Introduction
Multiple myeloma (MM) is a heterogeneous tumor
featured by infiltration of the bone marrow by malignant plasma
cells. It ranks as the second most prevalent disease among the yet
incurable hematologic malignancies, with survival durations ranging
from a few months to more than 10 years (1,2). Although
the mortality of MM has decreased because of the development of
high-dose therapy and innovative agents such as bortezomib,
thalidomide and lenalidomide, long-term prognosis remains poor
(3). Early diagnosis and treatment
can slow disease progression and enhance prognosis, but patients
with late-stage disease are usually unresponsive to therapeutic
interventions. Therefore, finding noninvasive biomarkers for the
detection of cancer may be beneficial for patient survival. Some
studies have revealed that molecular markers, including circulating
microRNAs (miRNAs), can be useful in diagnosing and monitoring
tumors (4,5). Therefore, a highly sensitive and
specific noninvasive biomarker that diagnoses MM and assists in
determining the prognosis for patients is needed.
miRNAs are noncoding RNA molecules approximately
19–25 nucleotides in length. miRNAs suppress mRNA translation and
gene expression by pairing with the 3′-untranslated region (3′UTR)
of target mRNAs (6). Since distinct
miRNA expression determines the signatures of various cancers,
miRNAs may have strong association with cancer pathogenesis and
progression (7). Indeed, abnormal
miRNA expression, which has been found in MM, leads to
carcinogenesis and cancer progression by promoting oncogene
expression (8,9). Earlier studies have revealed that
miR-125b-5p, which is upregulated in different types of cancers,
may belong to a novel class of oncogenes (10,11).
However, most of those studies focused on tissue- or cell-based
miR-125b-5p expression. To make use of this noninvasive biomarker,
the diagnostic and prognostic value of circulating miRNA-125b-5p in
patients with MM must be ascertained.
The purpose of this study was to probe into the
differential expression (DE) of global miRNAs in the plasma of MM
patients and to compare with miRNAs in healthy individuals. We used
an miRNA microarray analysis to determine the expression of eight
miRNAs (miR-125b-5p, miR-483-3p, miR-4326, miR-6894-3p, miR-4498,
miR-490-3p, miR-7155-5p, and miR-937-3p) that are notably increased
in MM patients and compared them to the levels in healthy
individuals. By quantitatively analyzing these miRNAs in a second
clinical study, we discovered that the levels of five miRNAs
(miR-125b-5p, miR-4326, miR-4498, miR-490-3p, and miR-7155-5p) were
increased in the plasma of all patients. Receiver operating
characteristic (ROC) analysis illustrated that miR-125b-5p and
miR-490-3p displayed considerable diagnostic accuracy for MM.
Moreover, plasma miR-125b-5p levels were correlated with shorter
event-free survival (EFS). Therefore, this study determined the
diagnostic value of circulating miRNAs and identified whether
miRNAs could be potentially prognostic biomarkers for MM.
Materials and methods
Tissue specimens and patient
characteristics
The specimens in this study were obtained from 41
patients who were diagnosed in accordance with the National
Comprehensive Cancer Network (NCCN) clinical practice guidelines
for MM at the 1st Affiliated Hospital of Nanchang University from
October 2015 to July 2017 (12).
Among the 41 plasma specimens from MM patients, 6 were used for
microarray analysis, and 35 served as the validation set. All
patients who receiving chemotherapy and/or biotherapy were
excluded, and patients with other types of malignant tumors were
also eliminated. From healthy controls, we used 20 plasma samples
in the validation set, who range in age from 17 to 63. 20 healthy
individuals were recruited to the 1st Affiliated Hospital of
Nanchang University between October 2015 to July 2017, including 8
males and 12 females. Venous blood was collected into tubes with
EDTA (BD Biosciences, Franklin Lakes, NJ, USA). Plasma was
transferred to a sterile tube and then immediately stored at −80°C
after being frozen in liquid nitrogen. The study was approved by
the Medical Research Ethics Committee of The First Affiliated
Hospital of Nanchang University (Nanchang, China), and written
informed consent was obtained from all study subjects.
