Introduction
Sinonasal squamous cell carcinoma (SN-SCC) is a
malignant tumor derived from the respiratory epithelium of the
sinonasal cavity, which accounts for approximately 3–6% of all head
and neck cancers (1,2). SN-SCC appears mainly in nasal cavity and
approximately 30% of the patients are related to contact with
leather, textile or wood (3,4). Currently, the best treatment is
postoperative radiotherapy, accompanied by adjuvant chemotherapy.
Local recurrence of SN-SCC is the main reason for low survival rate
(5,6).
Therefore, looking for the additional options for treatment of
SN-SCC and knowing the underlying mechanism of SN-SCC progression
is necessary.
miRNAs have been reported to regulate gene
expression by binding to the 3′-UTR mRNA (7). Many miRNAs are thought to be
tissue-specific and the expression varies in different human cancer
cells. They regulate tumor cell development, such as lung, ovary,
breast cancer and cancer of the esophagus (8–11).
However, there are fewer results showing the role of miRNAs in head
and neck cancer including SN-SCC.
Abundant evidence shows that miR-34a expression was
lower in tumors and showed an inhibitory effect on the progression
of various cancers. For example, miR-34a exhibited suppression
effect on esophageal squamous cell carcinoma via regulating PLCE1
(12) and inhibited the progress of
gastric cancer via regulating HK1 (13). Also, miR-34a mimic inhibited ovarian
cancer and cervical cancer cell proliferation by targeting BCL-2
(14,15), whereas inhibiting miR-34a promoted
colorectal cancer and lung cancer development (16,17). A
study found that miR-34a expression was lower in SN-SCC and related
to the poor prognosis of patients (18). However, the biological function of
miR-34a in SN-SCC progression and whether miR-34a targeted BCL-2 in
regulating SN-SCC migration and invasion has not been previously
reported.
B-cell lymphoma-2 (BCL-2) plays an important role in
cell apoptosis. In all tumors, apoptosis is an important target for
treatment intervention. Previous studies had reported that BCL-2
induced apoptosis of mitochondrial cells, indicating that BCL-2
played a role in tumor development by blocking apoptosis (19). BCL-2 expression was higher in multiple
cancers and regulated by various miRNAs, such as pancreatic
adenocarcinoma regulated by miR-126 (20), breast cancer regulated by miR-27a
(21), gastric cancer by miR-711
(22), pancreatic cancer by miR-1180
(23). A previous study reported that
BCL-2 expression was obviously increased in SN-SCC (24,25) and it
is involved in the tumor cell proliferation, migration, invasion
and apoptosis. However, the biological mechanism of BCL-2 in SN-SCC
cell regulated by miR-34a has not been reported.
Here, we studied the potential links of miR-34a and
SN-SCC. Firstly, we found a low expression of miR-34a in SN-SCC
cells and tissues by RT-PCR, suggesting miR-34a inhibitory effect
in SN-SCC. Secondly, miR-34a inhibited SN-SCC cell migration and
invasion. Thirdly, we found that the potential mechanism of miR-34a
in SN-SCC was achieved by targeting BCL-2. We finally found that
inhibitory effect of miR-34a in SN-SCC could be attenuated by
BCL-2. Our research indicated that miR-34a/BCL-2axis was a
potential target for treating SN-SCC.
Materials and methods
SN-SCC samples
Fifty-two SN-SCC specimens were obtained from
patients who underwent surgery at Linyi People's Hospital (Linyi,
China) from 2011 to 2017. Experienced pathologists confirmed the
tumor and normal tissues through HE staining. Finally, tissues were
stored in a refrigerator at −80°C. The study was approved by the
Ethics Committee of Linyi People's Hospital. Signed written
informed consents were obtained from the patients or guardians.
