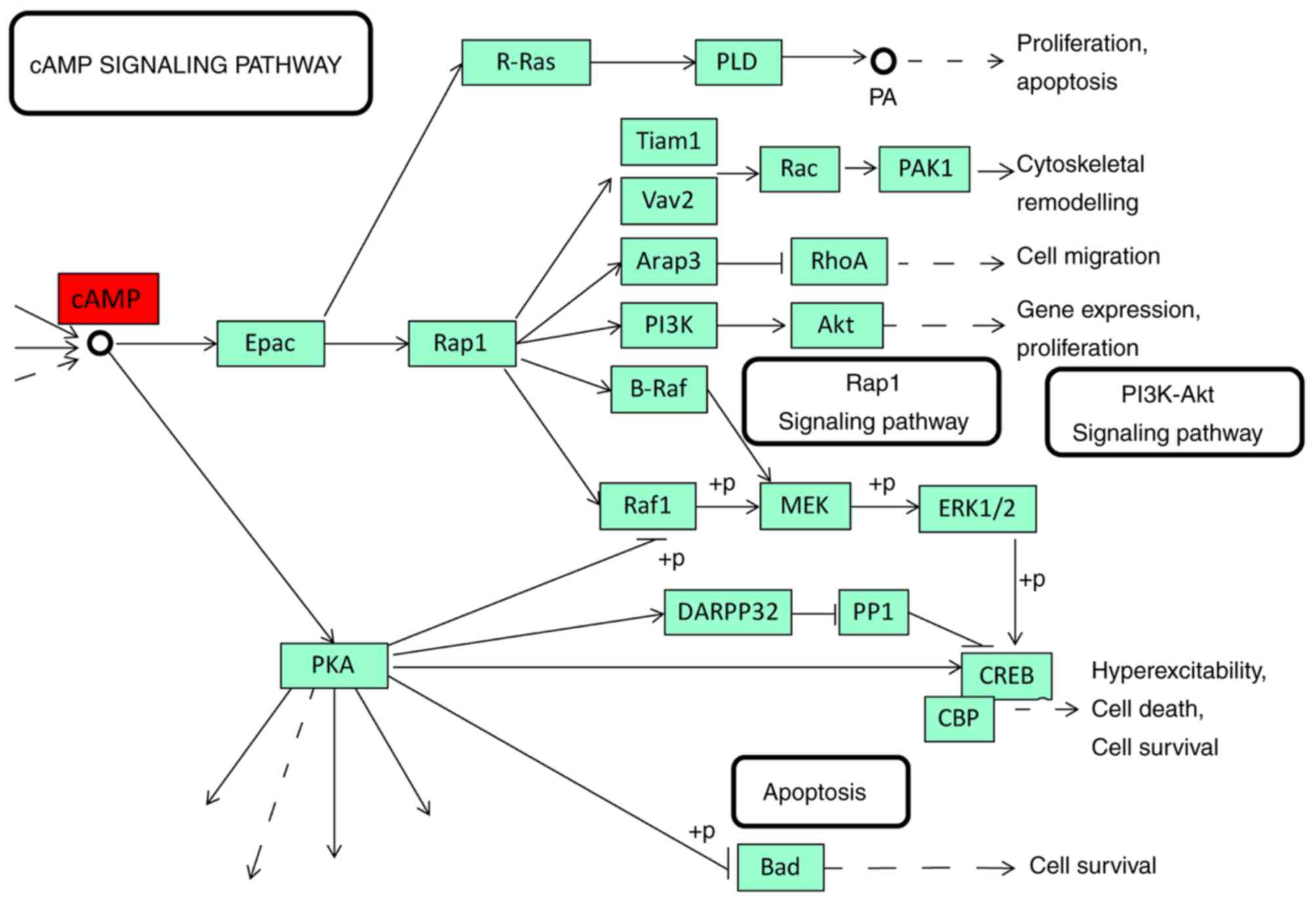
to identify the top 20 biological processes of CRH gene enrichment
(Table III) and a part of an
important signaling pathway of cAMP was generated on KEGG database
(Fig. 6).
 | Table III.Top 20 biological processes
significantly associated with CRH expression. |
Table III.