Additionally, FISH analysis was performed in 35 patients. Purified
MM patient PCs (~105 cells) were classified using the
FISH technique. The following probe sets obtained from Vysis
(Abbott Diagnostics, Berkshire, UK) were used for classification.
D13S319 Spectrum-Orange/LSI-13q34-Spectrum-Green (del(13q) probe),
IgH Spectrum-Green/FGFR3 Spectrum-Orange (t(4;14) fusion probe),
IgH Spectrum-Green/CCND1 Spectrum-Orange (t(11;14) fusion probe),
IgH Spectrum-Green/MAF Spectrum-Orange (t(14;16) fusion probe), and
IgH dualcolor (Sperctrum-Green/Spectrum-Orange) break-apart probe.
At least 100 nuclei were scored for each probe. 35 MM patients were
cytogenetically classified using FISH, then we investigate the
miR-125b-5p expression in the different genetic subtypes of MM
patients. The 35 MM patient samples were from the original cohort
of 41 patients with MM that were recruited to the study. The
patients details are described in Table
I.
 | Table I.Clinical characteristics of patients
with MM used for reverse transcription-quantitative polymerase
chain reaction analysis of plasma samples (n=35). |
Table I.
Clinical characteristics of patients
with MM used for reverse transcription-quantitative polymerase
chain reaction analysis of plasma samples (n=35).
| Characteristic | n (%) |
|---|
| Sex |
|
| Male | 23 (65.7) |
|
Female | 12 (34.3) |
| Age range
(years) | 35–75 |
| Mean age (years) | 59 |
| Durie-salmon
stage |
|
| I | 10 (28.57) |
| II | 6 (17.14) |
| III | 19 (54.29) |
| Karyotype |
|
|
t(4:14) | 9 (25.71) |
|
t(11:14) | 5 (14.28) |
|
t(14:16) | 6 (17.14) |
| Del(13q)
as a unique abnormality | 10 (28.57) |
| Normal
FISH | 5 (14.29) |
miRNA array
Total RNA was extracted from samples from 6 MM
patients and 6 healthy controls using phenol-chloroform (TRIzol;
Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA). The
quality of RNA was assessed by capillary electrophoresis (Agilent
Technologies, Inc., Santa Clara, CA, USA). Libraries for small RNA
sequencing were prepared using NEB kits (New England Biolabs, Inc.,
Ipswich, MA, USA). Libraries were quantified using Agilent
Bioanalyzer 2100 system by DNA high sensitivity chips. Clean reads
21–22 nucleotides in length were screened for miRNAs and mapped
onto a reference genome with Bowtie. The functions of new miRNAs
were analyzed by miRDeep2 software. DE sequence was used to
quantify differentially expressed miRNAs and to assess statistical
significance.
Validation of plasma miRNAs by
qRT-PCR
TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) was used to extract total RNA. cDNA was reverse
transcribed with the M-MLV Reverse Transcriptase kit (GeneCopoeia,
Rockville, MD, USA) following the manufacturer's protocol.