Cell line establishment and cell
transfection
All SN-SCC cell lines (RPMI-2650, SCCNC2 and SCCNC7)
used in this study were derived from the previously untreated
primary SCC, which originated in the maxillary sinus. Cells were
cultured in DMEM, containing 10% FBS, penicillin (100 U/ml) and
streptomycin (100 µg/ml) (Beijing Solarbio Science & Technology
Co., Ltd., Beijing, China), incubated at 37°C with 5%
CO2.
Synthetic miR-34a mimic/inhibitor was provided by
GenePharma Co., Ltd. (Shanghai, China). We transfected miR-34a
mimic into SN-SCC cells to overexpress miR-34a or miR-34a inhibitor
to silence miR-34. SCCNC2 and SCCNC7 cells were added into 24-well
plates 24 h before transfection. The Lipofectamine 2000™ reagent
(Invitrogen; Thermo Fisher Scientific, Inc., Carlsbad, CA, USA) was
used to perform the transfection into SN-SCC cell lines the next
day. The transfected cells were divided into several groups.
Transwell assay
The cells of each group were digested at the
logarithmic stage of growth and the number of cells was calculated
after transfected with miR-34a for 48 h. We seeded SN-SCC cells
(1×105 cells) onto the upper chamber membrane with the
medium being serum free. The RPMI-1640 medium contained 20% fetal
calf serum was seeded into the lower chamber as a chemoattractant
and incubated for 24 h at 37°C. Then, the upper chamber cells
migrated into the lower chamber and 0.1% crystal violet was used to
stain the migrated cells for another 30 min, eventually, images of
the migration cells in eight random views were photographed under a
microscope (Olympus, Tokyo, Japan) for records. For invasion assay,
except for coating the filter in the upper chamber with Matrigel,
it was similar to the Transwell migration assay.
Reverse Transcription-quantitative PCR
(RT-qPCR)
TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) was used to extract total RNA and Nanodrop 1000
(Thermo Fisher Scientific, Inc.) was carried out to quantify RNA
expression. The stem-loop real-time PCR miRNA kit (Guangzhou
RiboBio Co., Ltd., Guangzhou, China) was carried out to perform the
specific reverse transcription. SYBR Premix Ex Taq II (Takara Bio,
Inc., Otsu, Japan) was used to perform quantitative PCR. U6 and
GAPDH were used as an internal control. The 2−ΔΔCq
method was used to detect the mRNA expression. The sequences of the
primers used were: miR-34a-F: GCGGCCAATCAGCAAGTATACT, and R: GTG
CAGGGTCCGAGGT. BCL-2-F: GAAGCACAGATGGTT GATGC, and R:
CACCTCACAAGGTTCCAAT. U6-F: ATT GGAACGATACAGAGAAGATT, and R:
GGAACGCTT CACGAATTTG. GAPDH-F: TGGTATCGT GGAAGGACTC, and R:
AGTAGAGGCAGGGATGATG.
Western blot analysis
RIPA lysis buffer containing proteinase inhibitors
(Beyotime Institute of Biotechnology, Haimen, China) were used to
extract total protein from the SN-SCC cells or tissues. Protein
concentration measured using BCA reagent kit (Beyotime Institute of
Biotechnology). We analyzed the amount of protein by using
immunoblotting method. Protein in each group (50 mg) was added into
the well of polyacrylamide gel and separated by 10% SDS-PAGE and
then transferred to nitrocellulose filter (NC) membrane (EMD
Millipore, Billerica, MA, USA). Then, skim milk (5–10%) was used to
block the membrane at room temperature for 2 h and incubated with
the primary antibodies: Bcl-2 (10C4) mouse monoclonal IgG1, cat.
no. sc-23960, 1:500 from Santa Cruz Biotechnology, Inc. (Dallas,
TX, USA). GAPDH (D16H11) XP® Rabbit mAb; cat. no. 5174;
1:1,000; Cell Signaling Technology, Inc. (Danvers, MA, USA)
followed by the secondary antibodies: Goat anti-rabbit IgG-HRP,
cat. no. sc-2004; 1:2,000 and goat anti-mouse IgG-HRP; cat. no.
sc-2005; 1:2,000 (both from Santa Cruz Biotechnology, Inc.).