Top 20 biological processes
significantly associated with CRH expression.
| Rank | Pathway ID | Pathway
description | Associated
proteins |
|---|
| 1 | GO.0030819 | Positive regulation
of camp biosynthetic process | ADCYAP1, AVP, CRH,
CRHR1, CRHR2, GCG, GHRH, MC2R, MC4R |
| 2 | GO.0007187 | G-protein coupled
receptor signaling pathway, coupled to cyclic nucleotide second
messenger | ADCYAP1, CRHR1,
CRHR2, GCG, GHRH, MC2R, MC4R |
| 3 | GO.0007188 | Adenylate
cyclase-modulating G-protein coupled receptor signaling
pathway | ADCYAP1, CRHR1,
CRHR2, GCG, GHRH, MC4R |
| 4 | GO.0045935 | Positive regulation
of nucleobase-containing compound metabolic process | ADCYAP1, AVP, CRH,
CRHR1, CRHR2, GCG, GHRH, MC2R, MC4R, POMC |
| 5 | GO.0031328 | Positive regulation
of cellular biosynthetic process | ADCYAP1, AVP, CRH,
CRHR1, CRHR2, GCG, GHRH, MC2R, MC4R, POMC |
| 6 | GO.0050795 | Regulation of
behavior | CRH, CRHR1, CRHR2,
GHRH, MC4R, NPS |
| 7 | GO.2000852 | Regulation of
corticosterone secretion | CRH, CRHR1,
POMC |
| 8 | GO.0007218 | Neuropeptide
signaling pathway | ADCYAP1, CRHR1,
MC2R, NPS, POMC |
| 9 | GO.0007267 | Cell-cell
signaling | ADCYAP1, AVP, CRH,
CRHR1, GHRH, MC2R, MC4R, POMC |
| 10 | GO.0045937 | Positive regulation
of phosphate metabolic process | ADCYAP1, CRH,
CRHR1, CRHR2, GCG, GHRH, MC2R, MC4R |
| 11 | GO.1903532 | Positive regulation
of secretion by cell | AVP, CRH, CRHR1,
CRHR2, GCG, GHRH |
| 12 | GO.0032811 | Negative regulation
of epinephrine secretion | CRH, CRHR1,
CRHR2 |
| 13 | GO.0044060 | Regulation of
endocrine process | CRH, CRHR1, CRHR2,
POMC |
| 14 | GO.2000252 | Negative regulation
of feeding behavior | CRHR1, CRHR2,
MC4R |
| 15 | GO.0048521 | Negative regulation
of behavior | CRH, CRHR1, CRHR2,
MC4R |
| 16 | GO.0051952 | Regulation of amine
transport | AVP, CRH, CRHR1,
CRHR2 |
| 17 | GO.0021536 | Diencephalon
development | CRH, CRHR1, CRHR2,
GHRH |
| 18 | GO.0042753 | Positive regulation
of circadian rhythm | CRH, GHRH, NPS |
| 19 | GO.0046883 | Regulation of
hormone secretion | CRH, CRHR2, GCG,
GHRH, POMC |
| 20 | GO.0042749 | Regulation of
circadian sleep/wake cycle | CRH, GHRH, NPS |
Discussion
Oncogenesis is a multi-factorial process, and
numerous genes have been identified to be involved in HCC
oncogenesis (22). However, the role
of CRHBP expression in HCC remains unclear. The aim of the present
study was to investigate the association between CRHBP expression
and HCC.
Low expression of CRHBP was associated with large
tumor size (P=0.013) and high Edmondson Grade (P=0.002). This
supports previous research in human kidney cancer which suggested
that low expression of CRHBP in kidney tumor cell lines could
promote proliferation (16). In the
present study, microvascular invasion and metastasis had no
association with CRHBP expression; however, Tezval et al
(15) reported that clear cell renal
cell cancer with low expression of CRHBP had strong invasion and
metastasis. This may be due to the different sources of the tissues
or the change from in vivo to ex vivo
microenvironment. Besides, as CRHBP has a high affinity for CRH and
urocortin (23), we could infer that
when CRHBP expression was downregulated, the stimulate signal from
CRH would be decreased, and the physiological of the hepatocyte
might be changed. As it has been previously reported that CRH
promotes the proliferation of human colon cancer cells and
upregulates VEGF expression (24).