Approximately 1,200 µl plasma was employed for miRNA isolation. We
employed quantitative reverse transcription PCR (qRT-PCR) with
SYBR-Green (Takara, Osaka, Japan) to detect miR-125b-5p,
miR-483-3p, miR-4326, miR-6894-3p, miR-4498, miR-490-3p,
miR-7155-5p, and miR-937-3p expression levels, U6 was used as an
internal control. The reaction was performed via 40 amplification
cycles using the following protocol: 95°C for 3 min, 95°C for 45
sec, 55°C for 15 sec, and 72°C for 50 sec. There was no significant
difference in the U6 Ct value between control and MM samples. The
ΔCt=average Ct (miRNA of target)-average Ct (U6). The primers used
in PCR were as follows: miR-125b-5p forward,
5′-TGCGCTCCCTGAGACCCTAAC-3′ and reverse,
5′-CCAGTGCAGGGTCCGAGGTATT-3′; miR-483-3p forward,
5′-TGCGCTCACTCCTCTCCTCCC-3′ and reverse,
5′-CTCAAGTGTCGTGGAGTCGGCAA-3′; miR-4326 forward,
5′-GCCCGCTGTTCCTCTGTCTCCC-3′ and reverse, 5′-GTGCAGGGTCCGAGGT-3′;
miR-6894-3p forward, 5′-TGCGCTTGCCTGCCCTCTTCC-3′ and reverse,
5′-CTCAAGTGTCGTGGAGTCGGCAA-3′; miR-4498 forward,
5′-TGCGCTGGGCTGGTAGGGCAAG-3′ and reverse,
5′-CTCAAGTGTCGTGGAGTCGGCAA-3′; miR-490-3p forward,
5′-TGCGCCAACCTGGAGGATCCA-3′ and reverse,
5′-CTCAAGTGTCGTGGAGTCGGCAA-3′; miR-7155-5p forward,
5′-TGCGCTCTGGGGTCTTGGG-3′ and reverse,
5′-CTCAAGTGTCGTGGAGTCGGCAA-3′; miR-937-3p forward,
5′-TGCGCATCCGCGCTCTGACTCT-3′ and reverse,
5′-CTCAAGTGTCGTGGAGTCGGCAA-3′; and U6 forward,
5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCG-3′.
Samples were analyzed in triplicate, and gene expression was
quantified by normalizing target gene expression to that of the
internal control using the 2-ΔΔCt formula.
Statistical analysis
Data were analyzed using SPSS 19.0 software. All
assays were repeated three times, and data were represented as the
mean ± SD. Two-tailed t-tests were performed to compare the
difference in plasma miRNA expression levels. Student's t-test was
used to determine the significance of difference between two
groups, and ROC curves were constructed to evaluate the prediction
ability of the miRNAs for MM patients. EFS curves were constructed
using the Kaplan-Meier method and compared using the log-rank test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
miRNA microarray analysis
We carried out an miRNA microarray analysis to
screen candidate plasma miRNAs in the treatment group (MM patients)
and the control group. After data processing and analysis, we
identified a set of 12 miRNAs that were differentially expressed
between the treatment group (MM patients) and the control group,
among which, 8 were significantly upregulated, and 4 were
significantly downregulated (Fig.
1).
Detection of differentially expressed
miRNA by qRT-PCR
To further validate the differentially expressed
miRNA identified by the miRNA microarray analysis, we used qRT-PCR
to confirm the eight elevated miRNAs (miR-125b-5p, miR-483-3p,
miR-4326, miR-6894-3p, miR-4498, miR-490-3p, miR-7155-5p, and
miR-937-3p) in plasma samples from 35 MM patients and 20 healthy
individuals. The results suggested that the plasma levels of five
miRNAs, namely, miR-125b-5p, miR-4326, miR-4498, miR-490-3p and
miR-7155-5p (Fig. 2), were
significantly increased between the two groups. In addition,
miR-125b-5p expression was associated with extramedullary
infiltration and was markedly higher in stage III patients
(1.76±0.03) than in stage I/II patients (1.62±0.04) (both
P<0.05). However, miR-125b-5p expression was not correlated with
patient age, sex or karyotype (all P>0.05) (Table II).
 | Table II.Association between miR-125b-5p
expression and the clinical pathological characteristics of
patients with MM. |
Table II.
Association between miR-125b-5p
expression and the clinical pathological characteristics of
patients with MM.