Subsequently, the membrane was detected using an ECL
chemiluminescence detection kit. GAPDH served as the loading
control.
Dual luciferase reporter assay
The relative luciferase ability was performed using
the recombinant pMIR-REPORT luciferase vector. The wild-type and
mut-type BCL-2 3′UTR were constructed downstream of pMIR-REPORT
luciferase vector. We used Lipofectamine 2000 to transfect SN-SCC
cells with control mimic and miR-34a mimic. The dual luciferase
reporter assay system (Promega Corporation, Madison, WI, USA) was
then used to measure the luciferase activity values.
Statistical analysis
All independent experiments were conducted three
times. Data are presented as mean ± SD. SPSS 19.0 software (IBM
Corp., Armonk, NY, USA) and GraphPad Prism 5.02 Software (GraphPad
Software, Inc., La Jolla, CA, USA) were used to perform statistical
analysis and complete graph presentations, respectively. One-way
analysis of variance (ANOVA) was used and the post hoc test was
Tukey's post hoc, to compare the differences of the groups. The
correlation was analyzed by Pearson's regression analysis.
P<0.05 was considered to indicate a statistically significant
difference.
Results
miR-34a was overexpressed and PAQR3
was lower expressed in SN-SCC
miR-34a and BCL-2 expression in 52 paired SN-SCC
tissues and normal tissues was detected by RT-PCR. As Fig. 1A and C shows, the miR-34a average
expression in SN-SCC tissues was reduced markedly, while BCL-2
expression was increased obviously. We then analyzed miR-34a
expression in SN-SCC cells: RPMI-2650, SCCNC2 and SCCNC7 cells.
miR-34a expression was decreased in all cells while PAQR3 was
increased (Fig. 1B and D), which was
similar to SN-SCC tissues. Based on the results above, we further
analyzed the correlation between miR-34a and PAQR3 expression, as
Pearson's regression analysis showed that they were negatively
correlated (Fig. 1E).
miR-34a suppressed SN-SCC cell
migration and invasion