Gene enrichment analysis of the biological processes associated
with CRH indicated that CRH may regulate cytobiological state via
the cAMP signaling pathway. We hypothesize that CRHBP may prevent
CRH from activating CRHRs in incorrect locations or in excess.
Although HBV infection was demonstrated to be
associated with CRHBP expression (P=0.020), the molecular mechanism
behind downregulation of CRHBP following HBV infection remains
uncharacterized. Previous research suggests that CRBHP expression
was decreased following long-term stimulation by HBV (13,25,26), and
that hypermethylation of the CRHBP gene caused its downregulation
(15).
The results of the present study may have valuable
clinical application. Upregulation of CRHBP expression may inhibit
the proliferation of hepatocellular carcinoma cells as well as
tumor progression. Clinicians should be aware of the significance
of HBV infection when considering therapeutic strategies.
In conclusion, CRHBP expression was demonstrated to
be associated with high AFP level and overall survival rate in HCC,
and may be a potential biomarker for HCC diagnosis, treatment and
prognosis.
Acknowledgements
Not applicable.
Funding
This study was supported by the National Science
Foundation of China (grant nos. 81672474, 81672430, 81602706 and
81602174), Zhejiang Provincial Natural Science Foundation of China
(grant nos. LY15H160051, LQ16H160017 and LY16H160042), Funds of
Science Technology Department of Zhejiang Province (grant no.
2016C33055) and the Zhejiang Province Bureau of Health (grant nos.
WKJ-ZJ-1502 and 2015ZA009).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HX and HW wrote the manuscript and conducted the
experiments; LF, SW and LL performed data analysis. GR and XH were
in charge of sample processing. XM designed the present study and
revised the manuscript. XT and DH collected clinical data and
provided funding.
Ethics approval and consent to
participate
The research was approved by Review Board of
Hospital Ethics Committee, and the informed consent from each
participant was obtained before data collection.
Patient consent for publication
Written informed consent was obtained from the
patient for the publication of this paper together with the
accompanying images.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Elsegood CL, Tirnitz-Parker JE, Olynyk JK
and Yeoh GC: Immune checkpoint inhibition: Prospects for prevention
and therapy of hepatocellular carcinoma. Clin Transl Immunol.
6:e1612017. View Article : Google Scholar
|
|
4
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen W, Zheng R, Zeng H, Zhang S and He J:
Annual report on status of cancer in China, 2011. Chin J Cancer
Res. 27:2–12. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Llovet JM and Bruix J: Systematic review
of randomized trials for unresectable hepatocellular carcinoma:
Chemoembolization improves survival. Hepatology. 37:429–442. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chaffer CL and Weinberg RA: A perspective
on cancer cell metastasis. Science. 331:1559–1564. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ketchesin KD, Stinnett GS and Seasholtz
AF: Corticotropin-releasing hormone-binding protein and stress:
From invertebrates to humans. Stress. 20:449–464. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Behan DP, Linton EA and Lowry PJ:
Isolation of the human plasma corticotrophin-releasing
factor-binding protein. J Endocrinol. 122:23–31. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Peto CA, Arias C, Vale WW and Sawchenko
PE: Ultrastructural localization of the corticotropin-releasing
factor-binding protein in rat brain and pituitary. J Comp Neurol.