| Charactaristic | Cases | miR-125b-5p
expression | P-value |
|---|
| Age (years) |
|
| 0.241 |
| ≤50 | 16 | 1.66±0.03 |
|
|
>50 | 19 | 1.72±0.05 |
|
| Sex |
|
| 0.439 |
|
Male | 23 | 1.68±0.03 |
|
|
Female | 12 | 1.76±0.03 |
|
| Durie-salmon
stage |
|
| 0.009 |
|
I/II | 16 | 1.62±0.04 |
|
|
III | 19 | 1.76±0.03 |
|
| Extramedullary
infltration |
|
| 0.012 |
|
Yes | 8 | 1.82±0.05 |
|
| No | 27 | 1.66±0.03 |
|
| Karotype |
|
| 0.455 |
| Normal
FISH | 5 | 1.65±0.12 |
|
|
Abnormal FISH | 30 | 1.72±0.03 |
|
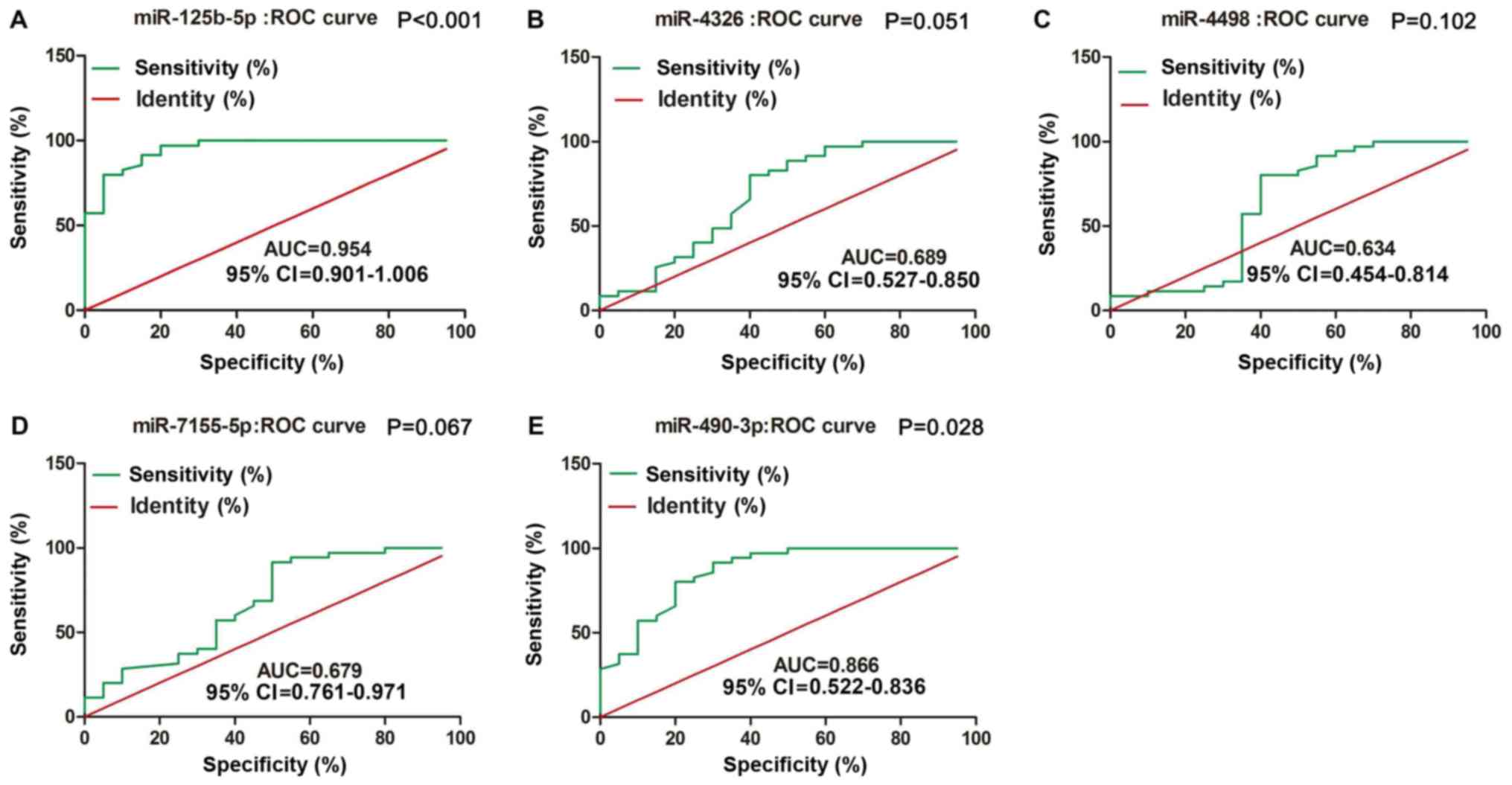
miR-125b-5p and miR-490-3p can serve
as diagnostic tools in MM
To compare the diagnostic value of miRNAs with that
of typical tumor markers such as carcinoembryonic antigen (CEA) and
cancer antigen 199 (CA199), we constructed ROC curves to validated
plasma miRNAs in identifying MM patients. As shown in Fig. 3, miR-125b-5p and miR-490-3p were
accurate in distinguishing MM patients from healthy controls. The
area under the curve (AUC) for miR-125b-5p was 0.954, with 86%
sensitivity and 96% specificity [95% confidence interval (CI)
0.901–1.006; P<0.001]; the AUC for miR-490-3p was 0.866, with
60% sensitivity and 85% specificity (95% CI 0.522–0.836;
P=0.028).