We then investigated miR-34a effect on SN-SCC.
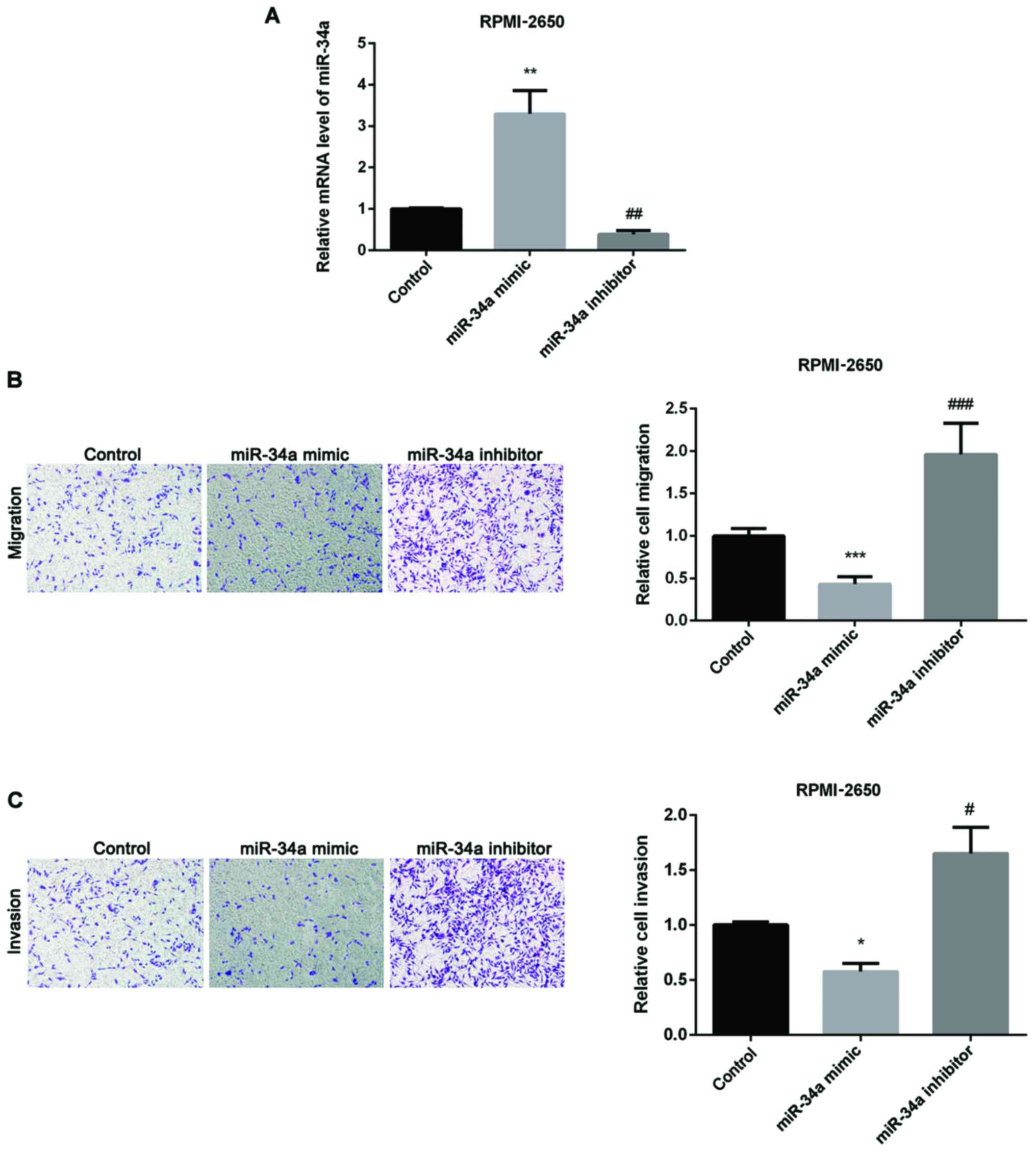
Firstly, we overexpressed or silenced miR-34a through transfecting
miR-34a mimic or inhibitor into RPMI-2650 cells. As seen in
Fig. 2A, the transfection was
successful. Secondly, we used Transwell assay to detect how miR-34a
affected SN-SCC cell migration and invasion and found that
upregulation of miR-34a inhibited SN-SCC cell migration, whereas
knockdown of miR-34a had the opposite effect (Fig. 2B). Fig.
2C shows inhibitory effect of cell invasion by miR-34a mimic
and promotion effect by miR-34a inhibitor in SN-SCC.
Silencing BCL-2 curbs the development
of SN-SCC
As previous study proved that BCL-2 was
overexpressed in SN-SCC, we chose to silence the expression of
BCL-2 to explore the role of BCL-2 in SN-SCC. The efficiency of
silencing BCL-2 is shown in Fig. 3A.
Then, we examined the relative cell migration and invasion when
silencing BCL-2 using Transwell assay. As the results show, BCL-2
siRNA inhibited SN-SCC cell migration and invasion (Fig. 3B and C).
BCL-2 was verified as the target of
miR-34a in SN-SCC
As previous reported BCL-2 functioned as a tumor
promoter and targeted by miR-34a in osteosarcoma. We suspected that
miR-34a targeted BCL-2 in regulating SN-SCC. We used
TargetScanHuman 7.1 to validate this prediction. The binding sites
of BCL-2 and miR-34a are presented in Fig. 4A. Then, we used luciferase reporter
assay to detect the luciferase ability in RPMI-2650 cells to
further determine the accuracy of this prediction. The results
showed that miR-106b mimic group had a significantly reduced
luciferase activity compared to control group in wild-type,
whereas, there was no effect in mut-type in RPMI-2650 cells
(Fig. 4B). Finally, we detected
whether miR-34a could affect BCL-2 expression in SN-SCC cells. As
shown in Fig. 4C, miR-34a mimic
decreased both miR-34a mRNA and protein level, while increased by
miR-34a inhibitor in RPMI-2650 cells.