413:241–254. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sutton SW, Behan DP, Lahrichi SL, Kaiser
R, Corrigan A, Lowry P, Potter E, Perrin MH, Rivier J and Vale WW:
Ligand requirements of the human corticotropin-releasing
factor-binding protein. Endocrinology. 136:1097–1102. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Borowski KS, Clark EA, Lai Y, Wapner RJ,
Sorokin Y, Peaceman AM, Iams JD, Leveno KJ, Harper M, Caritis SN,
et al: Neonatal genetic variation in steroid metabolism and key
respiratory function genes and perinatal outcomes in single and
multiple courses of corticosteroids. Am J Perinatol. 32:1126–1132.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kolasa M, Faron-Górecka A, Kuśmider M,
Szafran-Pilch K, Solich J, Żurawek D, Gruca P, Papp M and
Dziedzicka-Wasylewska M: Differential stress response in rats
subjected to chronic mild stress is accompanied by changes in
CRH-family gene expression at the pituitary level. Peptides.
61:98–106. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nan H, Dorgan JF and Rebbeck TR: Genetic
variants in hypothalamic-pituitary-adrenal axis genes and breast
cancer risk in Caucasians and African Americans. Int J Mol
Epidemiol Genet. 6:33–40. 2015.PubMed/NCBI
|
|
15
|
Tezval H, Dubrowinskaja N, Peters I, Reese
C, Serth K, Atschekzei F, Hennenlotter J, Stenzl A, Kuczyk MA and
Serth J: Tumor specific epigenetic silencing of corticotropin
releasing hormone-binding protein in renal cell carcinoma:
Association of hypermethylation and metastasis. PloS One.
11:e01638732016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tezval H, Atschekzei F, Peters I, Waalkes
S, Hennenlotter J, Stenzl A, Becker JU, Merseburger AS, Kuczyk MA
and Serth J: Reduced mRNA expression level of
corticotropin-releasing hormone-binding protein is associated with
aggressive human kidney cancer. BMC Cancer. 13:1992013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wurmbach E, Chen YB, Khitrov G, Zhang W,
Roayaie S, Schwartz M, Fiel I, Thung S, Mazzaferro V, Bruix J, et
al: Genome-wide molecular profiles of HCV-induced dysplasia and
hepatocellular carcinoma. Hepatology. 45:938–947. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen X, Cheung ST, So S, Fan ST, Barry C,
Higgins J, Lai KM, Ji J, Dudoit S, Ng IO, et al: Gene expression
patterns in human liver cancers. Mol Biol Cell. 13:1929–1939. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Roessler S, Jia HL, Budhu A, Forgues M, Ye
QH, Lee JS, Thorgeirsson SS, Sun Z, Tang ZY, Qin LX and Wang XW: A
unique metastasis gene signature enables prediction of tumor
relapse in early-stage hepatocellular carcinoma patients. Cancer
Res. 70:10202–10212. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mas VR, Maluf DG, Archer KJ, Yanek K, Kong
X, Kulik L, Freise CE, Olthoff KM, Ghobrial RM, McIver P and Fisher
R: Genes involved in viral carcinogenesis and tumor initiation in
hepatitis C virus-induced hepatocellular carcinoma. Mol Med.
15:85–94. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Burkhart RA, Ronnekleiv-Kelly SM and
Pawlik TM: Personalized therapy in hepatocellular carcinoma:
Molecular markers of prognosis and therapeutic response. Surg
Oncol. 26:138–145. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Inda C, Armando NG, Dos Santos Claro PA
and Silberstein S: Endocrinology and the brain:
Corticotropin-releasing hormone signaling. Endocr Connect.
6:R99–R120. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fang X, Hong Y, Dai L, Qian Y, Zhu C, Wu B
and Li S: CRH promotes human colon cancer cell proliferation via
IL-6/JAK2/STAT3 signaling pathway and VEGF-induced tumor
angiogenesis. Mol Carcinog. 56:2434–2445. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gouraud SS, Takagishi M, Kohsaka A, Maeda
M and Waki H: Altered neurotrophic factors' expression profiles in
the nucleus of the solitary tract of spontaneously hypertensive
rats. Acta Physiol (Oxf). 216:346–357. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Linnstaedt SD, Bortsov AV, Soward AC, Swor
R, Peak DA, Jones J, Rathlev N, Lee DC, Domeier R, Hendry PL and
McLean SA: CRHBP polymorphisms predict chronic pain development
following motor vehicle collision. Pain. 157:273–279. 2016.
View Article : Google Scholar : PubMed/NCBI
|