Prognostic value of plasma miRNA
expression in MM
We further assessed the correlation between miRNA
expression and MM prognosis by Kaplan-Meier analysis. MM patients
receive the chemotherapy of bortezomib, thalidomide plus
dexamethasone (VTD) regimen, followed by thalidomide maintenance
therapy. MM patients were divided into either high or low
miR-125b-5p/miR-490-3p expression subgroups. Using this approach,
we compared EFS between high- and low-expression subgroups. EFS
durations were calculated from the time of diagnosis to the date of
relapse or death. After a median follow-up 9.5 months (range, 1–26
months) from diagnosis, the median EFS duration in patients
expressing high levels of miR-125b-5p was 8 months, and that in
patients expressing low levels of miR-125b-5p was 13 months. Our
results indicated that patients with higher miR-125b-5p expression
had a shorter EFS than patients with lower miR-125b-5p expression
(P=0.02, Fig. 4). However, there was
no difference between high and low miR-490-3p expression groups,
with median EFS of 12 and 13 months, respectively (P=0.23; Fig. 4).
Discussion
Plasma miRNAs have been previously reported in the
context of some cancers (4,13,14). In
the present study, we performed an miRNA microarray analysis to
elucidate the expression profiles of plasma miRNA in six MM
patients and six healthy individuals. In accordance with the
microarray analysis, we found that the levels of eight
differentially expressed miRNAs were significantly higher in MM
patients than in healthy individuals. We then validated the
elevated levels of five of those (miR-125b-5p, miR-4326, miR-4498,
miR-490-3p and miR-7155-5p) in a second clinical study. Most of the
targets of these miRNAs are cancer-associated genes such as APC2,
IGF2, and IRF4 (15–17) that are involved in cell proliferation,
cell cycle, apoptosis, invasion and metastasis.
Furthermore, ROC analysis demonstrated that
miR-125b-5p and miR-490-3p were accurate in distinguishing MM
patients from healthy subjects, resulting in AUCs of 0.954
(sensitivity=86%, specificity=96%, 95% CI 0.901–1.006, P<0.001)
and 0.866 (sensitivity=60%, specificity=85%, 95% CI 0.522–0.836,
P=0.028), respectively. However, no significant association was
found between miR-490-3p and EFS of MM patients. Moreover, our data
showed that patients with higher miR-125b-5p expression had a
shorter EFS. Additionally, miR-125b-5p levels were correlated with
international staging system (ISS) stage, thereby indicating the
diagnostic value of miR-125b-5p.