BCL-2 reverses the inhibitory effect
of miR-34a in SN-SCC
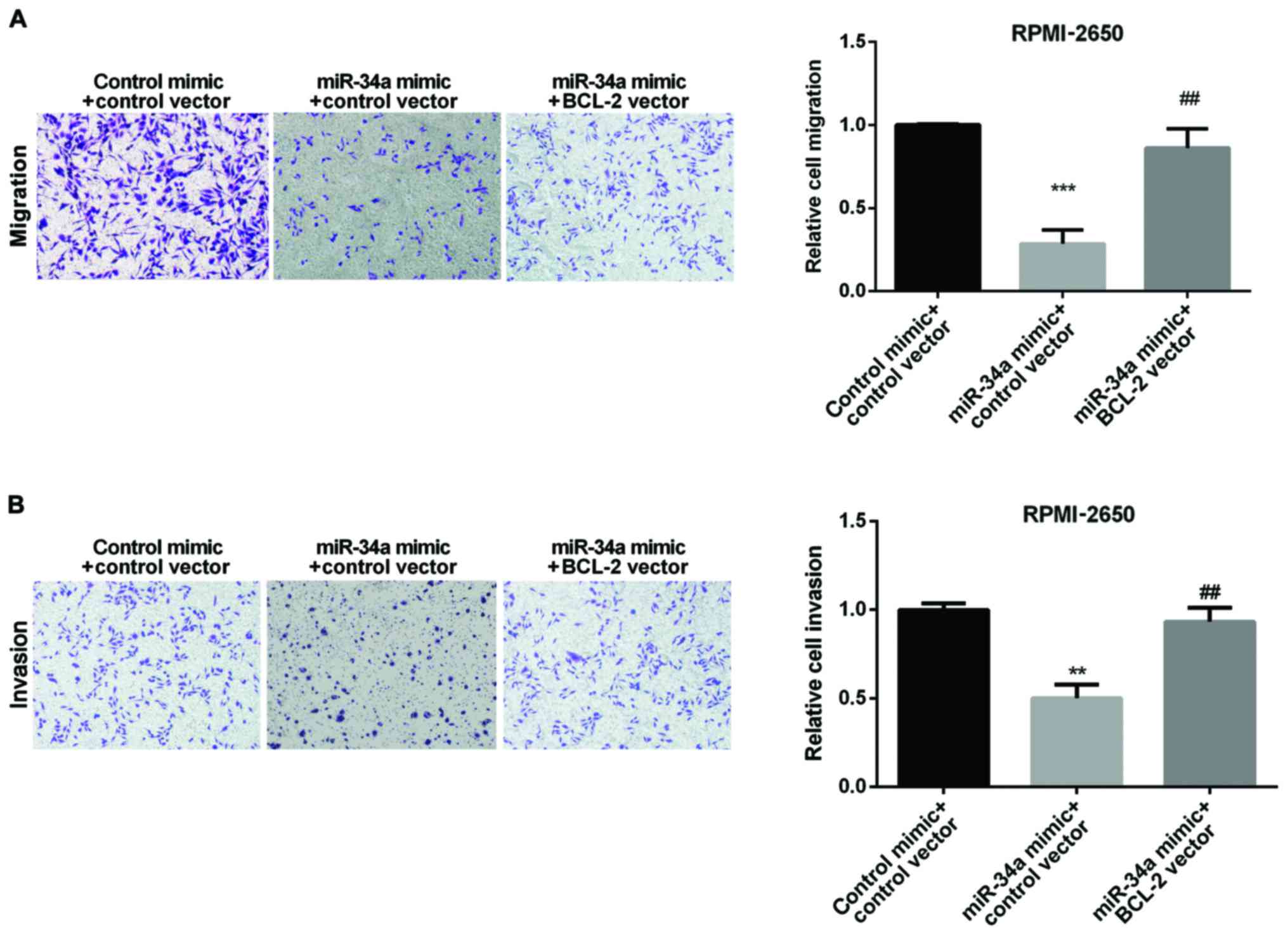
We explored whether BCL-2 could affect the miR-34a
on SN-SCC cell migration and invasion. We divided the cells into
three groups: Control, miR-34a mimic and miR-34a mimic + BCL-2
vector. As the results show, the migrated cells of RPMI-2650 were
increased when co-overexpression of both BCL-2 and miR-34a compared
with the cells overexpressied with miR-34a alone (Fig. 5A). Also, the same effect of the
invasive cells of RPMI-2650 was found (Fig. 5B). These data indicated that BCL-2
could reverse the inhibitory effect of miR-34a in regulating SN-SCC
cell invasion and migration.