Deregulation of miR-490-3p has been observed in many
types of cancers, such as lung cancer (18), colorectal cancer (19) and gastric cancer (20). Zhang et al demonstrated that
the urinary level of miR-490 is positively associated with focal
segmental glomerulosclerosis (FSGS) disease activity, and thus
miR-490 might be a promising biomarker for evaluating FSGS disease
activity (21). In this study,
miR-490 was overexpressed in MM patient samples and could thus be
used as an early tumor biomarker. miR-125b-5p has been demonstrated
to act as either cancer promoter or cancer suppressor in different
types of cancer. Upregulation of miR-125b-5p has been detected in
pancreatic cancer (22), prostate
cancer (23), and acute myeloid
leukemia (24), while downregulation
of miR-125b-5p has been observed in thyroid cancer (25) and oral squamous cell carcinoma
(26). miR-125b-5p has also been
found to promote tumor cell proliferation and inhibit p53-dependent
apoptosis in human neuroblastoma cells (27). Several studies indicate that p53 is
the direct target of miR-125b, and MM cells responding to
dexamethasone exhibit enhanced expression of the oncogenic miR-125b
(28,29). However, the mechanism by which
miR-125b regulates MM remains unknown. Importantly, circulating
miR-125b-5p has been reported to be correlated with colorectal
cancer (30), breast cancer (31) and hepatocellular carcinoma (32). Circulating and serum-derived miRNAs
have also been identified in MM patients (33). In our study, plasma miR-125b-5p levels
were correlated with extramedullary infiltration and ISS stage in
MM patients. Therefore, we concluded that plasma miR-125b-5p may be
correlated with poor prognosis in MM patients.
Circulating serum/plasma miRNA expression profiles
have been identified and used as noninvasive biomarkers for tumor
identification and prognosis (34).
Furthermore, some circulating miRNAs may be correlated with
chemotherapeutic resistance in tumors (31,35).
Therefore, the expression profiles of circulating miRNAs can be
used to monitor disease progression. To the best of our knowledge,
this is the first study to explore the relevance of plasma
miR-125b-5p to MM. We demonstrated that miR-125b-5p can be used as
a feasible biomarker with significant accuracy in diagnosing
patients with MM. Nevertheless, there are several limitations in
our study. First, our sample size is small, and long-term follow-up
is required to validate the relationship between miR-125b-5p levels
and patient outcomes. In addition, large-scale and in-depth studies
are necessary. Second, the potential mechanisms of miR-125b-5p in
MM were not elucidated.
In conclusion, this study demonstrated that
circulating miR-125b-5p levels can be used as a diagnostic and
predictive biomarker in MM. miR-125b-5p can be used to identify MM
patients with poor prognosis and aid in the selection of timely
comprehensive therapy.
Acknowledgements
Not applicable.
Funding
International Collaboration Fund from National
Science and Technology Committee of China (grant no. 2011DFA32820).
The National Natural Science Fund Project (grant no. 81460037 and
81760040). The National Science Foundation for Young Scientists of
China (grant no. 81600180). Innovation Fund Project in Jiangxi
Province (grant no. YC2016-B018).
Availability of data and materials
The datasets used during the current study are
available from the corresponding author on reasonable request.
Authors' contributions
YJ and YL performed the molecular biology
experiments and drafted the manuscript. HC performed the
statistical analysis. GC conceived the study and participated in
its design and helped to draft the manuscript.
Ethics approval and consent to
participate
The present study was approved by the Medical
Research Ethics Committee of The First Affiliated Hospital of
Nanchang University (Nanchang, China) and written informed consent
was obtained from all study participants.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sonneveld P, Avet-Loiseau H, Lonial S,
Usmani S, Siegel D, Anderson KC, Chng WJ, Moreau P, Attal M, Kyle
RA, et al: Treatment of multiple myeloma with high-risk
cytogenetics: A consensus of the International myeloma working
group. Blood. 127:2955–2962. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Abdi J, Chen G and Chang H: Erratum: Drug
resistance in multiple myeloma: Latest findings and new concepts on
molecular mechanisms. Oncotarget. 6:73642015.PubMed/NCBI
|
|
3
|
Mimura N, Hideshima T and Anderson KC:
Novel therapeutic strategies for multiple myeloma. Exp Hematol.
43:732–741. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liao TL, Chen YM, Hsieh CW, Chen HH, Lee
HC, Hung WT, Tang KT and Chen DY: Upregulation of circulating
microRNA-134 in adult-onset Still's disease and its use as
potential biomarker. Sci Rep. 7:42142017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pilli T, Cantara S, Marzocchi C, Cardinale
S, Santini C, Cevenini G and Pacini F: Diagnostic value of
circulating microRNA-95 and −190 in the differential diagnosis of
thyroid nodules: A validation study in 1000 consecutive patients.