Discussion
SN-SCC is a common malignant tumor. The occurrence
and progression of SN-SCC are not only related to abnormal
proliferation of cells, but are also associated with the abnormal
apoptosis.
miR-34a was proved to express abnormally in multiple
cancers and to act as a tumor inhibitor (26). Increasing evidence stated that miR-34a
had an inhibitory effect on tumor progression, such as inhibiting
medulloblastoma cell proliferation and promoting cell apoptosis.
Moreover, it had the same effect on gastric cancer (27), prostate cancer (28), hepatocellular carcinoma cells
(29), head and neck squamous cell
carcinoma (30), laryngeal squamous
cell carcinoma (31). Our study
showed a decreased expression of miR-34a in SN-SCC and inhibited
cell migratory and invasive ability, which is consistent with a
previous study that miR-34a expression was lower in SN-SCC and
associated with the poor prognosis of patients (18).
BCL-2 acted as an apoptosis factor and was proven to
take part in regulating some cancers targeted by miR-34a. It was
reported by Wen et al that miR-34a inhibited osteosarcoma
cell invasion and migration via targeting BCL-2 and C-IAP2
(32), which is similar to the study
that miR-34a suppressed the viability and migration of breast
cancer by inhibiting BCL-2 (33).
Wang et al showed that miR-34a mimic curbed cell viability
and facilitated the apoptosis of cervical cancer cell through
targeting BCL-2 (15). In our study,
we found that miR-34a curbed SN-SCC cell migratory and invasive
ability by targeting BCL-2.
In conclusion, miR-34a expression was lower while
BCL-2 was higher in SN-SCC and their correlation was negative. We
proved for the first time that BCL-2 was a direct target of miR-34a
in regulating the progress of SN-SCC and BCL-2 could attenuate
miR-34a inhibition effect on SN-SCC, indicating that miR-34a/BCL-2
axis has potential application value in SN-SCC diagnosis and
therapy.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
YZ designed this study and collected the data. YZ
and XW performed the experiments and interpreted the results. XW
wrote and finalized the manuscript. Both authors read and approved
the final manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
Linyi People's Hospital (Linyi, China). Patients who participated
in this research had complete clinical data. Signed written
informed consents were obtained from the patients or guardians.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ansa B, Goodman M, Ward K, Kono SA,
Owonikoko TK, Higgins K, Beitler JJ, Grist W, Wadsworth T, El-Deiry
M, et al: Paranasal sinus squamous cell carcinoma incidence and
survival based on Surveillance, Epidemiology, and End Results data,
1973 to 2009. Cancer. 119:2602–2610. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Turner JH and Reh DD: Incidence and
survival in patients with sinonasal cancer: A historical analysis
of population-based data. Head Neck. 34:877–885. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bonzini M, Battaglia P, Parassoni D, Casa
M, Facchinetti N, Turri-Zanoni M, Borchini R, Castelnuovo P and
Ferrario MM: Prevalence of occupational hazards in patients with
different types of epithelial sinonasal cancers. Rhinology.
51:31–36. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sanghvi S, Khan MN, Patel NR, Yeldandi S,
Baredes S and Eloy JA: Epidemiology of sinonasal squamous cell
carcinoma: A comprehensive analysis of 4994 patients. Laryngoscope.
124:76–83. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dulguerov P and Allal AS: Nasal and
paranasal sinus carcinoma: How can we continue to make progress?
Curr Opin Otolaryngol Head Neck Surg. 14:67–72. 2006.PubMed/NCBI
|
|
6
|
Youlden DR, Cramb SM, Peters S, Porceddu
SV, Møller H, Fritschi L and Baade PD: International comparisons of
the incidence and mortality of sinonasal cancer. Cancer Epidemiol.
37:770–779. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Scaria V, Hariharan M, Pillai B, Maiti S
and Brahmachari SK: Host-virus genome interactions: Macro roles for
microRNAs. Cell Microbiol. 9:2784–2794. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhu W, Shan X, Wang T, Shu Y and Liu P:
miR-181b modulates multidrug resistance by targeting BCL2 in human
cancer cell lines. Int J Cancer. 127:2520–2529. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yang H, Kong W, He L, Zhao JJ, O'Donnell
JD, Wang J, Wenham RM, Coppola D, Kruk PA, Nicosia SV, et al:
MicroRNA expression profiling in human ovarian cancer: miR-214
induces cell survival and cisplatin resistance by targeting PTEN.
Cancer Res. 68:425–433. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Pogribny IP, Filkowski JN, Tryndyak VP,
Golubov A, Shpyleva SI and Kovalchuk O: Alterations of microRNAs
and their targets are associated with acquired resistance of MCF-7
breast cancer cells to cisplatin. Int J Cancer. 127:1785–1794.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hamano R, Miyata H, Yamasaki M, Kurokawa
Y, Hara J, Moon JH, Nakajima K, Takiguchi S, Fujiwara Y, Mori M, et
al: Overexpression of miR-200c induces chemoresistance in
esophageal cancers mediated through activation of the Akt signaling
pathway. Clin Cancer Res. 17:3029–3038. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cui XB, Peng H, Li RR, Mu JQ, Yang L, Li
N, Liu CX, Hu JM, Li SG, Wei Y, et al: MicroRNA-34a functions as a
tumor suppressor by directly targeting oncogenic PLCE1 in Kazakh
esophageal squamous cell carcinoma. Oncotarget. 8:92454–92469.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhou Y, Ding BZ, Lin YP and Wang HB:
miR-34a, as a suppressor, enhance the susceptibility of gastric
cancer cell to luteolin by directly targeting HK1. Gene. 644:56–65.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ding N, Wu H, Tao T and Peng E: NEAT1
regulates cell proliferation and apoptosis of ovarian cancer by
miR-34a-5p/BCL2. OncoTargets Ther. 10:4905–4915. 2017. View Article : Google Scholar
|
|
15
|
Wang X, Xie Y and Wang J: Overexpression
of microRNA-34a-5p inhibits proliferation and promotes apoptosis of
human cervical cancer cells by downregulation of Bcl-2. Oncol Res.
Aug 30–2017.(Epub ahead of print). View Article : Google Scholar
|
|
16
|
Zhang X, Ai F, Li X, Tian L, Wang X, Shen
S and Liu F: MicroRNA-34a suppresses colorectal cancer metastasis
by regulating Notch signaling. Oncol Lett. 14:2325–2333. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li Y, Zhang H, Dong Y, Fan Y, Li Y, Zhao
C, Wang C, Liu J, Li X, Dong M, et al: miR-146b-5p functions as a
suppressor miRNA and prognosis predictor in non-small cell lung
cancer. J Cancer. 8:1704–1716. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ogawa T, Saiki Y, Shiga K, Chen N,
Fukushige S, Sunamura M, Nagase H, Hashimoto S, Matsuura K, Saijo
S, et al: miR-34a is downregulated in cis-diamminedichloroplatinum
treated sinonasal squamous cell carcinoma patients with poor
prognosis. Cancer Sci. 103:1737–1743. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xiong S, Zheng Y, Jiang P, Liu R, Liu X
and Chu Y: MicroRNA-7 inhibits the growth of human non-small cell
lung cancer A549 cells through targeting BCL-2. Int J Biol Sci.