Thyroid. 27:1053–1057. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang B, Pan X, Cobb GP and Anderson TA:
microRNAs as oncogenes and tumor suppressors. Dev Biol. 302:1–12.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zheng P, Guo H, Li G, Han S, Luo F and Liu
Y: PSMB4 promotes multiple myeloma cell growth by activating
NF-κB-miR-21 signaling. Biochem Biophys Res Commun. 458:328–333.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shen X, Guo Y, Yu J, Qi J, Shi W, Wu X, Ni
H and Ju S: miRNA-202 in bone marrow stromal cells affects the
growth and adhesion of multiple myeloma cells by regulating B
cell-activating factor. Clin Exp Med. 16:307–316. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shi XB, Xue L, Yang J, Ma AH, Zhao J, Xu
M, Tepper CG, Evans CP, Kung HJ and deVere White RW: An
androgen-regulated miRNA suppresses Bak1 expression and induces
androgen-independent growth of prostate cancer cells. Proc Natl
Acad Sci USA. 104:19983–19988. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bousquet M, Quelen C, Rosati R, Mansat-De
Mas V, La Starza R, Bastard C, Lippert E, Talmant P,
Lafage-Pochitaloff M, Leroux D, et al: Myeloid cell differentiation
arrest by miR-125b-1 in myelodysplastic syndrome and acute myeloid
leukemia with the t(2;11) (p21;q23) translocation. J Exp Med.
205:2499–2506. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Anderson KC: Progress and paradigms in
multiple myeloma. Clin Cancer Res. 22:5419–5427. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Riazalhosseini B, Mohamed R, Apalasamy YD,
Langmia IM and Mohamed Z: Circulating microRNA as a marker for
predicting liver disease progression in patients with chronic
hepatitis B. Rev Soc Bras Med Trop. 50:161–166. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wu L, Hu B, Zhao B, Liu Y, Yang Y, Zhang L
and Chen J: Circulating microRNA-422a is associated with lymphatic
metastasis in lung cancer. Oncotarget. 8:42173–42188.
2017.PubMed/NCBI
|
|
15
|
Xu G, Zhang Z, Zhang L, Chen Y, Li N, Lv
Y, Li Y and Xu X: miR-4326 promotes lung cancer cell proliferation
through targeting tumor suppressor APC2. Mol Cell Biochem.
443:151–157. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bertero T, Gastaldi C, Bourget-Ponzio I,
Imbert V, Loubat A, Selva E, Busca R, Mari B, Hofman P, Barbry P,
et al: miR-483-3p controls proliferation in wounded epithelial
cells. FASEB J. 25:3092–3105. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Morelli E, Leone E, Cantafio ME, Di
Martino MT, Amodio N, Biamonte L, Gullà A, Foresta U, Pitari MR,
Botta C, et al: Selective targeting of IRF4 by synthetic
microRNA-125b-5p mimics induces anti-multiple myeloma activity in
vitro and in vivo. Leukemia. 29:2173–2183. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gu H, Yang T, Fu S, Chen X, Guo L and Ni
Y: MicroRNA-490-3p inhibits proliferation of A549 lung cancer cells
by targeting CCND1. Biochem Biophys Res Commun. 444:104–108. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zheng K, Zhou X, Yu J, Li Q, Wang H, Li M,
Shao Z, Zhang F, Luo Y, Shen Z, et al: Epigenetic silencing of
miR-490-3p promotes development of an aggressive colorectal cancer
phenotype through activation of the Wnt/β-catenin signaling
pathway. Cancer Lett. 376:178–187. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhou B, Wang Y, Jiang J, Jiang H, Song J,
Han T, Shi J and Qiao H: The long noncoding RNA colon
cancer-associated transcript-1/miR-490 axis regulates gastric
cancer cell migration by targeting hnRNPA1. IUBMB Life. 68:201–210.
2016. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang W, Zhang C, Chen H, Li L, Tu Y, Liu
C, Shi S, Zen K and Liu Z: Evaluation of microRNAs miR-196a,
miR-30a-5P, and miR-490 as biomarkers of disease activity among
patients with FSGS. Clin J Am Soc Nephrol. 9:1545–1552. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bloomston M, Frankel WL, Petrocca F,
Volinia S, Alder H, Hagan JP, Liu CG, Bhatt D, Taccioli C and Croce
CM: MicroRNA expression patterns to differentiate pancreatic
adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA.
297:1901–1908. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ma Q, Chen Z, Jia G, Lu X, Xie X and Jin
W: The histone demethylase PHF8 promotes prostate cancer cell
growth by activating the oncomiR miR-125b. Onco Targets Ther.
8:1979–1988. 2015.PubMed/NCBI
|
|
24
|
Liu J, Guo B, Chen Z, Wang N, Iacovino M,
Cheng J, Roden C, Pan W, Khan S, Chen S, et al: miR-125b promotes
MLL-AF9-driven murine acute myeloid leukemia involving a
VEGFA-mediated non-cell-intrinsic mechanism. Blood. 129:1491–1502.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Visone R, Pallante P, Vecchione A,
Cirombella R, Ferracin M, Ferraro A, Volinia S, Coluzzi S, Leone V,
Borbone E, et al: Specific microRNAs are downregulated in human
thyroid anaplastic carcinomas. Oncogene. 26:7590–7595. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Veerla S, Lindgren D, Kvist A, Frigyesi A,
Staaf J, Persson H, Liedberg F, Chebil G, Gudjonsson S, Borg A, et
al: MiRNA expression in urothelial carcinomas: important roles of
miR-10a, miR-222, miR-125b, miR-7 and miR-452 for tumor stage and
metastasis, and frequent homozygous losses of miR-31. Int J Cancer.
124:2236–2242. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xia HF, He TZ, Liu CM, Cui Y, Song PP, Jin
XH and Ma X: MiR-125b expression affects the proliferation and
apoptosis of human glioma cells by targeting Bmf. Cell Physiol
Biochem. 23:347–358. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Le MT, Teh C, Shyh-Chang N, Xie H, Zhou B,
Korzh V, Lodish HF and Lim B: MicroRNA-125b is a novel negative
regulator of p53. Genes Dev. 23:862–876. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Murray MY, Rushworth SA, Zaitseva L,
Bowles KM and Macewan DJ: Attenuation of dexamethasone-induced cell
death in multiple myeloma is mediated by miR-125b expression. Cell
Cycle. 12:2144–2153. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen H and Xu Z:
Hypermethylation-associated silencing of miR-125a and miR-125b: A
potential marker in colorectal cancer. Dis Markers.
2015:3450802015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang H, Tan G, Dong L, Cheng L, Li K, Wang
Z and Luo H: Circulating MiR-125b as a marker predicting
chemoresistance in breast cancer. PLoS One. 7:e342102012.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liu W, Hu J, Zhou K, Chen F, Wang Z, Liao
B, Dai Z, Cao Y, Fan J and Zhou J: Serum exosomal miR-125b is a
novel prognostic marker for hepatocellular carcinoma. Onco Targets
Ther. 10:3843–3851. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wong KY, Li Z, Zhang X, Leung GK, Chan GC
and Chim CS: Epigenetic silencing of a long non-coding RNA KIAA0495
in multiple myeloma. Mol Cancer. 14:1752015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Jairajpuri DS, Malalla ZH, Mahmood N and
Almawi WY: Circulating microRNA expression as predictor of
preeclampsia and its severity. Gene. 627:543–548. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Niu J, Xue A, Chi Y, Xue J, Wang W, Zhao
Z, Fan M, Yang CH, Shao ZM, Pfeffer LM, et al: Induction of
miRNA-181a by genotoxic treatments promotes chemotherapeutic
resistance and metastasis in breast cancer. Oncogene. 35:1302–1313.
2016. View Article : Google Scholar : PubMed/NCBI
|