7:805–814. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Feng SD, Mao Z, Liu C, Nie YS, Sun B, Guo
M and Su C: Simultaneous overexpression of miR-126 and miR-34a
induces a superior antitumor efficacy in pancreatic adenocarcinoma.
OncoTargets Ther. 10:5591–5604. 2017. View Article : Google Scholar
|
|
21
|
Wu J, Sun Z, Sun H and Li Y: MicroRNA 27a
promotes tumorigenesis via targeting AKT in triple negative breast
cancer. Mol Med Rep. 17:562–570. 2018.PubMed/NCBI
|
|
22
|
Liao A, Tan G, Chen L, Zhou W and Hu H:
RASSF1A inhibits gastric cancer cell proliferation by miR-711-
mediated downregulation of CDK4 expression. Oncotarget.
7:5842–5851. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gu L, Zhang J, Shi M and Peng C: The
effects of miRNA-1180 on suppression of pancreatic cancer. Am J
Transl Res. 9:2798–2806. 2017.PubMed/NCBI
|
|
24
|
Lu H, Wang C, Hao L, Yin G and Hao R: The
expression and significance of programmed cell death 5 and B-cell
lymphoma/leukemia-2 in sinonasal squamous cell carcinoma. Lin Chung
Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 28:1301–1304. 2014.(In
Chinese). PubMed/NCBI
|
|
25
|
Katori H, Nozawa A and Tsukuda M: Cell
proliferation, apoptosis, and apoptosis inhibition in malignant
transformation of sinonasal inverted papilloma. Acta Otolaryngol.
127:540–546. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cao W, Yang W, Fan R, Li H, Jiang J, Geng
M, Jin Y and Wu Y: miR-34a regulates cisplatin-induce gastric
cancer cell death by modulating PI3K/AKT/survivin pathway. Tumour
Biol. 35:1287–1295. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cao W, Fan R, Wang L, Cheng S, Li H, Jiang
J, Geng M, Jin Y and Wu Y: Expression and regulatory function of
miRNA-34a in targeting survivin in gastric cancer cells. Tumour
Biol. 34:963–971. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hagman Z, Haflidadottir BS, Ansari M,
Persson M, Bjartell A, Edsjö A and Ceder Y: The tumour suppressor
miR-34c targets MET in prostate cancer cells. Br J Cancer.
109:1271–1278. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li N, Fu H, Tie Y, Hu Z, Kong W, Wu Y and
Zheng X: miR-34a inhibits migration and invasion by downregulation
of c-Met expression in human hepatocellular carcinoma cells. Cancer
Lett. 275:44–53. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kumar B, Yadav A, Lang J, Teknos TN and
Kumar P: Dysregulation of microRNA-34a expression in head and neck
squamous cell carcinoma promotes tumor growth and tumor
angiogenesis. PLoS One. 7:e376012012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Shen Z, Zhan G, Ye D, Ren Y, Cheng L, Wu Z
and Guo J: MicroRNA-34a affects the occurrence of laryngeal
squamous cell carcinoma by targeting the antiapoptotic gene
survivin. Med Oncol. 29:2473–2480. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wen J, Zhao YK, Liu Y and Zhao JF:
MicroRNA-34a inhibits tumor invasion and metastasis in osteosarcoma
partly by effecting C-IAP2 and Bcl-2. Tumour Biol.
39:10104283177057612017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li L, Yuan L, Luo J, Gao J, Guo J and Xie
X: miR-34a inhibits proliferation and migration of breast cancer
through downregulation of Bcl-2 and SIRT1. Clin Exp Med.
13:109–117. 2013. View Article : Google Scholar : PubMed/NCBI
|